Abstract
Risk classification for prostate cancer (PCa) aggressiveness and underlying mechanisms remain inadequate. Interactions between single nucleotide polymorphisms (SNPs) may provide a solution to fill these gaps. To identify SNP–SNP interactions in the four pathways (the angiogenesis-, mitochondria-, miRNA-, and androgen metabolism-related pathways) associated with PCa aggressiveness, we tested 8587 SNPs for 20,729 cases from the PCa consortium. We identified 3 KLK3 SNPs, and 1083 (P < 3.5 × 10–9) and 3145 (P < 1 × 10–5) SNP–SNP interaction pairs significantly associated with PCa aggressiveness. These SNP pairs associated with PCa aggressiveness were more significant than each of their constituent SNP individual effects. The majority (98.6%) of the 3145 pairs involved KLK3. The 3 most common gene–gene interactions were KLK3-COL4A1:COL4A2, KLK3-CDH13, and KLK3-TGFBR3. Predictions from the SNP interaction-based polygenic risk score based on 24 SNP pairs are promising. The prevalence of PCa aggressiveness was 49.8%, 21.9%, and 7.0% for the PCa cases from our cohort with the top 1%, middle 50%, and bottom 1% risk profiles. Potential biological functions of the identified KLK3 SNP–SNP interactions were supported by gene expression and protein–protein interaction results. Our findings suggest KLK3 SNP interactions may play an important role in PCa aggressiveness.
Similar content being viewed by others
Introduction
Prostate cancer (PCa) accounts for over 10% of all cancer-related deaths, making it the second leading cause of cancer-related deaths among American men in 20211. Because of the substantial clinical heterogeneity of this disease, physicians often have difficulty distinguishing at the time of diagnosis between patients who will develop indolent tumors and those who will develop aggressive PCa2. For PCa patients considered to be at a low risk for aggressive PCa, conservative management and treatment are presently recommended. However, ~ 20% of PCa patients who are classified as a low risk using the known classification features (such as prostate specific antigen [PSA], tumor stage, and Gleason score) still die during conservative treatment3. This demonstrates an unmet need to identify better biomarkers for predicting PCa aggressiveness.
Genetic association studies have primarily focused on the effects of individual single-nucleotide polymorphisms (SNPs), which are insufficient to explain the complexity of disease susceptibility. However, the majority of SNPs identified by genome-wide association studies (GWAS) are for PCa risk, and only a few are for PCa progression. In fact, only 41 SNPs have been suggested to be associated with PCa progression related phenotypes (such as aggressiveness, early-onset, and PCa survival) in GWAS from the GWAS catalog4. These 41 SNPs are located across 53 genes (some are intergenic SNPs), including KLK3, ADGRG1, ARHGAP6, CASC8, and TCF44. We and others reported several polygenic risk scores (PRS) for PCa risk based on multiple individual SNP effects5,6,7, but the prediction model of PCa aggressiveness remains underdeveloped.
It has been established that gene–gene/SNP–SNP interactions may play a larger role in the causality of complex diseases8. Although SNP–SNP interactions have received more attention in the past decade, few have been validated, and most are without known biological functions. The limited SNP–SNP interaction findings may be due to insufficient statistical methods. The conventional approach for testing 2-way SNP–SNP interactions is the Additive–Additive full interaction (AA_Full) approach, the full or hierarchical interaction model (2 main effects + interaction) with the additive SNP inherited mode. AA_Full is the most complicated interaction pattern with the 9 distinct risk-groups, so a large sample size will be needed for detecting this complicated pattern. Using AA_Full to detect SNP–SNP interactions tends to lead to false-negative findings because it only tested one complicated interaction pattern. In order to overcome this issue, we developed two statistical methods: SNP Interaction Pattern Identifier (SIPI) and Additive-Additive 9 Interaction-Model approach (AA9int)9,10. The AA9int approach, which treats all SNPs in an additive inheritance mode, tests nine interaction patterns of pairwise SNP–SNP interactions associated with an outcome10. The SIPI approach is an extended version of AA9int and tests 45 biologically meaningful SNP–SNP interaction patterns by considering three SNP inheritance modes (additive, dominant and recessive)9.
