Abstract
We discovered a new Crocidura species of shrew (Soricidae: Eulipotyphla) from Narcondam Island, India by using both morphological and molecular approaches. The new species, Crocidura narcondamica sp. nov. is of medium size (head and body lengths) and has a distinct external morphology (darker grey dense fur with a thick, darker tail) and craniodental characters (braincase is rounded and elevated with weak lambdoidal ridges) in comparison to other close congeners. This is the first discovery of a shrew from this volcanic island and increases the total number of Crocidura species catalogued in the Indian checklist of mammals to 12. The newly discovered species shows substantial genetic distances (12.02% to 16.61%) to other Crocidura species known from the Indian mainland, the Andaman and Nicobar Archipelago, Myanmar, and from Sumatra. Both Maximum-Likelihood and Bayesian phylogenetic inferences, based on mitochondrial (cytochrome b) gene sequences showed distinct clustering of all included soricid species and exhibit congruence with the previous evolutionary hypothesis on this mammalian group. The present phylogenetic analyses also furnished the evolutionary placement of the newly discovered species within the genus Crocidura.
Similar content being viewed by others
Introduction
The terrestrial, insectivorous mammalian species in the genus Crocidura Wagler 1832 (subfamily Crocidurinae of the family Soricidae, order Eulipotyphla) are commonly referred to as white-toothed shrews1. Crocidura is a widespread and speciose genus with 198 species occurring in Africa, Europe, and Asia2,3, making it the most species-rich genus of mammals4,5. It is characterized by a small to medium-sized body (head and body length 35–100 mm) with usually short dense grey fur, a first-unicuspid tooth which is large, protrudes forward and is hooked, with a small cusp present behind the main cusp, unpigmented teeth, and no zygomatic arches6. The genus Crocidura (three upper unicuspids) can be differentiated from the nearest genus Suncus (four upper unicuspids) through dental formula1,7 (Supplementary Fig. S1). So far, 20 species of Crocidura are known from the Indian mainland, the Andaman and Nicobar (AN) Archipelago, Myanmar, and the Sundaic continental shelf island of Sumatra8 (Supplementary Table S1). Although these regions house members of 10 genera (Anourosorex, Blarinella, Chimarrogale, Crocidura, Episoriculus, Feroculus, Nectogale, Sorex, Soriculus, and Suncus) of the family Soricidae2,3, the AN Archipelago is known to house only a single genus Crocidura, with four species, namely the Andaman shrew C. andamanensis, the Andaman spiny shrew C. hispida, Jenkin’s shrew C. jenkinsi, and the Nicobar shrew C. nicobarica9.
In the first record of the genus Crocidura from India Miller (1912)10 described C. andamanensis and C. nicobarica, each based on a single specimen collected on the South Andaman Island and Great Nicobar Island, respectively. Subsequently, again based upon a single specimen Thomas11 described C. hispida from the northern middle Andaman Island and Chakraborty12 described C. jenkinsi from Mt. Harriet National Park, South Andaman Island4.
Due to their secretive behaviour and conservative external morphological characters, shrews are regarded as the least studied mammalian group5,13. Consequently, in the last two decades between 2000 and 2020, a total of 24 Crocidura species have been newly discovered throughout the world, of which 15 species were discovered in the Indo-Malayan region and Sundaland, especially from the continental islands2,5,8,14,15,16,17,18,19,20,21,22,23. Molecular studies were also consecutively used to discriminate shrew species, detect cryptic diversity, or study phylogenetic evolution, biogeographic origin and radiation, and phylogeography8,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43. Only a single study aimed to assess the genetic signature of two endemic species, C. andamanensis and C. nicobarica from the AN Archipelago44. Therefore, an understanding of the species diversity on this group of islands remains incomplete. The present study is based on the assumption that a few hitherto unreported shrew species exist beyond the known biogeographic distribution of the group in the AN Archipelago, which warrants further investigation through integrative approaches. We performed both morphological and molecular assessments to confirm a new shrew species from the volcanic Narcondam Island in the AN Archipelago, which is herein described as Crocidura narcondamica sp. nov. (Fig. 1). The newly discovered species is validated by a morphometric and molecular comparison with 13 and 15 species, respectively, distributed in the AN Archipelago, on the mainland of India, and the close biogeographic realms of Myanmar and Sumatra. We estimated the genetic divergence from related species and performed phylogenetic analyses to corroborate the taxonomic identity and evolutionary relationships of this novel species.
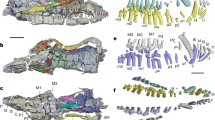
Crocidura narcondamica sp. nov. (holotype ZSI-29313, female). (a) Dorso-lateral view of female (adult alive). Views of the cranium (b) dorsal, (c) ventral, and (d) lateral. Views of the mandible (e) lateral and (f) occlusal (BC braincase, I1 first incisor/first unicuspid). The photographs were captured by the first (b–f) and fourth (a) authors using a Nikon D7000 camera and edited manually in Adobe Photoshop CS 8.0.
