Abstract
Cushing’s syndrome is an endocrine disease in dogs that negatively impacts upon the quality-of-life of affected animals. Cushing’s syndrome can be a challenging diagnosis to confirm, therefore new methods to aid diagnosis are warranted. Four machine-learning algorithms were applied to predict a future diagnosis of Cushing's syndrome, using structured clinical data from the VetCompass programme in the UK. Dogs suspected of having Cushing's syndrome were included in the analysis and classified based on their final reported diagnosis within their clinical records. Demographic and clinical features available at the point of first suspicion by the attending veterinarian were included within the models. The machine-learning methods were able to classify the recorded Cushing’s syndrome diagnoses, with good predictive performance. The LASSO penalised regression model indicated the best overall performance when applied to the test set with an AUROC = 0.85 (95% CI 0.80–0.89), sensitivity = 0.71, specificity = 0.82, PPV = 0.75 and NPV = 0.78. The findings of our study indicate that machine-learning methods could predict the future diagnosis of a practicing veterinarian. New approaches using these methods could support clinical decision-making and contribute to improved diagnosis of Cushing’s syndrome in dogs.
Similar content being viewed by others
Introduction
Cushing’s syndrome (or hyperadrenocorticism) is an endocrine disease in dogs that occurs due to a chronic excess of circulatory glucocorticoids that ultimately produce the classical clinical signs in affected dogs1. Affected dogs typically show various combinations of polyuria, polydipsia, polyphagia, a potbellied appearance, muscle weakness, bilateral alopecia, panting and lethargy1,2,3,4. These clinical signs, along with potential consequential complications of the disease such as diabetes mellitus, pancreatitis and hypertension, highlight the importance of timely diagnosis and optimal control of Cushing’s syndrome for ongoing health and good quality-of-life5, 6. However, Cushing’s syndrome can be a challenging diagnosis to confirm due to non-pathognomonic clinical features making it difficult to distinguish from other possible diseases, low disease prevalence within the general dog population estimated at 0.28% and the absence of highly accurate diagnostic tests2, 7,8,9. Obtaining a correct and timely diagnosis of Cushing’s syndrome is crucial for early commencement of appropriate treatment to improve the quality-of-life of affected dogs10. Additionally, an incorrect diagnosis of Cushing’s syndrome could lead to unnecessary treatment which could be potentially harmful. Therefore, new methods to aid the diagnosis of Cushing’s syndrome are warranted.
A number of epidemiological studies have provided evidence for associations for several risk factors with Cushing’s syndrome such as increasing age, specific breeds and sex2, 7. Additionally one study has demonstrated the predictive ability of demographic and clinical features in dogs with Cushing’s syndrome under primary veterinary care in the UK, using stepwise logistic regression to develop a risk score4. Alternative advanced statistical and machine-learning methods are available and could offer an alternative, improved approach to standard prediction modelling11. Machine-learning methods have been demonstrated to outperform conventional risk models for disease prediction due to their ability to model complex, non-linear interactions between features (variables) and to handle higher numbers of features11, 12. Machine-learning based classification algorithms have been increasingly described in the human and veterinary medical literature, and have been applied to specific clinical problems such as using laboratory data to identify dogs with Addison’s disease and to identify cats with chronic kidney disease11, 13,14,15,16,17. A machine-learning tool to predict dogs with Cushing’s syndrome could aid veterinarians within the practice setting and could facilitate timely commencement of treatment for affected dogs.
This study aimed to explore whether novel applications of machine-learning methods to UK primary-care veterinary electronic patient records could predict a veterinarian’s recorded diagnosis of Cushing’s syndrome using clinical information at the point of first suspicion of disease.
Results
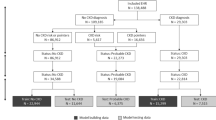
Anonymised data were collected from 886 primary-care UK veterinary practices participating within the VetCompass programme. The study population contained 905,544 dogs attending practices in 2016, of which 10,141 were identified to have a mention of Cushing’s syndrome within their electronic patient records (EPRs). Manual revision of 62% (6287) of these EPRs identified dogs meeting the study inclusion criteria; 419 cases (recorded as having Cushing’s syndrome) and 581 non-cases (suspected of having Cushing’s syndrome but ruled out after further investigation). Dogs with no recorded information regarding clinical signs within the two-week period of first suspicion were excluded from the study, retaining 398/419 (95.0%) cases and 541/581 (93.1%) non-cases for analysis. Thirty features (variables) were extracted from the EPRs of dogs included in the study.
Data pre-processing
Three features were removed from analysis due to near zero variance within the dataset: current administrations of insulin, l-thyroxine supplementation and anti-hypertensive agents. Six features were removed due to large proportions of missing data; body temperature (67.9% missing), heart rate (61.8%), alkaline phosphatase (ALKP) measurements (67.9%), urine specific gravity (USG) measurements (61.3%), presence of proteinuria (64.0%) and dilute USG (59.9%) at first suspicion. No high correlation between variables was identified, retaining twenty-one features (Table 1). Following one-hot encoding of breed, sex-neuter and weight change, 40 features were included in the modelling process.