Accumulating evidence suggests that interplay among angiogenesis-, mitochondria-, miRNA-, and androgen metabolism-related pathways may play a critical role in PCa11,12,13,14. Androgen expression, epigenetic factors, and oxygen levels in the tumor microenvironment regulate angiogenesis, which leads to metastatic PCa15,16. Therapies targeting both angiogenesis and androgen for recurrent PCa patients could increase survival rates17. These findings suggest that the relationship between androgen and angiogenesis and how their interaction can impact on PCa aggressiveness. Genes involved in androgen metabolism pathways also lead to oncogenic metabolic phenotypes, such as mitochondrial respiration and cell proliferation, in PCa cells. In addition, androgen repression in PCa cells decreases mitochondrial activity18. We and others have reported that miRNAs (such as miR-221, miR-222, and miR-155) are involved in regulating various aspects of angiogenesis and PCa progression12,19. This evidence suggests that the interplay among genes in these 4 pathways may impact PCa aggressiveness.
This study was inspired by the shortage of SNP findings associated with PCa aggressiveness. Only a small number of SNPs were identified to be associated with PCa aggressiveness, and there is no PRS for PCa aggressiveness. In addition, most of the PRS is based on the sum of several individual SNP effects without considering SNP–SNP interactions. Because of the inspiration, this study's objective is to identify SNP–SNP/gene–gene interactions and to build a SNP-interaction based PRS (SNPint-PRS) associated with PCa aggressiveness. These SNPs were selected based on genes in the four PCa biological related pathways (angiogenesis, mitochondria, miRNA, and androgen metabolism).
Results
Individual SNP effects
The process of identifying the significant effects of individual SNPs and SNP–SNP interactions associated with PCa aggressiveness is described in Fig. 1. The individual SNP effect analyses were performed for both the discovery and validation sets. The criterion of a P < 0.001 was applied in both the discovery and validation sets to declare validated results. As shown in Table 1, there were 3 KLK3 SNPs (rs17632542, rs62113212, and rs2569735) with a P < 0.001 in both study sets, and all of these three SNPs reached the Bonferroni significance level (P < 1.9 × 10–6) in the combined set. The best inheritance mode for these 3 SNPs was an additive mode. PCa cases with the minor G allele in rs17632542 tended to have a higher risk of PCa aggressiveness (odds ratio [OR]= 1.45; P = 1.7 × 10–13). Cases with the minor A allele in rs62113212 (OR per A allele 1.45; P = 2.1 × 10–13) and rs2569735 (OR= 1.23; P = 1.4 × 10–8) also had a higher risk of PCa aggressiveness. The relationships for these KLK3 SNPs are shown in Supplementary Fig. S1. rs17632542 and rs62113212 were highly correlated (linkage disequilibrium [LD] r2 = 0.98), and the association between rs17632542 and rs2569735 was moderate (LD r2 = 0.42).
Selection procedure of SNPs and SNP–SNP interaction pairs associated with prostate cancer aggressiveness. aP < 5.8 × 10–6 is based on Bonferroni correction; candidate SNPs for interaction analyses: 5345 SNPs; 3 SNPs are rs17632542, rs62113212, and rs2569735 in KLK3. bFor SNP interactions, the 2-stage AA9int + SIPI was applied. Using AA9int, 66,619 pairs were selected with a P < 10–3 in the discovery set, then 66,321 pairs were selected in the second stage using the SIPI approach. SIPI was applied for the validation and combined set analyses. AA9int Additive–Additive 9 interaction-model approach, p_individual p-value of individual SNP effect, p_int p-value of interaction, SIPI SNP interaction pattern identifier, SNP single nucleotide polymorphism. cAA_Full Additive–additive full interaction model.
SNP–SNP interactions
By applying the AA_Full approach for 1.4 × 107 SNP pairs associated with PCa aggressiveness, only one pair (rs390993 + rs473640 in RIPK2 and NOS1) qualified the selection criteria: P < 0.001 in the discovery, validation, and combined sets (Fig. 1). Using the AA_Full approach, 13,127 SNP pairs had P < 0.001 in the discovery set. Among them, only 1 pair (rs390993 + rs473640 in RIPK2 and NOS1) satisfied the validation criteria. The interaction p-values of rs390993 + rs473640 were 4.5 × 10–5, 9.2 × 10–4, and 2.8 × 10–7 for the discovery, validation, and combined sets. The interaction patterns for this SNP pair were shown in Supplementary Fig. S2.