Materials and methods
Study area
Narcondam Island (13.45° N 94.27° E; Fig. 2) is located about 130 km east of North Andaman, and about 446 km off the west coast of Myanmar45,46,47. The island covers an area of 6.8 km2 and the highest peak (volcanic cone) is 710 m above sea level; however, the base lies approximately 1500 m beneath the sea45. This isolated island is part of a volcanic arc that continues northward from Sumatra to Myanmar48. The climatic condition of this small, conical island can be defined as a humid, tropical, and coastal. The island is thickly vegetated, bordered by cliffs on the southern side and crested by three peaks. The forest types can broadly be categorized as three zones: wet evergreen on the slopes and highest zones of the volcano, moist deciduous or semi-evergreen at lower elevations, and littoral forest along the coastline47.
Map showing the Narcondam Island (marked by a red colour box) in the Andaman group of Island, India (marked by a blue colour box) along with the habitat where the new species was encountered. Map prepared using QGIS 2.6.1 (http://www.qgis.org) and edited manually in Adobe Photoshop CS 8.0. The habitat photographs were captured by the fourth author using a Nikon D7000 camera and edited manually in Adobe Photoshop CS 8.0.
Ethics statement, sampling, and morphological examination
To conduct the field survey and sampling, prior permission was acquired from the office of the Principal Chief Conservator of Forests (Wildlife), Andaman and Nicobar Islands, Port Blair (Letter no. CWLW/WL/24/339, dated 04 February 2020). The experimental protocols were approved by the Zoological Survey of India and were carried out in accordance with relevant guidelines in compliance with the ARRIVE 2.0. guidelines (https://www.arriveguidelines.org)49. Two individuals of this unique shrew species were captured from the same location (13° 27.290′ N 94° 16.436′ E) by the pitfall method. Their external measurements were taken in the field and include head and body length (HB), tail length (TL), ear length (E), and hindfoot length including claw (HF). Photographs of fresh specimens were taken by the fourth author in the field. The collected specimens were preserved in 70% molecular grade ethanol for further investigation. Skulls were extracted later, cleaned, and prepared in the museum (Fig. 1a–e) and deposited in the National Zoological Collections (NZC-Mammal and Osteology section) of the Zoological Survey of India (ZSI), Kolkata, India, under the registration numbers 29313 and 29314. The nomenclature of external and craniodental characters follows Hutterer et al.22. Field methods followed the guidelines approved by the American Society of Mammalogists50. The craniodental measurements were taken by the first author with a digital caliper accurate to the nearest 0.01 mm. The measurements include condylo-incisive length (CIL), braincase height (BH), rostrum width (RW), hard palatine length (PL), maxillary breadth (MB), least interorbital breadth (LIOB), braincase breadth (BB), upper toothrow length (UTR), maximum breadth across the outer anterobuccal margins of the parastyles of the second upper molars (M2-M2), length of the anterior tip of the fourth premolar (P4) to posterior border of M3 (P4-M3), braincase length (BL), postglenoid width (PGL), the maximum length from the anterior face of the first upper incisor to the posterior margin of the third unicuspid (I-UN3), length of 1st upper incisor (in lateral view) from tip to upper margin of cingulum (LI1), mandibular toothrow length (MTR), length of mandible from the tip of incisor to the posterior edge of the condyle (ML), length of lower molar series (m1-m3), length of 1st lower incisor from tip to posterior margin of the cingulum (Li1), and height of the coronoid process (COR) (Table 1, Supplementary Table S2). Only adult specimens were included, as determined by fully erupted molars and fused basioccipital suture5,51. Photographs of cranial and dental views were taken by the first author using a Nikon D7000 camera. The collected specimens of the possible new species of shrew were compared with the morphometric data of 13 congeners reported from the AN Archipelago, the mainland of India and from Myanmar (Supplementary Table S2). The external and craniodental characters were also compared with the archival specimens of C. attenuata and C. jenkinsi available in the NZC of ZSI, Kolkata (Fig. 3, Supplementary Figs. S2, S3).
Cranium and mandible of (I) C. narcondamica sp. nov. (holotype, ZSI-28000) (II) C. attenuata (ZSI-16129) and (III) C. jenkinsi (ZSI-19860). From top to bottom, (a) dorsal, (b) ventral, (c) lateral views of the cranium, and (d) lateral and (e) occlusal views of the mandible. Distinct morphological features are labelled as BC brain case, LR lambdoidal ridge, FO foramen ovale, and I1 first incisor/first unicuspid. The photographs were captured by the first author using a Nikon D7000 camera and edited manually in Adobe Photoshop CS 8.0.
Comparative materials
Morphometric measurements of other known congeners distributed in the AN Archipelago, the mainland of India and in Myanmar were also acquired from published literature5,10,52,53,54 (Supplementary Table S2). The morphology and craniodental measurements along with other collateral information of the museum and other comparative species are given in Supplementary Table S2.