Data were split randomly with two-thirds of the data incorporated into a training dataset, used to optimise the prediction model (n = 626; cases = 259 and non-cases = 367). The remaining one-third of the data formed a testing dataset, used to independently evaluate the model performance (n = 313; cases = 139 and non-cases = 174).
Model training and optimisation
Four models were trained and optimised:
-
(i)
A Least Absolute Shrinkage and Selection Operator (LASSO) model was optimised with a penalty (lambda) of 0.014 during tenfold cross-validation. The application of this penalty term to the likelihood being maximised results in feature selection at the time of model training. The features retained in the final model included: age, specified breeds, sex, clinical signs and laboratory features (Table 2). The model demonstrated good discrimination when examining the confusion matrix during cross-validation of the training dataset with an area under the receiver operating characteristic (AUROC) curve of 0.83 (95% confidence interval (CI): 0.80–0.86) (Table 3).
-
(ii)
A random forest (RF) model was optimised by tuning the ‘mtry’ hyperparameter to include 3 features per node split and the ‘ntree’ hyperparameter to grow 200 trees. The optimum selected threshold for the diagnosis of Cushing’s syndrome was ≥ 0.50 predicted probability which obtained a maximum PPV of 0.71 and NPV 0.72. The training dataset performance had an area under the receiver operating characteristic curve of 0.77 (95% CI 0.73–0.81). Variable importance analysis illustrated that the majority of features had small contributions to improved prediction accuracy within the RF model. Clinical signs of a potbelly and polyuria, and laboratory features (ALT and ALKP) had the greatest influence on the model.
-
(iii)
The linear support vector machine (SVM) model was optimised using tenfold cross-validation on the training set. A range of 0.25 to 16 was searched for the cost hyperparameter and the optimal value was 0.5. The training dataset performance had an area under the receiver operating characteristic curve of 0.83 (95% CI 0.80–0.87).
-
(iv)
The non-linear SVM model with a radial basis function (RBF) kernel was optimised by tenfold cross-validation using a grid search, tuning the cost hyperparameter to 4 (searched between 0.25 and 16) and the gamma hyperparameter to 0.02 (searched between 0.01 and 32)18. The training dataset performance had an area under the receiver operating characteristic curve of 0.84 (95% CI 0.81–0.87).
The four models indicated good performance in the training dataset all with an AUROC ≥ 0.77 (Table 3). The non-linear RBF SVM model indicated the best performance during model training with the highest AUROC.
Final model performance on test dataset
Final performance of the models was assessed on the independent test dataset. All models indicated good discrimination (Fig. 1) and calibration (Fig. 2). The LASSO model indicated the best performance when applied to the test dataset (Table 4) with an AUROC = 0.85 (95% CI 0.80–0.89) which is consistent with the training cross-validation performance (Table 3). The calibration plot of the LASSO model demonstrated good model calibration with narrow 95% confidence intervals that crossed the 45 degree line of perfect calibration. The calibration plot indicates slightly poorer calibration of the lower probability predictions in the model.
Receiver operating characteristic curve for the final prediction models for a diagnosis of Cushing’s syndrome evaluated in an independent test dataset, applied to dogs attending primary-care veterinary practices in the UK (n = 313; cases = 139 and non-cases = 174). LASSO, least absolute shrinkage and selection operator; RF, random forest; SVM, support vector machine; RBF, radial basis function.
Calibration plots of the final prediction models for a diagnosis of Cushing’s syndrome, applied to dogs attending primary-care veterinary practices in the UK (n = 313; cases = 139 and non-cases = 174). The plot describes the mean observed proportions of dogs with a diagnosis of Cushing’s compared to the mean predicted probabilities, by deciles of predictions. The 45 degree line denotes perfect calibration.
The SVM models demonstrated a drop in performance when applied to the test set: Linear SVM AUROC = 0.73 (95% CI 0.67–0.78); RBF SVM AUROC = 0.72 (95% CI 0.66–0.78) (Table 3). The RF model maintained a reasonable performance (AUROC = 0.74; 95% CI 0.68–0.79). Calibration plots suggested good calibration for the RF and SVM models (Fig. 2).