We applied the 2-stage Additive–Additive 9 interaction model approach and SNP Interaction Pattern Identifier (AA9int + SIPI) approach to search for SNP–SNP interactions associated with PCa aggressiveness in the discovery set to reduce computation burden and maintain prediction power (Fig. 1). When testing 1.4 × 107 SNP pairs using the AA9int approach, 66,619 pairs had a P < 0.001. Then, we applied SIPI for further interaction analyses and found 66,321 pairs with a P < 0.001 in the discovery set, of which 11,449 pairs were validated. In the combined set, 9492 pairs were promising because they had a P < 1 × 10–5, which was the criterion we selected based on the plot of the − log10 P values in Supplementary Fig. S3. Further, 3795 of these SNP pairs also met the stringent Bonferroni criterion (P < 3.5 × 10–9). Among 3795 SNP pairs, 1083 pairs’ interaction had a lower p-value than their 2 constituent SNP effects. Of the 9492 promising pairs, 3144 SNP interaction pairs were more significant than their constituent SNPs. All 1083 SNP pairs and 98.6% of the top 3144 promising pairs associated with PCa aggressiveness were involved with KLK3 SNPs. Among the top 3144 SNP pairs associated with PCa aggressiveness, 4 KLK3 clusters contributed 91% of these pairs (n = 2856/3144). These 4 KLK3 clusters were rs17632542 (769 pairs), rs2569735 (1132 pairs), rs1058205 (601 pairs), and rs174776 (350 pairs). The most significant 5 SNP pairs within these 4 KLK3 clusters are listed in Table 2, and the top non-KLK3 pairs with P < 1 × 10–6 are listed in Supplementary Table S1.
The most significant SNP pair was rs4783709 + rs17632542 in CYB5B: LOC105371325 and KLK3, with a P value of 4.6 × 10–16 in the combined set, 2.2 × 10–8 in the discovery set, and 3.1 × 10–9 in the validation set. As shown in Table 2 and Fig. 2A, the identified interaction pattern for the rs4783709 + rs17632542 SNP pair in the discovery, validation, and combined sets was AA_int_ro, which indicates the additive–additive interaction-only pattern with the reverse and original mode for the first and second SNPs, respectively. PCa cases with the major allele G in rs4783709 and the minor allele G in rs17632542 in KLK3 had a higher risk of PCa aggressiveness (P = 4.6 × 10–16; OR= 1.29 per one unit of the G × G allele with a coding of 0, 1, 2 for each allele) in the combined set. In the SNP pair of rs4783709 + rs17632542, the prevalence of PCa aggressiveness was 20%, 26%, 33%, and 41% for PCa cases with the genotype combinations of AA + AA, GA + AG, GG/AG, and GG + GG, respectively, with a respective coding of 0, 1, 2, and 4. The rs2569735 + rs4802754 (2 KLK3 SNP interactions) pair in the combined set (Fig. 2B) had a DD_int_or pattern, which indicates a dominant–dominant interaction-only pattern with the original mode for SNP1 and reverse mode for SNP2. The PCa cases with the GA/AA + GG genotype in rs2569735 + rs4802754, respectively, had a higher risk of PCa aggressiveness (OR= 1.36; P = 4.2 × 10–11) compared with the group with other genotypes for this SNP pair. As shown in Fig. 2B, the prevalence of PCa aggressiveness was 23% for the entire cohort and 27–32% for the GA/AA + GG group in the SNP pair of rs2569735 + rs4802754. Although the validation set had a different interaction pattern of AA_int_or compared with the discovery and combined sets, their trends were similar.
Selected SNP–SNP interactions associated with prostate cancer aggressiveness. PCa aggr (n): prevalence of prostate cancer aggressiveness (sample size in the genotype combination); SNP–SNP Interaction Pattern: “(SNP1 mode)(SNP2 mode)_int_(SNP1 code)(SNP2 code)”; OR of PCa aggressiveness adjusted for study sites and six principal components for population stratification. The darker color indicates a higher chance of prostate cancer aggressiveness. A additive inheritance modes, aggr aggressiveness, D dominant inheritance modes, o original coding based on the minor allele, OR odds ratio, PCa prostate cancer, R recessive inheritance modes, r reverse code direction, SNP single-nucleotide polymorphism. These heat tables were generated using the plot3by3 function in the SIPI R package.