DNA extraction, PCR amplification, and sequencing
The genomic DNA was extracted from both the holotype and the paratype specimen by the standard phenol–chloroform isoamyl alcohol method55. The extracted DNA was visualized through 1% agarose gel electrophoresis. The published primer pair (mcb 398: 5′-TACCATGAGGACAAATATCATTCTG-3′ and mcb 869: 5′-CCTCCTAGTTTGTTAGGGATTGATCG-3′)56 was used to amplify the widely applied mitochondrial Cytochrome b (mtCytb) gene segment for the identification of shrew species7,57. The 25 ml PCR mixture comprises 10 pmol of each primer, 20 ng of DNA template, 1X PCR buffer, 1.0–1.5 mM of MgCl2, 0.25 mM of each dNTPs, and 1 U of Platinum Taq DNA Polymerase High fidelity (Invitrogen). The PCR reaction was performed in Veriti Thermal Cycler (Applied Biosystems) with the published thermal profile. The PCR products were purified using a QIAquick Gel Extraction Kit (QIAGEN) with standard protocol. The cycle sequencing was executed by using BigDye Terminator ver. 3.1 Cycle Sequencing Kit (Applied Biosystems) and 3.2 pmol of each primer on Veriti Thermal Cycler. The products were cleaned by BigDye X-terminator kit (Applied Biosystems) with standard protocol and subsequently bidirectional sequenced by the 48 capillary 3730 Genetic Analyzer (Applied Biosystems).
Sequence quality control and dataset preparation
The study obtained both forward and reverse chromatograms from the holotype and paratype samples. The noisy parts of each chromatogram were trimmed at both ends and quality value (> 40) was used to screen for making the consensus sequences through the SeqScanner Version 1.0 (Applied Biosystems). The sequences were translated through the online platform ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) to validate the sequence by comparison to the vertebrate consensus. The annotated sequences were contributed to GenBank. A total of 54 mtCytb sequences were acquired from GenBank (Supplementary Table S3). A total of 58 sequences were aligned by ClustalX software58 to form a combined dataset (473 bp) for further genetic distance and phylogenetic analysis. The sequences (accession no. KP061993 and KP062007) of Crocidura monax endemic to Tanzania, East Africa were used as an out-group in the present phylogenetic analyses.
Genetic distance and phylogenetic analysis
The Kimura-2-parameter (K2P) genetic distance was calculated in MEGAX59. The most suitable model for the present dataset was estimated by using JModelTest v2 with the lowest BIC (Bayesian Information Criterion) score60. The maximum-Likelihood (ML) phylogenetic tree was constructed using the IQ-Tree web server (http://iqtree.cibiv.univie.ac.at.) with General Time Reversible (GTR) model including a proportion of invariable sites (+ I) and rate of variation across sites and 1000 bootstrap support61. The Bayesian (BA) tree was constructed in Mr. Bayes 3.1.2 by selecting nst = 6 for the GTR + G + I with one cold and three hot chains of metropolis-coupled Markov Chain Monte Carlo (MCMC); it was run for 1,000,000 generations with 25% burn-in with trees saving at every 100 generations62. The MCMC analysis was used to generate the convergence metrics, until the standard deviation (SD) of split frequencies attained to 0.01 and the potential scale reduction factor (PSRF) for all parameters neared 1.0. The web-based iTOL tool (https://itol.embl.de/) was used for better illustration of the BA phylogenetic tree63.
Results
Mammalia Linnaeus, 1758
Eulipotyphla Waddell et al., 1999
Soricidae G. Fischer, 1814
Crocidurinae Milne-Edwards, 1872
Crocidura Wagler 1832
Crocidura narcondamica sp. nov.
Etymology and nomenclatural acts
The new species is named for the type locality, Narcondam Island, where the type specimens were collected. The specific epithet is feminine latinized adjective. Taxonomic nomenclature published in this article follows the amended International Code of Zoological Nomenclature (ICZN, version effective from 1 January 2012). The ZooBank LSID (Life Science Identifier) for this publication can be accessed through urn:lsid:zoobank.org:pub:7EDEF162-C85B-499B-A855-D26E6EB9763F. Suggested common name: Narcondam shrew.
Type specimens
Holotype
Adult female, ZSI 29313, collected at Narcondam Island (13° 27.290′ N, 94° 16.436′ E), Andaman and Nicobar Archipelago, in the Bay of Bengal, India (Fig. 2); 11 m elevation; collected by G. Gokulakrishnan on April 17, 2020. The specimen consists of a fluid-preserved carcass and a cleaned skull. Paratype: Locality and collector same as holotype. Adult male, ZSI 29314. The specimen consists of a fluid-preserved skin and cleaned skull. Skull extracted by the first author.
Distribution and habitat
The new species is presently known only from its type locality, Narcondam Island, Andaman and Nicobar Archipelago, in the Bay of Bengal, India. It was collected from a littoral forest along the coastline at 11 m elevation. No anthropogenic disturbances were observed in the habitat except for a security post (Fig. 2).
Diagnosis and description
The new species C. narcondamica sp. nov. (Fig. 1) is assigned to the genus Crocidura by the presence of three upper unicuspids and clearly distinguished from all other AN Archipelago shrews by its body size and tail length, which are considerably shorter (Supplementary Table S2). The new species possesses a darker-grey dense fur dorsally and a thick and darker tail (Fig. 1a), whereas the species known from the AN Archipelago possesses a different dorsal pelage and tail; C. jenkinsi (Supplementary Fig. S3a) and C. hispida have a spiny dorsal fur with a slender tail, C. andamanensis has a bluish-grey dorsal fur washed with brown and a darker brown tail, C. nicobarica has a bristly sooty brown dorsal fur with a slender tail, and C. attenuata has a soft brownish-grey dorsal fur with a slender tail2,51 (Supplementary Fig. S2a).