Discussion
This study demonstrates the ability of machine-learning methods to correctly classify the recorded veterinarian diagnosis of Cushing’s syndrome in dogs from the point of first suspicion, using electronic patient records of dogs under primary veterinary care in the UK. Our study assessed four classification machine-learning models, all with good predictive performance. Of our four models, the LASSO penalised regression was the best performing model for support of a diagnosis of Cushing’s syndrome with the highest AUROC in the test set validation. The LASSO aims to selects a model that achieves a trade-off between goodness of fit and model complexity, from a large list of potential models19. This has been used in other prediction methods and is recommended for use in the consensus paper for medical prediction models12, 20, 21. Little overfitting was observed in the calibration plot of the LASSO model; however greater uncertainty was observed at the lowest predictions, with 95% confidence intervals narrowly missing the 45 degree line of perfect calibration. LASSO performs feature selection at the same time as model training, therefore requires fewer features to be considered and could be easily implemented for use in practice22. Another benefit of the LASSO is that it works well in low-dimensional, binary data which could be a reason for its superior performance in the classification of Cushing’s syndrome diagnosis23.
The RBF SVM model had the superior performance to all optimised models during the cross-validation of the training dataset with an AUROC of 0.84, however performance dropped to 0.72 in the test dataset. The RF model had the poorest performance when applied to the training datasets and retained a reasonable performance in the test dataset. There are many machine-learning methods that can be used for classification problems, each with their own advantages and drawbacks12, 24. A review paper examined the performance of different machine-learning methods for disease prediction and found the methods performed differently depending on the types of data used. SVM and RF models were found to perform less well than simpler models, such as regression models, when clinical and demographic data were used, which reflects the findings in the current study24. SVM has advantages in high dimensional datasets (considering large numbers of features) as well as for features with small predictive effects (such as for genome-wide associations)25. The gamma hyperparameter of the RBF SVM model affects the complexity of the model with higher values of gamma increasing the flexibility of the SVM hyperplane. In the current study, performance of the non-linear RBF SVM model was similar to the linear SVM model in the test dataset suggesting the non-linear model could have largely learnt a linear relationship. This could be due to the predominant inclusion of binary features in our dataset22. In our study a low gamma hyperparameter for the non-linear model was identified during model optimisation suggesting a less complex relationship was being modelled by the RBF SVM model26.
The drop in performance of the RBF SVM model when applied to the test dataset could indicate overfitting of the models to the training data or could be as a result of randomly splitting the data into a single training and testing group. A single train-test split is dependent on which data are randomly allocated to either group and can result in high variability between the two datasets and is less reliable at inferring generalisability of model performance27, 28. Other methods such as nested cross-validation can be used to reduce test set variability and could provide a less biased estimate of model generalisation performance27, 29. This could be used as an alternative strategy in future studies. The poorer performance of the RF model could be due to the inclusion of predominantly binary categorical variables, resulting in the model growing sparse decision trees22, 30. When examining the importance plot of the features in the RF model, the majority of features had a low mean decrease in accuracy and Gini which could have resulted in a model that is not highly robust. However the features with most importance were the presence of polyphagia, and polyuria as well as abnormal ALT and ALKP laboratory findings, which are clinical features frequently reported in dogs with Cushing’s syndrome1, 9.
Automated prediction of Cushing’s syndrome in dogs could support veterinarian decision-making and contribute to improved diagnosis of the disease. The currently available tests used for the diagnosis of Cushing’s syndrome in primary-care practice have varying sensitivities and specificities. The ACTH stimulation test is the most commonly used test in primary-care practice and has an estimated sensitivity between 57 and 83% and a specificity between 59 and 95%2, 31,32,33,34. The test characteristics vary according to the study referenced with superior test specificity estimates stemming from test populations which include healthy controls or controls without a clinical suspicion of Cushing’s syndrome. When applied to comparison non-case populations, similar to those used in our study, the ACTH stimulation test specificity falls between 59 and 61%31, 32, 35. The LDDST has an estimated sensitivity between 85 and 97% and a specificity between 70 and 73%8, 34, 36. In primary-care practice, these tests may be performed in dogs with a low suspicion of Cushing’s syndrome which can add to the uncertainty of interpretation for veterinarians and multiple tests are often performed, increasing the financial cost during the diagnostic process to the dog owner4. A prediction tool with good reliability that could be used from the point of first suspicion would offer a minimally invasive and low cost diagnostic method to support the veterinarian. Insured dogs are four times more likely to be diagnosed with Cushing’s syndrome compared to non-insured dogs, suggesting a high level of under-diagnosis related to the financial burden of gaining a diagnosis for this disease2. Future application of the LASSO predictive algorithm could be used to develop a computer application for mobile devices or implement it within a clinical practice management system to provide automated prediction within the consultation room37.
The models in this study included the information available to veterinarians during the initial stages of disease investigation; therefore these data largely include the dog’s demographic factors and presenting clinical signs. The good performance of these models suggests that discrimination of dogs with and without Cushing’s syndrome can be correctly determined at the point of first suspicion based on these factors. Due to some laboratory tests performed at external laboratories, specific measurements were therefore not routinely captured within VetCompass unless laboratory results were manually recorded within the free text clinical notes. Inclusion of specific laboratory measurement data into our study was limited. The predictive ability of these models could be improved with inclusion of additional features, with further laboratory factors offering an opportunity of future model adjustment and improved predictive performance.