The majority of the identified SNP interactions are interaction-only models with only 1 interaction term in each model; only a few SNP interactions had a complicated model with > 1 terms (such as M1_int or M2_int). Some examples of these complicated interaction patterns are listed in Table 2 and Supplementary Fig. S4. The interaction pattern for the SNP pair of rs10520259 in HAND2 + rs174776 in KLK3, which was associated with PCa aggressiveness, was AA_M1_int_o1. This is an additive–additive model plus the main effect of rs10520259 (rs10520259 + rs10520259 × rs174776) with an original mode for both SNPs. Among PCa cases with the rs174776 GG genotype, an A allele of rs10520259 was associated with a lower risk of PCa aggressiveness (OR= 0.91; P = 0.001). For PCa cases with the rs174776 GA/AA genotypes, the rs10520259 A allele had an opposite effect (OR= 1.31; P = 1.2 × 10–9).
Examples of SNP–SNP interaction pairs with a large effect size
There were 8 significant SNP pairs with a large effect size (OR ≥ 2.00) associated with PCa aggressiveness for the binary-mode interaction models, including the DD_, DR_, RD_, and RR_ models. To identify a large effect size, different OR criteria should be applied for the binary-mode interaction models and AA models because they have different coding scales: (0, 1) for the binary-mode interaction models and (0, 1, 2, and 4) for the AA models. For AA models, there were 20 pairs with OR ≥ 1.3, and the top 5 pairs with OR ≥ 1.35 are shown in Table 3. All 5 of these pairs are involved with KLK3 rs17632542.
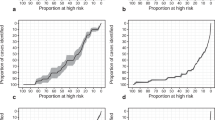
As shown in Table 3, the pair with the largest effect size was rs3775202 + rs174776 with the RR_int_oo pattern, a recessive–recessive interaction-only pattern with an original mode. The interaction pattern in the discovery set was not the same but had a similar trend. Though the prevalence of aggressive PCa for the entire cohort was 23% (Fig. 3A), the prevalence among the cases with the AA + AA genotype in rs3775202 + rs174776 in VEGFC and KLK3 was 54% (OR= 4.12 for AA + AA genotype compared with others; P = 5.9 × 10–7) in the combined set. Another example of an SNP pair with a large effect size was rs1250240 + rs174776 (FN1 + KLK3) with the DR_int_ro pattern, a dominant–recessive interaction with a reverse mode in the validation and combined sets (Fig. 3B). PCa cases with the GG + AA genotype in rs1250240 + rs174776 had a higher risk of being aggressive (OR= 3.22 and 2.96 in the validation and combined sets, respectively). The interaction pattern of this pair was similar in the discovery set. As shown in Fig. 3A,B, PCa prevalence for a specific SNP genotype would be altered when interacting with another SNP. Our results demonstrated that these SNP–SNP interaction patterns could explain the PCa aggressiveness profile better than their constituent SNPs’ individual effects.
Selected SNP–SNP interactions with a large effect size associated with prostate cancer aggressiveness. Note: PCa aggr (n): prevalence of prostate cancer aggressiveness (sample size in the genotype combination); SNP–SNP Interaction Pattern: “(SNP1 mode)(SNP2 mode)_int_(SNP1 code)(SNP2 code)”; OR: odds ratio of PCa aggressiveness adjusted for study sites and 6 principal components for population stratification. The darker color indicates a higher risk of prostate cancer aggressiveness. A additive inheritance mode, aggr aggressiveness, D dominant inheritance mode, o original coding based on the minor allele, OR odds ratio, PCa prostate cancer, R recessive inheritance mode, r reverse code direction, SNP single-nucleotide polymorphism. These heat tables were generated using the plot3by3 function in the SIPI R package.
The pair with the second largest effect size was rs2317676 + rs7802277 (ITGB3 + MTCYBP42) with the DR_M2_int_o2 pattern, a dominant–recessive interaction with the main effect of the second SNP (rs7802277) and an original mode for both SNPs. As shown in Table 3 and Supplementary Fig. S4, the prevalence of aggressive PCa was 44% for cases with the AG/GG + AA genotype (OR= 3.13; P = 6.5 × 10–5) and only 16% for cases with the AA + AA genotype (OR= 0.62; P = 8.3 × 10–4), whereas the overall prevalence of aggressive PCa was 23% in the combined set. In addition, there were 88 SNP pairs associated with PCa aggressiveness, with a medium effect size (0.5 ≤ OR < 0.67 or 1.5 ≤ OR < 2) for SNP pairs with binary modes.