The new species also differs from other congeners occurring in the mainland of India, and in Myanmar (Table 1, Supplementary Table S2). The head and body length of the new species (holotype: 67 and paratype: 63 mm) is considerably smaller, than C. fuliginosa and C. pullata, but overlapping with other species such as, C. attenuata (60–89 mm), C. cranbrooki (65–86 mm), C. horsfieldii (49–71 mm), C. indochinensis (53–71 mm), C. pergrisea (65–86 mm), C. rapax (56–70 mm) and C. vorax (54–90 mm). However, the tail length (TL) of the new species (58.5 and 55.6 mm) is longer than C. horsfieldii (30–48 mm), C. indochinensis (40–50 mm), C. pergrisea (39–53 mm), C. pullata (37–51 mm), C. rapax (38–47 mm) and C. vorax (41–51 mm), and shorter than C. cranbrooki (65–88 mm), C. fuliginosa (62–89 mm). Although the HB and TL of the newly discovered species overlap with C. attenuata, the morphological characters are significantly different (soft brownish-grey dorsal pelage with a brownish slender tail; Supplementary Fig S2a).
The length of the hindfoot of C. narcondamica sp. nov. (holotype: 13.4 and paratype: 12.4 mm) also differs from three species, C. pullata (14–16 mm), C. cranbrooki (14–16 mm), and C. fuliginosa (15–19 mm), which are distributed in India and Myanmar. However, the rest of the congeners known from the same biogeographic region show an overlapping length of the hindfoot (Supplementary Table S2).
The new species was further examined and compared with the closest congener C. attenuata (Fig. 3, Supplementary Fig. S2). The braincase (BC) of C. narcondamica sp. nov. is rounded and elevated (Fig. 1b,d), with weaker lambdoidal ridges (LR; Fig. 1b) than in C. attenuata (slightly flattened with developed LR; Supplementary Fig. S2c,e). The foramen ovale (FO) is more prominent than in C. attenuata (Fig. 3b). In C. narcondamica sp. nov., the condylobasal length (holotype: 19.6 and paratype: 18.9 mm), palatal length (7.4 and 7 mm), upper toothrow (8.8 and 7.9 mm), maxillary toothrow (7.7 and 7 mm) and mandible length (11.6 and 10.8 mm) are significantly either higher or lower than in other congeners (Supplementary Table S2). The first incisor (I1) of C. narcondamica sp. nov. is less sharp and slightly protruded from the rostrum, than in C. attenuata (Figs. 1d, 3c).
Molecular identification and phylogenetic interpretation
Partial mitochondrial mtCytb gene sequences (Accession Nos. MW417367 and MW417368) were generated from both the holotype and paratype of the new species and submitted to GenBank. The BLAST search results showed a 90% similarity with the available sequence of a specimen Crocidura sp. (MN691031) collected from Xizang, China. The next closest results of the similarity search revealed a 89.57% similarity with a specimen of Crocidura sp. (MN691019) collected from Yunnan, China and with C. attenuata (MK765768) collected from Jiangxi, China. The present dataset of 21 Crocidura species, including the new one shows an overall mean inter-species genetic distance of 11.7%. The new species revealed a substantial mean genetic distance (12.02% to 16.61%) to other Crocidura species (Supplementary Table S4). The new species is genetically distant to C. andamanensis (16.61%) and C. nicobarica (15.09%) distributed in the same group of islands, AN archipelago. The new species also genetically distant (12.02% to 16.57%) from other congeners known from the mainland of India and Myanmar. The new species also show substantial genetic distance (13.13% to 16.44%) to other congeners distributed in Sumatra. Both ML and BA phylogenetic trees showed similar topologies with high posterior probabilities and bootstrap supports (Fig. 4, Supplementary Fig. S4) and showed that the three Crocidura species from the AN archipelago included in the analysis do not form a monophyletic group but are related to entirely different branches of the evolutionary tree.
The Bayesian phylogenetic analysis of the mitochondrial Cytb gene depicted a distinct clustering of C. narcondamica sp. nov. in comparison with other Crocidura species distributed in the AN Archipelago, the mainland of India, Myanmar, and in Sumatra. The posterior probability supports are noted with each node. The GenBank accession numbers and species name are marked as per clade pattern. The distinct clade of the new species is marked by a red colour box. The figure was prepared in web-based iTOL tool (https://itol.embl.de/) and edited manually in Adobe Photoshop CS 8.0.
Discussion
Island ecosystems are regarded as discrete biogeographic units and as a significant model for evolutionary studies8,32,64. In the Miocene–Pliocene, volcanic eruption produced many new islands and their sporadic land connections during the Pleistocene, allowed both geographic and temporal processes of species diversification in Southeast Asia32. The Indian plate separated from Africa-Madagascar- Seychelles and drifted towards the Eurasian plate, which also allowed multiple opportunities for animal oversea dispersal and biological connections between the India mainland and Southeast Asia65. Due to the remoteness and inaccessibility throughout the year, the smaller islands of the AN archipelago and other tropical islands are little explored in comparison to larger islands.