There are some limitations to this study. This study used supervised machine-learning methods that require structured data for model training. In veterinary EPRs there are some standardised coding systems in place, such as VeNom coding systems, however these are not commonly used in clinical practice with the majority of information recorded as clinical free text38. Clinical features in this study were extracted through manual revision of the clinical notes, restricting the sample size. Future work for feature extraction using natural language processing methods or classification of clinical features could be beneficial for the clinical application of such predictive algorithms to optimise the analysis of large datasets, like VetCompass39. Due to the retrospective collection of the data, there is a possibility of feature misclassification and an introduction of noise, which could have diluted some predictive effects. The sample size included in this study is comparable to similar studies. However it is possible that additional training examples would support further improvements in prediction13, 16. Finally, further investigation on an independent dataset from a different cohort of dogs could examine the external validation of these models40.
In conclusion, this study applied four machine-learning models to predict the diagnosis of Cushing’s syndrome in dogs from the point of first suspicion of disease. The LASSO penalised regression model was the best performing model when applied to a held-out test dataset. The findings indicate machine-learning aided diagnosis could predict the diagnosis of a practising veterinarian and that utilising machine-learning methods as decision support tools, may contribute to improved diagnosis in Cushing’s syndrome in dogs. This study has shown that is it feasible to apply machine-learning methods to clinical data available within primary veterinary care EPRs for disease prediction and could open up the opportunities for further development in this area through application to other clinical problems.
Methods
Data were collected from the VetCompass programme, which collates EPRs from primary-care veterinary practices in the UK. To be included in the study, dogs in the VetCompass cohort were required to have been under veterinary care in 2016 which was defined as having at least one EPR recorded during 2016 and/or at least one EPR recorded both in 2015 and 2017. To identify dogs where Cushing’s syndrome was considered as a clinical diagnosis, search terms were applied to the EPRs: ‘Cushing*, HAC, hyperadren*, hyperA, trilos*, Vetory*’. A random selection of dogs identified by the search terms were reviewed through manual revision of the EPRs. Dogs were included as a case if, (i) an initial diagnosis of Cushing’s syndrome was recorded within their EPR between 1 January 2016 and 1 June 2018 and (ii) a record was present of a low dose dexamethasone suppression test (LDDST) or adrenocorticotropic hormone (ACTH) stimulation test being performed within the EPR prior to diagnosis. Dogs were excluded as a case if a diagnosis was made prior to their first available patient record during the study period or dogs were considered to have iatrogenic Cushing’s syndrome (had glucocorticoid administration in the 30 days prior to first suspicion). Dogs were included as a comparison reference population of non-cases if, (i) there was a recorded suspicion of Cushing’s syndrome within the EPR, (ii) they subsequently had Cushing’s syndrome ruled out after undergoing a urine cortisol-creatinine ratio (UCCR) test, LDDST and/or an ACTH stimulation test between 1 January 2016 and 1 June 2018 and (iii) an alternative diagnosis was made within the EPR. Dogs with no recorded information regarding clinical signs were excluded from analysis.
Multiple features (variables) were extracted for analysis. Demographic features including breed, sex, neuter status, date of birth and bodyweight were routinely recorded within the EPRs. Breeds were categorised according to a standardised breed list adapted from the VeNom Coding Group system (Venom Coding Group 2019). Individual breeds were specified if at least 10 dogs of that breed had been included as a case or non-case. All other purebreds were grouped into a ‘purebreed other’ category. Dogs classified as a crossbreed (e.g. poodle X) or a designer breed (e.g. cockapoo) were classified into a ‘crossbreed’ category. Sex was categorised to include neuter status: female-entire, female-neuter, male-entire or male-neuter. Age at first suspicion (years) was calculated by using the date of birth and date of first suspicion of Cushing’s syndrome. Bodyweight (kg) was the bodyweight value recorded closest to the date of first suspicion. A change in weight was calculated using the recorded weight at the date of first suspicion and that recorded one year previously, where available.
Additional data were extracted manually from the EPRs. Date of first suspicion was the earliest date with evidence in the EPRs that Cushing’s syndrome was being considered as a diagnosis, and subsequently led to the veterinarian to pursue the diagnosis through further investigation. Clinical signs and routine laboratory measurements present at first suspicion (recorded one week prior and one week after the date of first suspicion) were extracted. Individual clinical signs were recorded as binary features: ‘present’ or ‘not present’ (‘not present’ was recorded if the clinical sign was specifically recorded as not present or if no information was recorded). ALKP and ALT were recorded as ‘elevated’, ‘not elevated’ or ‘unknown’ (either no test was performed or results were not reported). Proteinuria (based on a urine dipstick, including a trace recording or a urine protein-creatinine ratio) was recorded as ‘present’, ‘not present’ or ‘unknown. USG was recorded as ‘dilute’ (≤ 1.020), ‘not dilute’ (> 1.020) or ‘unknown’. Continuous data for recorded ALKP enzyme activities and USG measurements were also extracted. Treatment data (currently being received when first suspected of Cushing’s syndrome) for insulin, l-thyroxine supplementation and anti-hypertensive agents (amlodipine, benazepril, enalapril or telmisartan) were extracted41. Additionally clinical management data on whether dogs were hospitalised in the previous 12 months before first suspicion was included9.