Non-KLK3 SNP–SNP interaction pairs
Only 43 SNP pairs among the top 3144 SIPI identified pairs did not involve KLK3, and 21 out of these 43 pairs were involved with rs7446 in KPNA3. The top 17 SNP pairs with a P < 10–6 are listed in Supplementary Table S1. For the most significant SNP pair of rs2266967 (MAPK1) + rs7446 (KPNA3), PCa cases with the CA/AA + AG/AA genotype had a higher risk of PCa aggressiveness (OR= 1.21; P = 3.6 × 10–8) compared to cases with other genotypes for this SNP pair. The SNP pair with the largest effect size was rs2317676 (ITGB3) + rs7802277 (MTCYBP42).
SNP-interaction PRS
Among the 3144 candidate pairs selected using the AA9int + SIPI approach, the majority were in the 11 clusters identified in Supplementary Table S2; only 23 pairs were not involved in these clusters. Some SNP pairs showed up in 2 clusters (such that SNPA–SNPB showed up in both cluster A and cluster B), and so we dropped the duplicated pairs for model building. Nine of these 11 clusters were involved with KLK3. By deleting the highly correlated SNP pairs and performing variable selection associated with PCa aggressiveness within each cluster (Supplementary Table S2), the number of candidate pairs for modeling was reduced from 3144 to 96 pairs. The largest cluster was for rs2569735 in KLK3 with 1132 pairs, of which only 23 were low-correlated pairs (r < 0.7). After applying stepwise selection for pairs associated with PCa aggressiveness with P < 0.1 as the selection criteria within a cluster, only 14 SNPs pairs were selected. The number of the selected pairs for other clusters are listed in Supplementary Table S2. The candidate set for the multipair model was composed of 96 pairs, including 76 selected pairs within the 11 clusters and 20 other SNP pairs.
We applied stepwise selection in logistic model for the SIPI selected pairs in the combined set. Using this approach, 23 pairs and 12 pairs were selected based on a criterion of P < 0.01 and P < 1 × 10–5, respectively. We evaluated the 24-pair model by adding 1 SNP pair (rs390993 + rs473640 in RIPK2 and NOS1) selected using the AA_Full approach to the list of 23 SNP pairs. The 24-pair model comprised interactions between 42 SNPs (Supplementary Table S3), and 9 of these pairs were involved with the KLK3 SNPs. The 12-pair model was made of interactions between 24 total SNPs, and 3 pairs were involved with the KLK3 SNPs. Based on these 2 models, we built the SNPint-PRS, then classified PCa cases into 7 groups based on their risk profiles (see Supplementary Methods).
For the 24-pair model (Supplementary Table S4A), the OR of aggressiveness for PCa cases in the top 1% high-risk group was 3.65 (95% CI= 2.71–4.91), and the OR for cases in the second highest risk group was 1.97 by using the 50% PCa cases with an average risk as to the reference group after adjusting for study site and the 6 principal components of population stratification as suggested by the PRACTICAL study20. In addition, the ORs of the cases with the lowest 1% and the 1–10% risk profile were 0.27 and 0.55, respectively. As shown in Fig. 4, the prevalence of PCa aggressiveness for cases with the top 1%, middle 50%, and bottom 1% risk profiles were 49.8%, 21.9%, and 7.0%, respectively. For the 12-pair model (Supplementary Table S4B), the ORs of PCa aggressiveness for the top 1% and bottom 1% were 3.06 and 0.51, respectively.
For the individual-effect model, only rs17632542 was selected based on the stepwise selection with a significance level of 1 × 10–5 based on the 3 verified KLK3 SNPs in Table 1. As shown in Supplementary Table S5, we compared the performance of these 2 SNPint-PRS with this individual-effect model using the area under the receiver operator characteristics curve (AUC). The AUC values for the 24-pair model, 12-pair model and individual-effect model were 0.696, 0.687, and 0.668, respectively. The 24-pair model performed significantly better than the individual-effect model with rs17632542 and the 12-pair model (AUC comparison P = 6.5 × 10–27 and 2.7 × 10–8, respectively). We also performed an internal validation using the bootstrap approach. The AUC’s 95% confidence interval for the 24-pair interaction model was 0.686–0.702 based on 1000 bootstrap runs.