The political boundaries of India include a number of islands in both the Arabian Sea and the Bay of Bengal. The islands of the Arabian Sea are primarily built up by coral reefs, whereas the Bay of Bengal islands are distinguished by habitable submarine mountains45. Narcondam Island is one of 836 islands of the AN Archipelago, and is a small dormant volcanic island with almost 80% forest cover45,66 (Fig. 2). It is believed that the volcano was active during Holocene eruptions66,67. However, bathymetric study revealed that a number of seamounts have been well-developed on the Andaman seafloor, which changes the trend of the volcanic arc from northeast to southeast to Sumatra Island. Hence, it is evidenced that there is a direct connection between the submerged volcanic arc-chain of the Andaman Sea from Sumatra to Barren and Narcondam volcanic islands68,69. The island has been recognised by UNESCO as a World Heritage site due to its sensitive ecosystem and the occurrence of an endangered species, the Narcondam hornbill (Rhyticeros narcondami). Further, this island has been also designated as Wildlife Sanctuary under the provisions of the Indian Wildlife (Protection) Act, 1972.
The unparalleled biogeography of oceanic islands provides a suitable habitat for many Crocidura species discovered in the recent past. The known distribution of the new shrew species is restricted to Narcondam Island, a very small island. Isolated islands appear to provide a suitable habitat for many endemic Crocidura species, such as C. canariensis, which is restricted to the Canary Islands, C. fingui and C. thomensis, restricted to the Sao Tome and Principe Islands, C. orii, restricted to the Amamioshima group of the Ryukyu Islands, and C. trichura, restricted to Christmas Island4,70. The present discovery of a new shrew from isolated Narcondam Island adds an interesting detail to this pattern.
Considering the molecular-based species identification, the new species is clearly genetically distinct. Previous studies already showed that the mtCytb gene can often be effectively used to discriminate shrew species and to detect cryptic diversity in different geographical regions7,42,43,57. The genetic assessment of Crocidura species also facilitates the description of their radiation and diversification in Southeast Asian countries32. The estimated K2P genetic distance, ML and BA phylogenies clearly discriminate all the studied shrew species with sufficient genetic distances and distinct clustering. The new species C. narcondamica sp. nov. shows a substantial genetic distance (12.02%) to C. rapax (distributed in China, India, Myanmar, and Taiwan), and even larger distances in comparison with other AN archipelago species such as C. andamanensis with 16.61%, and C. nicobarica with 15.09%. The initial phylogenetic analysis indicates that the numerous endemic Crocidura species are not the result of a local radiation, as could be initially expected, but appear to be derived from at least three independent colonization events. To understand this surprising pattern, we recommend generating more molecular data of this group of mammals from different geographical regions to clarify their in-depth phylogenetic relationships, provide estimates of the divergence events, and allow a better alignment with the biogeographical history of the Indo-Malayan and Sundaic realms.
So far, Narcondam Island is popularly known by the occurrence of the endemic Narcondam hornbill. The ecological study of animals on this island was initially restricted only to this bird. Later on, researchers focused on a faunal expedition and recorded 17 fishes, 8 reptiles, 28 birds, 2 mammals (Chinese Forest Rat, Rattus andamanensis and Island Flying Fox, Pteropus hypomelanus), 13 spiders, 8 butterflies, and 2 sea cucumbers from this isolated island47. However, the known diversity of the mammalian fauna is very sparse on Narcondam Island; especially because no data were reported on soricid fauna (shrews). The discovery of C. jenkinsi on South Andaman Island by Chakraborty12 was the latest discovery of a Crocidura species from India. With this new description of the Narcondam shrew, altogether twelve species of Crocidura are now known from India including the AN Archipelago; viz., C. andamanensis, C. attenuata, C. fuliginosa, C. hispida, C. horsfieldii, C. jenkinsi, C. nicobarica, C. pergrisea, C. pullata, C. rapax, C. vorax, and C. narcondamica sp. nov. Among the Indian Crocidura shrews, four species known from the AN Archipelago have been categorized as threatened by the IUCN Red List of Threatened Species (2020-3) due to their remarkable endemism70. Habitat loss due to selective logging, anthropogenic activities, and natural disasters have been identified as the major threats for this group of animals in the AN Archipelago71,72. As the Narcondam Island is uninhabited, the new species C. narcondamica sp. nov. may not face the anthropogenic disturbances, but the extremely restricted insular habitat and the associated limited population size will automatically result in increased vulnerability of the species73. Raptors may be assumed to be potential natural predators of this species, but this threat is probably negligible compared to the generally precarious position of any small-island endemic with respect to natural disasters and stochastic population fluctuations. Further studies on the taxonomy, ecology, and distribution of the new species will help to understand its present status in more detail.
Data availability
The following information was supplied regarding the accessibility of DNA sequences: The generated partial fragment of mitochondrial Cytochrome b gene sequences are deposited in GenBank of NCBI under accession number MW417367 and MW417368.
References
Lekagul, B. & McNeely, J. A. Mammals of Thailand 758 (Association for the Conservation of Wildlife, 1977).
Burgin, C. J., Colella, J. P., Kahn, P. L. & Upham, N. S. How many species of mammals are there? J. Mamm. 99, 1–14 (2018).
Wilson, D. E. & Mittermeier, R. A. Handbook of the Mammals of the World, Vol 8: Insectivores, Sloths and Colugos 709 (Lynx Edicions, 2018).