Data pre-processing
All analyses were performed in R version 4.0.042. Features were descriptively analysed with categorical data assessed using the counts and corresponding percentages. For continuous data, normally distributed data were summarised using the mean (standard deviation (SD)) and non-normally distributed data using the median (interquartile range (IQR) and range). Variance of the features for all dogs was assessed and those with zero or near-zero variance (proportion of unique values over the sample size was < 10%) were excluded from analysis22. Pairwise correlations between predictor features were explored to identify collinearity using correlation coefficients; correlations (r) > 0.80 were considered highly correlated43. When pairs of highly correlated predictor features were identified, the variable considered to be most complete within the dataset and most clinically relevant was selected for modelling43. Data were assessed for missing values with features excluded if > 50% of the data was missing44.
The selected data were randomly split into two parts. Two-thirds (67%) of the data were allocated to a training dataset and one-third (33%) to the test dataset. Features with ≤ 50% missing data were imputed separately for training and test sets using multiple imputation by chained equations using the mice package in R22, 45, 46. Continuous variables had a normal distribution and were standardised for analysis by converting to z-scores47. One-hot encoding was applied to nominal features; breed, neuter-status and weight change22.
Model training and optimisation
Four prediction models using different supervised machine-learning algorithms were applied to the training set: LASSO, RF, a linear SVM and a non-linear SVM. For each algorithm, hyperparameter tuning was conducted by cross-validation to optimise the models and to minimise model overfitting48. The hyperparameters tuned varied between the different algorithms.
-
(i)
LASSO is a penalised regression method49. This method adds a penalty (lambda) to the sum of the absolute coefficients which shrinks the coefficients towards the null, with each predictor coefficient shrunk differently. The addition of a penalty reduces the likelihood of the model overfitting the data to improve prediction accuracy22. Lambda was optimised by tenfold cross-validation50. The mean lambda from the cross-validation loops was applied to the training set to determine the final model coefficients and training set performance51. The model was applied using the glmnet package in R which automatically standardises the data for the estimation of predictor effects and back transforms the final regression coefficients on the original scale51.
-
(ii)
RF is an ensemble learning based classification method22. It uses training data to construct multiple decision-trees by bootstrap resampling and classifies unseen data using the mode of the tree output decisions30. These decision trees have small randomised differences in characteristics, which improves generalisation performance. Tuning of the model was performed by changing the number of decision trees grown within the ensemble (‘ntree’) and the number of features randomly sampled as candidates at each tree split (‘mtry’). Variable importance was determined for each tree within the final optimised random forest model by calculating the permutation importance index as well as measuring the decrease in node impurity22, 52. Importance was assessed by mean decrease accuracy, indicating the mean decrease in model accuracy due to the exclusion of that feature, and by mean decrease Gini, indicating the mean decrease in node purity achieved by each feature. The model was applied using the rpart package in R53.
-
(iii)
SVM models map training data into a multi-dimensional space and separate the binary outcome data by a hyperplane that is maximally distant from the two outcome groups26. This best separating hyperplane minimises classification error and maximises geometric margin of classification. Two models were assessed: a linear and a non-linear model (with a radial basis function (RBF) kernel). The non-linear kernel SVM model can learn more complex hyperplanes than a linear SVM model22. Model tuning was performed for the optimal cost hyperparameter for the linear SVM model using tenfold cross-validation. The optimal cost and kernel function (gamma) hyperparameters were tuned for the non-linear RBF SVM model, using a grid search with tenfold cross-validation. The two SVM models were applied using the e1071 package in R18.
Models were optimised through cross-validation by maximising the area under the receiver operating characteristic curve11. The optimum predictive thresholds for the LASSO and RF models were identified by maximising the PPV without a detrimental decrease in the NPV as this was deemed the most clinically valuable classification for clinicians. Once the hyperparameters had been optimised via cross-validation on the training dataset, the final model parameters were then applied to the whole training dataset11, 21, 22. The training performance was presented by plotting the ROC curve, calculating the AUROC curve and examining the confusion matrix (outlining sensitivity, specificity, PPV, NPV, kappa statistic and accuracy). Confidence intervals for AUROC were calculated using the DeLong method54 and exact binomial confidence intervals were presented for accuracy55. The best performing model in the training set was defined by having the highest AUROC.