Expression quantitative trait loci analyses
To evaluate the biological functions of the identified SNP pairs, we performed 2-way expression quantitative trait loci (eQTL) tests, which evaluated associations between 1 identified SNP pair (2 SNPs) and 1 gene expression in the 4 target pathways. The linear model-based AA9int approach was applied. As shown in Supplementary Table S5, there were 24 significant eQTL tests with a P value < 1.1 × 10–8 (Bonferroni criteria = 0.05/4.5 × 106). The distribution of − log10-transformed P values for the eQTL tests is shown in Supplementary Fig. S5. Among these 24 significant eQTL tests (Supplementary Table S5), all of them involved 1 of the 4 top KLK3 SNPs (rs17632542, rs2569735, rs1058205, and rs174776), and 16 tests involved SNP pairs within the top 3144 pairs. One SNP in FHIT, rs995633, interacted with 2 KLK3 SNPs (rs1058205 and rs2569735), which link to SLC25A21 expression. In addition, the genotype combinations of rs7224135 (CAVIN1) + rs174776 (KLK3) had a significant effect on SRD5A2 expression (Supplementary Fig. S6, P = 2.5 × 10–9). These promising eQTL results support that the KLK3 SNP pairs may have an impact on PCa aggressiveness by altering specific gene expression.
Gene interaction network with KLK3
For gene-level interactions, there were 6 common genes with > 50 SNP pairs from the top 3145 SNP pairs that interacted with KLK3 and were associated with PCa aggressiveness. As shown in Fig. 5, The 6 most common gene–gene interaction pairs were KLK3-COL4A1: COL4A2, KLK3-CDH13, KLK3-TGFBR3, KLK3-EGFR, KLK3-FGFR2, and KLK3-PRKCA. We applied the STRING database (https://string-db.org/)21 to analyze the gene–gene (protein–protein) interaction network. We also added 2 genes (FN1 and the androgen receptor [AR]) based on our literature review. Among the top 3145 pairs, there were 27 pairs for KLK3-FN1 and 4 pairs for KLK3-AR. This gene–gene interaction network supports the idea that the majority of our identified SNP–SNP interactions are directly or indirectly linked with KLK3. KLK3 had a direct link with FN1 and AR, which both link to EGFR. EGFR is also the hub for this gene–gene interaction network. For pathway-level analyses, the summary of the within- or between-pathway interactions for the top 3145 SNP pairs is shown in Supplementary Table S6. Among our 4 target pathways, the most common pathway-pathway interactions are androgen-angiogenesis (1543 SNP pairs) and androgen-mitochondria (1138 SNP pairs) interactions. Among the 3145 SNP pairs, 98.6% were involved with KLK3 in the androgen pathway. This explains why the androgen pathway is the hub for pathway–pathway interactions.
KLK3 gene–gene interaction network based on common genes interacted with KLK3. KLK3 and AR are in the androgen pathway. All other genes in the table are in the angiogenesis pathway. SNP single-nucleotide polymorphism. The gene–gene interaction network plot was generated using the STRING software (https://string-db.org/).
Discussion
Our study identified 3 KLK3 SNPs (rs17632542, rs62113212, and rs2569735) and 3145 SNP interaction pairs that were associated with PCa aggressiveness. The KLK3 SNP rs17632542, which is in a strong LD with rs62113212, had been previously identified in GWAS as being associated with several PCa-related outcomes, such as the patient’s PSA level, PCa risk, and age at PCa diagnosis and the tumor’s volume, aggressiveness, and Gleason score22. The SNP rs2569735 has been shown to influence miRNA functions23. The SNP rs2735839, which has a strong LD with rs2569735, is associated with PCa aggressiveness24.
The prostate is an androgen-dependent organ, and SNPs in genes involved in the androgen metabolism pathway, such as KLK3, have been previously implicated in PCa risk and progression22,25. KLK3 is a protein-coding gene on chromosome 19q13.4, and its protein product, PSA, is a serine protease exclusively secreted by the prostate gland into the seminal fluid, where it plays an important functional role in the normal physiology of the prostate26. PSA is not normally secreted into the bloodstream; therefore, serum PSA levels are used for screening, diagnosing, and prognosticating PCa27.
Our results were consistent with previously published literature. As shown in Supplementary Table S7, our literature review showed that the KLK3 individual SNPs rs17632542, rs2569735, and rs1058205 were significantly associated with PCa aggressiveness and PSA level. KLK3 was involved in 98.6% of the top 3145 SNP pairs associated with PCa aggressiveness; the 4 KLK3 SNPs (rs17632542, rs2569735, rs1058205, and rs174776) contributed to 90.8% (2856 pairs) of the identified SNP pairs. Interestingly, the 2 most common SNPs (rs17632542 and rs2569735), which interacted with many other SNPs, also had significant individual effects. According to the quality-controlled gene–gene (protein–protein) association network (Fig. 5), the majority of our identified genes had a direct or indirect link with KLK3. In this network, FN1 and AR had a direct link with KLK3. Both FN1 and AR are linked to EGFR, which is the hub for this gene–gene interaction network. KLK3 and AR are in the androgen pathway, and FN1 and EGFR are in the angiogenesis pathway.