Hutterer, R. Order Soricomorpha. In Mammal Species of the World: A Taxonomic and Geographic Reference (eds Wilson, D. E. & Reeder, D. A.) 220–311 (Johns Hopkins University Press, 2005).
Jenkins, P. D., Lunde, D. P. & Moncrieff, C. B. Chapter 10—Descriptions of new species of Crocidura (Soricomorpha: Soricidae) from mainland Southeast Asia, with synopses of previously described species and remarks on biogeography. In Systematic Mammalogy Contributions in Honour of Guy G. Musser Vol. 331 (eds Voss, R. S. & Carleton, M. C.) 356–405 (Bulletin of the American Museum of Natural History, 2009).
Martin, R. E., Pine, R. P. & DeBlase, A. F. A Manual of Mammalogy with Keys to Families of the World 333 (Waveland Press, Inc, 2011).
Ohdachi, S. D. et al. Molecular phylogenetics of soricid shrews (Mammalia) based on mitochondrial cytochrome b gene sequences: With special reference to the Soricinae. J. Zool. 270, 177–191 (2006).
Demos, T. C. et al. Local endemism and within-island diversification of shrews illustrate the importance of speciation in building Sundaland mammal diversity. Mol. Ecol. 25, 5158–5173 (2016).
Kamalakannan, M. & Venkatraman, C. A Checklist of Mammals of India (Zoological Survey of India, 2017).
Miller, G. S. Mammals of the Andaman and Nicobar Islands. Proc. U.S Nat. Mus. 24, 751–795 (1902).
Thomas, O. A new shrew (Crocidura hispida) from the Andaman Islands. Ann. Mag. Nat. His. 8, 468–469 (1913).
Chakraborty, S. A new shrew (Crocidura jenkinsi) from the Andaman Islands. Bull. Zool. Surv. India 1, 303 (1978).
Bannikova, A. A., Abramov, A. V., Borisenko, A. V., Lebedev, V. S. & Rozhnov, V. V. Mitochondrial diversity of the white-toothed shrews (Mammalia, Eulipotyphla, Crocidura) in Vietnam. Zootaxa 2812, 1–20 (2011).
Jenkins, P. D., Abramov, A. V., Rozhnov, V. V. & Makarova, O. V. Description of two new species of white-toothed shrews belonging to the genus Crocidura (Soricomorpha: Soricidae) from Ngoc Linh Mountain, Vietnam. Zootaxa 1589, 57–68 (2007).
Meegaskumbura, S., Meegaskumbura, M., Pethiyagoda, R., Manamendra-Arachchi, K. & Schneider, C. J. Crocidura hikmiya, a new shrew (Mammalia: Soricomorpha: Soricidae) from Sri Lanka. Zootaxa 1665, 19–30 (2007).
Jenkins, P. D., Abramov, A. V., Rozhnov, V. V. & Olsson, A. A new species of Crocidura (Soricomorpha: Soricidae) from southern Vietnam and north-eastern Cambodia. Zootaxa 2345, 60–68 (2010).
Esselstyn, J. A. & Goodman, S. M. New species of shrew (Soricidae: Crocidura) from Sibuyan Island, Philippines. J. Mamm. 91, 1467–1472 (2010).
Abramov, A. V., Bannikova, A. A. & Rozhnov, V. V. White-toothed shrews (Mammalia, Soricomorpha, Crocidura) of coastal islands of Vietnam. ZooKeys 207, 37–47 (2012).
Esselstyn, J. A., Achmadi, A. S. & Maharadatunkamsi, K. C. R. A new species of shrew (Soricomorpha: Crocidura) from West Java, Indonesia. J. Mamm. 95, 216–224 (2014).
Ceríaco, L. M. P. et al. Description of a new endemic species of shrew (Mammalia, Soricomorpha) from Príncipe Island (Gulf of Guinea). Mammalia 79, 1–18 (2015).
Demos, T. C., Achmadi, A. S., Handika, H., Maharadatunkamsi, K. C. R. & Esselstyn, J. A. A new species of shrew (Soricomorpha: Crocidura) from Java, Indonesia: Possible character displacement despite interspecific gene flow. J. Mamm. https://doi.org/10.1093/jmammal/gyw183 (2016).
Hutterer, R., Balete, D. S., Giarla, T. C., Heaney, L. R. & Esselstyn, J. A. A new genus and species of shrew (Mammalia: Soricidae) from Palawan Island, Philippines. J. Mamm. 99, 518–536 (2018).
Andino-Madrid, A. J., Colindres, J. E. M., Pérez-Consuegra, S. G. & Matson, J. O. A new species of long-tailed shrew of the genus Sorex (Eulipotyphla: Soricidae) from Sierra de Omoa, Honduras. Zootaxa 4809, 56–70 (2020).
Ruedi, M. Phylogenetic evolution and biogeography of Southeast Asian shrews (genus Crocidura: Soricidae). Biol. J. Linnean. Soc. 58, 197–219 (1996).
Ruedi, M., Auberson, M. & Savolainen, V. Biogeography of Sulawesian shrews: Testing for their origin with a parametric bootstrap on molecular data. Mol. Phylogenet. Evol. 9, 567–571 (1998).