Final model performance
Performance of the final, tuned models were assessed by applying the selected prediction model to the independent test dataset. Final model performance was assessed by a confusion matrix and AUROC curves to examine the discriminatory ability of the models (distinguishes between dogs that have the outcome and those that do not)22, 56. Calibration of the models (the agreement between the observed outcomes and predictions) was assessed by calibration plots to assess the reliability of the probability estimates of the final models57, 58. The plots compared the mean observed proportions of dogs with a diagnosis of Cushing’s to the mean predicted probabilities by deciles of predictions. Perfect predictions should lie on the 45 degree line56, 57. The best performing model in the test set was defined as having the highest AUROC and a corresponding calibration curve indicating good calibration.
Ethical approval
Ethical approval was granted by the Royal Veterinary College Ethics and Welfare Committee (URN SR2018-1652). All methods were performed in accordance with the relevant regulations and the ARRIVE guidelines.
Abbreviations
- CI:
-
Confidence interval
- EPR:
-
Electronic patient record
- LASSO:
-
Least absolute shrinkage and selection operator
- RF:
-
Random forest
- SVM:
-
Support vector machine
- RBF:
-
Radial basis function
- UCCR:
-
Urine cortisol-creatinine ratio
- LDDST:
-
Low dose dexamethasone suppression test
- AUROC:
-
Area under the receiver operating characteristic curve
References
Feldman, E. C. & Nelson, R. W. Canine and Feline Endocrinology and Reproduction (W.B. Saunders, 2004).
O’Neill, D. G. et al. Epidemiology of hyperadrenocorticism among 210,824 dogs attending primary-care veterinary practices in the UK from 2009 to 2014. J. Small Anim. Pract. 57, 365–373. https://doi.org/10.1111/jsap.12523 (2016).
Nagata, N., Kojima, K. & Yuki, M. Comparison of Survival times for dogs with pituitary-dependent hyperadrenocorticism in a primary-care hospital: treated with trilostane versus untreated. J. Vet. Intern. Med. 31, 22–28. https://doi.org/10.1111/jvim.14617 (2017).
Schofield, I. et al. Development and internal validation of a prediction tool to aid the diagnosis of Cushing’s syndrome in dogs attending primary-care practice. J. Vet. Intern. Med. (2020).
Cook, A. K., Breitschwerdt, E. B., Levine, J. F., Bunch, S. E. & Linn, L. O. Risk factors associated with acute pancreatitis in dogs: 101 cases (1985–1990). J. Am. Vet. Med. Assoc. 203, 673–679 (1993).
Miceli, D. D., Pignataro, O. P. & Castillo, V. A. Concurrent hyperadrenocorticism and diabetes mellitus in dogs. Res. Vet. Sci. 115, 425–431. https://doi.org/10.1016/j.rvsc.2017.07.026 (2017).
Carotenuto, G. et al. Cushing’s syndrome—an epidemiological study based on a canine population of 21,281 dogs. Open Vet. J. 9, 27–32 (2019).
Bennaim, M., Shiel, R. E., Forde, C. & Mooney, C. T. Evaluation of individual low-dose dexamethasone suppression test patterns in naturally occurring hyperadrenocorticism in dogs. J. Vet. Intern. Med. 32, 967–977. https://doi.org/10.1111/jvim.15079 (2018).
Behrend, E. N., Kooistra, H. S., Nelson, R., Reusch, C. E. & Scott-Moncrieff, J. C. Diagnosis of spontaneous canine hyperadrenocorticism: 2012 ACVIM consensus statement (small animal). J. Vet. Intern. Med. 27, 1292–1304. https://doi.org/10.1111/jvim.12192 (2013).
Schofield, I. et al. Development and evaluation of a health-related quality-of-life tool for dogs with Cushing’s syndrome. J. Vet. Intern. Med. 33, 2595–2604. https://doi.org/10.1111/jvim.15639 (2019).
Weng, S. F., Reps, J., Kai, J., Garibaldi, J. M. & Qureshi, N. Can machine-learning improve cardiovascular risk prediction using routine clinical data?. PLoS ONE 12, e0174944. https://doi.org/10.1371/journal.pone.0174944 (2017).
Christodoulou, E. et al. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J. Clin. Epidemiol. 110, 12–22. https://doi.org/10.1016/j.jclinepi.2019.02.004 (2019).
Reagan, K. L., Reagan, B. A. & Gilor, C. Machine learning algorithm as a diagnostic tool for hypoadrenocorticism in dogs. Domest. Anim. Endocrinol. https://doi.org/10.1016/j.domaniend.2019.106396 (2019).
Bradley, R. et al. Predicting early risk of chronic kidney disease in cats using routine clinical laboratory tests and machine learning. J. Vet. Intern. Med. 33, 2644–2656. https://doi.org/10.1111/jvim.15623 (2019).