FN1 is known as a ubiquitous multifunctional glycoprotein and is involved in cell growth, migration, and differentiation28. An 8-gene panel has reported FN1 to distinguish high-grade PCa from indolent PCa, with a sensitivity of 93% and specificity of 70%29. AR is known to play a vital role in PCa development and progression, and PCa cells rely on androgens for proliferation and survival30. KLK3 gene expression is regulated by AR through androgen response elements in the promoter of PSA. Both FN1 and AR are regulated by miRNA-1207-3p in PVT1, and these two genes are overexpressed in human PCa cell lines and tissues and are associated with PCa aggressiveness31. We also reported that miR-3162-5p is associated with an rs1058205-T allele in KLK3 and can regulate KLK3 and AR expression32. The bioinformatics analyses using STRING and GeneMANIA33 software show the links between KLK3, FN1, and AR.
As shown in Fig. 5, the 6 most common genes/regions interacting with KLK3 are all in the angiogenesis pathway. KLK3 expression may decrease PCa aggressiveness by inhibiting angiogenesis34. We previously reported that several angiogenesis genes influenced PCa aggressiveness35. Both COL4A1 and COL4A2 influence angiogenesis and tumor growth. COL4A1 was associated with cell invasion and movement as a gene in the epithelial-to-mesenchymal (EMT) transition process, and COL4A1 gene expression has been associated with the Gleason score36. COL4A2 provides a structural component for basement membranes, is known as an inhibitor of angiogenesis, and has been considered as a biomarker for screening for benign prostatic hyperplasia37.
CDH13 is located on chromosome 16q24 and is a well-known tumor suppressor gene involved in cell–cell adhesion. The expression of CDH13 has been associated with poor PCa prognosis and low proliferation rates of prostate tumor cells38. TGFBR3 is one of TGF-β receptors and is abundantly expressed in PCa cells. TGFBR3 is downregulated during the transition from benign to malignant and metastatic prostate tissues, especially in bone metastases39. Several studies consistently report downregulation of TGFBR3 in prostate tumors, suggesting that it has an important role as a tumor suppressor gene40,41.
KLK3 can attenuate the response to FGF2, an angiogenesis-stimulating factor that binds to FGFR2 and reduces endothelial cell proliferation and migration, indicating FGF2 suppressive effect during metastasis42. PRKCA is a key negative regulator of the TGF-β pathway and is downregulated in PCa tumors. Cell migration, invasion, and bone metastasis are suppressed by overexpression of PRKCA. Further, expression of PRKCA has been correlated with clinical variables, such as PSA levels, Gleason score, and metastasis status of patients with PCa39.
For the non-KLK3 SNP pairs, KPNA3, with 21 SNP pairs, was the most common gene, and there were 5 pairs for KPNA3-AR interaction. KPNA3 is one of the subunits of the nuclear pore complex and plays a role in nuclear protein import43. In addition, the expression of KPNA3 was inversely associated with survival. Baker et al. demonstrated that KPNA3 knockdown inhibited cell proliferation, migration, and invasion. KPNA3 may regulate protein transfer to promote colorectal cancer growth, metastasis, and relapse43.
We identified one additional SNP pair (rs390993 in RIPK2 + rs473640 in NOS1) using the AA_Full model. The receptor-interacting serine/threonine-protein kinase 2 (RIPK2) was one of the key regulators of the immune response44. NO synthases (NOS) generate nitric oxide (NO), which can regulate tumorigenesis. Increased nitric oxide level by NOS is cytotoxic to cancer cells45. NOS1 downregulation reduced the growth of chemokine expressing fibroblasts and their ability to promote tumor formation in prostate cancer cells46. Although these genes' role has not been studied in PCa aggressiveness, RIPK2 polymorphisms were associated with risk of gastric47 and breast48 cancers. NOS1 polymorphisms were associated with the risk of various cancers, such as pancreatic49, glioma50, colorectal cancer, and melanoma51.