Fumagalli, L. et al. Molecular phylogeny and evolution of Sorex shrews (Soricidae: Insectivora) inferred from mitochondrial DNA sequence data. Mol. Phylogenet. Evol. 1, 222–235 (1999).
Ohdachi, S. D. et al. Molecular phylogenetics of Crocidura shrews (Insectivora) in east and Central Asia. J. Mamm. 85, 396–403 (2004).
Dubey, S., Zaitsev, M., Cosson, J. F., Abdukadier, A. & Vogel, P. Pliocene and Pleistocene diversification and multiple refugia in a Eurasian shrew (Crocidura suaveolens group). Mol. Phylogenet. Evol. 38, 635–647 (2006).
Bannikova, A. A., Lebedev, V. S., Kramerov, D. A. & Zaitsev, M. V. Phylogeny and systematics of the Crocidura suaveolens species group: Corroboration and controversy between nuclear and mitochondrial DNA markers. Mammalia 70, 106–119 (2006).
Dubey, S. et al. Biogeographic origin and radiation of the old world Crocidurine shrews (Mammalia: Soricidae) inferred from mitochondrial and nuclear genes. Mol. Phylogenet. Evol. 48, 953–963 (2008).
Esselstyn, J. A. & Brown, R. M. The role of repeated sea-level fluctuations in the generation of shrew diversity in the Philippine Archipelago. Mol. Phylogenet. Evol. 53, 171–181 (2009).
Esselstyn, J. A., Timm, R. M. & Brown, R. M. Do geological or climatic processes drive speciation in dynamic archipelagos? The tempo and mode of diversification in southeast Asian shrews. Evolution 63, 2595–2610 (2009).
Esselstyn, J. A. & Oliveros, C. H. Colonization of the Philippines from Taiwan: A multi-locus test of the biogeographic and phylogenetic relationships of isolated populations of shrews. J. Biogeogr. 37, 1504–1514 (2010).
He, K. et al. A multi-locus phylogeny of Nectogalini shrews and influences of the paleoclimate on speciation and evolution. Mol. Phylogenet. Evol. 56, 734–746 (2010).
Jacquet, F., Nicolas, V., Bonillo, C., Cruaud, C. & Denys, C. Barcoding, molecular taxonomy and exploration of the diversity of shrews (Soricomorpha: Soricidae) on Mount Nimba (Guinea). Zool. J. Linn. Soc. 166, 672–687 (2012).
Esselstyn, J. A., Maharadatunkamsi, A., Siler, C. D. & Evans, B. J. Carving out turf in a biodiversity hotspot: Multiple, previously unrecognized shrew species co-occur on java island, Indonesia. Mol. Ecol. 22, 4972–4987 (2013).
Wan, T., He, K. & Jiang, X. L. Multilocus phylogeny and cryptic diversity in Asian shrew-like moles (Uropsilus, Talpidae): Implications for taxonomy and conservation. BMC Evol. Biol. 13, 232 (2013).
Yuan, S. L. et al. A mitochondrial phylogeny and biogeographical scenario for Asiatic water shrews of the genus Chimarrogale: Implications for taxonomy and low-latitude migration routes. PLoS ONE 8, 77156 (2013).
Giarla, T. C. & Esselstyn, J. A. The challenges of resolving a rapid, recent radiation: Empirical and simulated phylogenomics of Philippine shrews. Syst. Biol. 64, 727–740 (2015).
Stanley, W. T., Hutterer, R., Giarla, T. C. & Esselstyn, J. A. Phylogeny, phylogeography and geographical variation in the Crocidura monax (Soricidae) species complex from the montane islands of Tanzania, with descriptions of three new species. Zool. J. Linnean Soc. 174, 185–215 (2015).
Jacquet, F. et al. Phylogeography and evolutionary history of the Crocidura olivieri complex (Mammalia, Soricomorpha): From a forest origin to broad ecological expansion across Africa. BMC Evol. Biol. 15, 71 (2015).
Li, Y. Y. et al. A revision of the geographical distributions of the shrews Crocidura tanakae and C. attenuata based on genetic species identification in the mainland of China. ZooKeys 869, 147–160 (2019).
Chen, S. et al. Multilocus phylogeny and cryptic diversity of white-toothed shrews (Mammalia, Eulipotyphla, Crocidura) in China. BMC Evol. Biol. 20, 29 (2020).
Kundu, S. et al. Molecular investigation of non-volant endemic mammals through mitochondrial cytochrome b gene from Andaman and Nicobar archipelago. Mitochondrial DNA B 5, 1447–1452 (2020).
Yahya, H. S. A. & Zarri, A. A. Status, ecology and behaviour of Narcondam Hornbill (Aceros narcondami) in Narcondam Island, Andaman and Nicobar Islands, India. J. Bom. Nat. His. Soc. 99, 434–445 (2002).
Pal, T. et al. Dacite-andesites of Narcondam volcano in the Andaman Sea-An imprint of magma mixing in the inner arc of the Andaman-Java subduction system. J. Volcanol. Geotherm. Res. 168, 93–113 (2007).
Raman, T. S. R. et al. An expedition to Narcondam: Observations of marine and terrestrial fauna including the island-endemic hornbill. Curr. Sci. 105, 346–360 (2013).