Fenlon, C. et al. A comparison of 4 predictive models of calving assistance and difficulty in dairy heifers and cows. J. Dairy Sci. 100, 9746–9758 (2017).
Hyde, R. M. et al. Automated prediction of mastitis infection patterns in dairy herds using machine learning. Sci. Rep. 10, 4289. https://doi.org/10.1038/s41598-020-61126-8 (2020).
Jammeh, E. A. et al. Machine-learning based identification of undiagnosed dementia in primary care: a feasibility study. BJGP Open 2 (2018).
Meyer, D. et al. Package ‘e1071’. R J. (2019).
Bunea, F. et al. Penalized least squares regression methods and applications to neuroimaging. Neuroimage 55, 1519–1527 (2011).
Collins, G. S., Reitsma, J. B., Altman, D. G. & Moons, K. G. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ 350, g7594. https://doi.org/10.1136/bmj.g7594 (2015).
Kidd, A. C. et al. Survival prediction in mesothelioma using a scalable Lasso regression model: instructions for use and initial performance using clinical predictors. BMJ Open Respir. Res. 5, e000240. https://doi.org/10.1136/bmjresp-2017-000240 (2018).
Kuhn, M. & Johnson, K. Applied Predictive Modeling Vol. 26 (Springer, 2013).
Pavlou, M., Ambler, G., Seaman, S., De Iorio, M. & Omar, R. Z. Review and evaluation of penalised regression methods for risk prediction in low-dimensional data with few events. Stat. Med. 35, 1159–1177. https://doi.org/10.1002/sim.6782 (2016).
Uddin, S., Khan, A., Hossain, M. E. & Moni, M. A. Comparing different supervised machine learning algorithms for disease prediction. BMC Med. Inform. Decis. Mak. 19, 281. https://doi.org/10.1186/s12911-019-1004-8 (2019).
Yu, W., Liu, T., Valdez, R., Gwinn, M. & Khoury, M. J. Application of support vector machine modeling for prediction of common diseases: the case of diabetes and pre-diabetes. BMC Med. Inform. Decis. Mak. 10, 16 (2010).
Vapnik, V. The Nature of Statistical Learning Theory (Springer, 2013).
Varma, S. & Simon, R. Bias in error estimation when using cross-validation for model selection. BMC Bioinform. 7, 91. https://doi.org/10.1186/1471-2105-7-91 (2006).
Harrell, F. E. Jr., Lee, K. L. & Mark, D. B. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat. Med. 15, 361–387 (1996).
Raschka, S. Model evaluation, model selection, and algorithm selection in machine learning. arXiv preprint arXiv:1811.12808 (2018).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Monroe, W. E., Panciera, D. L. & Zimmerman, K. L. Concentrations of noncortisol adrenal steroids in response to ACTH in dogs with adrenal-dependent hyperadrenocorticism, pituitary-dependent hyperadrenocorticism, and nonadrenal illness. J. Vet. Intern. Med. 26, 945–952. https://doi.org/10.1111/j.1939-1676.2012.00959.x (2012).
Nivy, R. et al. The interpretive contribution of the baseline serum cortisol concentration of the ACTH stimulation test in the diagnosis of pituitary dependent hyperadrenocorticism in dogs. J. Vet. Intern. Med. 32, 1897–1902. https://doi.org/10.1111/jvim.15330 (2018).
Kaplan, A. J., Peterson, M. E. & Kemppainen, R. J. Effects of disease on the results of diagnostic tests for use in detecting hyperadrenocorticism in dogs. J. Am. Vet. Med. Assoc. 207, 445–451 (1995).
Van Liew, C. H., Greco, D. S. & Salman, M. D. Comparison of results of adrenocorticotropic hormone stimulation and low-dose dexamethasone suppression tests with necropsy findings in dogs: 81 cases (1985–1995). J. Am. Vet. Med. Assoc. 211, 322–325 (1997).
Reusch, C. E. & Feldman, E. C. Canine hyperadrenocorticism due to adrenocortical neoplasia. Pretreatment evaluation of 41 dogs. J. Vet. Intern. Med. 5, 3–10 (1991).
Rijnberk, A. & Mol, J. Assessment of two tests for the diagnosis of canine hyperadrenocorticism. Vet. Rec. 122, 178–180 (1988).
Noorbakhsh-Sabet, N., Zand, R., Zhang, Y. & Abedi, V. Artificial intelligence transforms the future of health care. Am. J. Med. 132, 795–801. https://doi.org/10.1016/j.amjmed.2019.01.017 (2019).
VeNom Coding Group. VeNom Veterinary Nomenclature (2018).
Kennedy, N., Brodbelt, D. C., Church, D. B. & O’Neill, D. G. Detecting false-positive disease references in veterinary clinical notes without manual annotations. NPJ Digit. Med. 2, 33. https://doi.org/10.1038/s41746-019-0108-y (2019).