Krishnan, M. S. Volcanic episodes in Indian geology. J. Madras. Univ. 27, 193–209 (1957).
Percie du Sert, N. et al. Reporting animal research: Explanation and elaboration for the ARRIVE guidelines 2.0. PLoS Biol. 18, e3000411 (2020).
Sikes, R. S. The Animal Care and Use Committee of the American Society of Mammalogists. Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. J. Mamm. 97, 663–688 (2016).
Pankakoski, E. Variation in the tooth wear of the shrews Sorex araneus and S. minutus. Ann. Zool. Fennici 26, 445–457 (1989).
Menon, V. Indian Mammals—A Field Guide 528 (Hachette Book Publishing India Pvt Limited, Gurgaon, 2014).
Jiang, X. L. & Hoffmann, R. S. A revision of the white-toothed shrews (Crocidura) of Southern China. J. Mamm. 82, 1059–1079 (2001).
Tez, C. & Kefelioglu, H. Does Crocidura pergrisea arispa Spitzenberger, 1971 occur in Turkey? Pak. J. Biol. Sci. 3, 2197–2198 (2000).
Sambrook, J. & Russell, D. W. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2001).
Verma, S. K. & Singh, L. Novel universal primers establish identity of an enormous number of animal species for forensic application. Mol. Ecol. Notes 3, 28–31 (2002).
Motokawa, M. et al. Phylogenetic relationships among east Asian species of Crocidura (Mammalia, Insectivora) inferred from mitochondrial cytochrome b gene sequences. Zool. Sci. 17, 497–504 (2000).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. JModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 9, 772 (2012).
Trifinopoulos, J., Nguyen, L.-T., von Haeseler, A. & Minh, B. Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235 (2016).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019).
Upham, N. S., Esselstyn, J. A. & Jetz, W. Ecological causes of uneven diversification and richness in the mammal tree of life. BioRxiv. https://doi.org/10.1101/504803 (2019).
Garg, S. & Biju, S. D. New microhylid frog genus from Peninsular India with Southeast Asian affinity suggests multiple Cenozoic biotic exchanges between India and Eurasia. Sci. Rep. 9, 1906 (2019).
Bandopadhyay, P. C. Chapter 12 Innerarc Volcanism: Barren and Narcondam Islands Vol. 47, 167–192 (Geological Society, 2017).
Pal, T. & Bhattacharya, A. Block-and-ash flow deposit of the Narcondam Volcano: product of dacite-andesite dome collapse in the Burma-Java subduction complex. Island Arcs. 20, 520–534 (2011).
Rodolfo, K. S. Bathymetry and marine geology of the Andaman basin, and tectonic implications for Southeast Asia. Geol. Soc. Am. Bull. 80, 1203–1230 (1969).
Tripathi, S. K. et al. Morphology of submarine volcanic seamounts from inner volcanic arc of Andaman Sea. Indian J. Geosci. 71, 451–470 (2017).
IUCN. The IUCN Red List of Threatened Species, Version 2020–3 (IUCN, 2020).
Molur, S. et al. Status of Non-volant Small Mammals: Conservation Assessment and Management Plan (CAMP) Workshop Report 618 (Zoo Outreach Organisation, 2005).
Mohan, A. V., Orozco-terWengel, P., Shanker, K. & Vences, M. The Andaman day gecko paradox: An ancient endemic without pronounced phylogeographic structure. Sci. Rep. 10, 11745 (2020).
MacArthur, R. H. & Wilson, E. O. The Theory of Island Biogeography 224 (Princeton University Press, Princeton, NJ, 2001).
Acknowledgements
The authors are grateful to the Ministry of Environment, Forest and Climate Change (MoEF&CC), Government of India for funding of this work and the Officials of the Department of Environment and Forests, Andaman and Nicobar Islands for the necessary permission to carry out the survey and sampling on the Narcondam Island. We are also thankful to the anonymous reviewers and Professor Rainer Breitling, Manchester Institute of Biotechnology, The University of Manchester for their critical comments and suggestions. The third author (S.K) acknowledges a fellowship grant received from the Council of Scientific and Industrial Research (CSIR) Senior Research Associateship (Scientists’ Pool Scheme) Pool No. 9072-A.
Funding
Ministry of Environment, Forest and Climate Change (MoEF&CC), Government of India (Zoological Survey of India Core Funding).
Author information
Authors and Affiliations
Contributions
Conceptualization: M.K. and S.K.; Data curation: G.G.; Formal analysis: M.K. and S.K.; Funding acquisition: C.S. and K.C.; Investigation: M.K., S.K. and C.V.; Methodology: M.K., S.K. and G.G.; Project administration: C.S. and K.C.; Resources: C.S. and K.C.; Software: M.K. and S.K.; Validation: M.K. and S.K.; Visualization: C.S., C.V. and K.C.; Writing—original draft: M.K. and S.K.; Writing—review & editing: M.K., S.K. and K.C.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kamalakannan, M., Sivaperuman, C., Kundu, S. et al. Discovery of a new mammal species (Soricidae: Eulipotyphla) from Narcondam volcanic island, India. Sci Rep 11, 9416 (2021). https://doi.org/10.1038/s41598-021-88859-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-88859-4
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.