Goldstein, B. A., Navar, A. M., Pencina, M. J. & Ioannidis, J. Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review. J. Am. Med. Inform. Assoc. 24, 198–208 (2017).
Acierno, M. J. et al. ACVIM consensus statement: Guidelines for the identification, evaluation, and management of systemic hypertension in dogs and cats. J. Vet. Intern. Med. 32, 1803–1822. https://doi.org/10.1111/jvim.15331 (2018).
R Core Team. (Vienna, Austria, 2013).
Dohoo, I. R., Martin, W. & Stryhn, H. Methods in Epidemiologic Research (VER Incorporated Charlottetown, 2012).
Rubin, D. B. Inference and missing data. Biometrika 63, 581–592 (1976).
Buuren, S. V. & Groothuis-Oudshoorn, K. mice: Multivariate imputation by chained equations in R. J. Stat. Softw. 1–68 (2010).
Graham, J. W. Missing data analysis: making it work in the real world. Annu. Rev. Psychol. 60, 549–576. https://doi.org/10.1146/annurev.psych.58.110405.085530 (2009).
Eager, C. standardize: Tools for standardizing variables for regression in R. R package version 0.2 1 (2017).
Molinaro, A. M., Simon, R. & Pfeiffer, R. M. Prediction error estimation: a comparison of resampling methods. Bioinformatics 21, 3301–3307 (2005).
Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B (Methodol.) 58, 267–288 (1996).
Kohavi, R. in Ijcai. 1137–1145 (Montreal, Canada).
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33, 1 (2010).
Strobl, C., Boulesteix, A.-L., Zeileis, A. & Hothorn, T. Bias in random forest variable importance measures: illustrations, sources and a solution. BMC Bioinform. 8, 25. https://doi.org/10.1186/1471-2105-8-25 (2007).
Therneau, T., Atkinson, B., Ripley, B. & Ripley, M. B. Package ‘rpart’. http://cran.ma.ic.ac.uk/web/packages/rpart/rpart.pdf. Accessed 20 April 2016 (2015).
DeLong, E. R., DeLong, D. M. & Clarke-Pearson, D. L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44, 837–845 (1988).
Clopper, C. J. & Pearson, E. S. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 26, 404–413. https://doi.org/10.2307/2331986 (1934).
Steyerberg, E. W. et al. Assessing the performance of prediction models: a framework for some traditional and novel measures. Epidemiology (Camb. Mass.) 21, 128 (2010).
Fenlon, C., O’Grady, L., Doherty, M. L. & Dunnion, J. A discussion of calibration techniques for evaluating binary and categorical predictive models. Prev. Vet. Med. 149, 107–114. https://doi.org/10.1016/j.prevetmed.2017.11.018 (2018).
Steyerberg, E. W. & Vergouwe, Y. Towards better clinical prediction models: seven steps for development and an ABCD for validation. Eur. Heart J. 35, 1925–1931. https://doi.org/10.1093/eurheartj/ehu207 (2014).
Acknowledgements
We are grateful to Dechra Veterinary Products Ltd for their funding of this research. We acknowledge the Medivet Veterinary Partnership, Vets4Pets/Companion Care, Goddard Veterinary Group, Independent Vet Care, CVS Group, Beaumont Sainsbury Animal Hospital, Vets Now and the other UK practices who collaborate in VetCompass™. We are grateful to The Kennel Club and The Kennel Club Charitable Trust for supporting VetCompass™.
Funding
I.S is supported at the RVC by an award from Dechra Veterinary Products Ltd. Dechra Veterinary Products Ltd did not have any input in the design of the study, the collection, analysis and interpretation of data or in writing the manuscript.
Author information
Authors and Affiliations
Contributions
I.S. designed and executed the study, undertook data extraction, statistical analysis and wrote the manuscript. D.C.B. assisted in study design, data preparation and manuscript preparation. N.K. assisted in data preparation, provided expertise in data interpretation and manuscript preparation. S.J.M.N. assisted in study design and manuscript preparation. D.B.C. assisted in data preparation and manuscript preparation. R.F.G. assisted in study design and manuscript preparation. D.G.O. oversaw data preparation, assisted in study design and manuscript preparation. Each author agrees to be accountable for all aspects of the accuracy or integrity of the work.
Corresponding author
Ethics declarations
Competing interests
I.S is supported at the RVC by an award from Dechra Veterinary Products Ltd. S.J.M.N has undertaken consultancy work for Dechra Veterinary Products Ltd. The remaining authors have no conflicts of interest to declare.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Schofield, I., Brodbelt, D.C., Kennedy, N. et al. Machine-learning based prediction of Cushing’s syndrome in dogs attending UK primary-care veterinary practice. Sci Rep 11, 9035 (2021). https://doi.org/10.1038/s41598-021-88440-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-88440-z
This article is cited by
-
Predicting health outcomes in dogs using insurance claims data
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.