Our validated SNP–SNP interactions have been supported by both the protein–protein interaction network and eQTL results. As shown in Supplementary Table S5 and Supplementary Fig. S6, SNP interactions between CAVIN1 and KLK3 influenced SRD5A2 expression. CAVIN1 has been shown to reduce lymphangiogenesis and angiogenesis52. As shown in Fig. 5, CAVIN1 links to KLK3 through EGFR and AR according to the protein–protein interaction network. SRD5A2 converts testosterone to dihydrotestosterone, which activates ARs53,54. SRD5A2 expression was significantly higher among patients with high-grade PCa vs. those with low-grade PCa55,56. In our previous study, we reported that the validated SNP–SNP interaction pairs of MMP16-EGFR, MMP16-ROBO1, and MMP16-CSF1 were significantly associated with PCa aggressiveness and that EGFR is the hub of these interactions13. We observed that MMP16, EGFR, ROBO1, and CSF1 interacted with KLK3 and were associated with PCa aggressiveness. Among the top 3145 SNP pairs, 62 pairs were KLK3-EGFR interactions, 9 pairs were KLK3-MMP16 interactions, 9 pairs were KLK3-ROBO1 interactions, and 1 pair was a KLK3-CSF1 interaction. Both proteins form MMP16 and EGFR have been implicated in PCa. Several cancers that involve EGFR signaling often show an abnormally high expression of MMP1657. Although 2 genes (CDH13 and TGFBR3) do not have a link to KLK3 according to the protein–protein interaction network (Fig. 5), several miRNAs (such as miR_6731_5p, miR_8085, and miR_3919) contribute to the links between KLK3 and these 2 genes58.
Importantly, our study demonstrated that SNP–SNP interactions could explain PCa aggressiveness better than individual SNP effects focusing on genes from the 4 selected pathways. Our study also showed that the 2-stage AA9int + SIPI approach is a powerful tool for identifying and validating SNP–SNP interactions associated with a selected phenotype. The AA9int + SIPI approach is powerful because it allows genotype subgroups with similar risk profiles or small sample sizes combined. Our risk classification system based on the model with 24 validated SNP interaction pairs is promising. Using this SNPint-PRS (score range 0–100), the prevalence of aggressive PCa cases from our cohort was 49.8%, 21.9%, and 7.0% with top 1%, middle 50%, and bottom 1% risk profiles, respectively. We observed that SNP pairs in the same cluster tended to be highly correlated. This may explain why we identified > 9000 significant SNP pairs. By dropping the highly correlated pairs within a cluster, we effectively reduced variable dimensions for building a multi-pair prediction model. Further biological studies will be needed to distinguish driver or passenger effects for the identified SNP pairs.
For pathway-level interactions related to PCa aggressiveness, the most common interactions were involved with the androgen pathway (such as KLK3), especially androgen-angiogenesis (49.1%) and androgen-mitochondria pathway interactions (36.2%, Supplementary Table S6). Angiogenesis is induced by overexpression of angiogenesis-related genes, which are regulated by many factors, including elevated androgen levels16. A previous study reported interaction between the regulation of angiogenesis-related genes and androgen59. Further, clinical therapies targeting genes involved in the androgen and angiogenesis pathways suggest an interaction between these 2 pathways17. In addition, recent studies have reported that androgen-related genes regulate mitochondrial respiration in PCa cells18. Androgen treatment leads to an increase in the activity of several metabolic pathways, including mitochondrial biogenesis and activity60. However, androgen repression in PCa cells decreases mitochondrial activity and cell proliferation18.
There are some limitations to our study. First, there is no external validation for the SNPint-PRS, and the identified SNP–SNP interactions may be influenced by the significance of their constituent SNP individual effects. Further analyses using large-scale studies will be needed for further verification. Second, it is challenging to identify causal SNP pairs because some SNP pairs are highly correlated, especially for SNP pairs in the same cluster. The downstream gene expression analyses or laboratory experiments will be needed to identify causal SNP pairs with biological functions. Lastly, this study only evaluated SNP–SNP interactions for the selected 4 pathways, so evaluation of SNP–SNP interactions in other pathways or genes will be required to gain a thorough understanding of the complicated gene–gene interactions associated with PCa prognosis.
In summary, this study demonstrates that KLK3 alone and interactions between KLK3 and other identified genes play an important role in PCa aggressiveness. As shown in Fig. 5, KLK3 directly links with FN1 and AR, and other genes are indirectly linked to KLK3 through these two genes. The KLK3 SNP–SNP interactions can explain PCa aggressiveness better than individual SNPs. The identified SNP–SNP and gene–gene interactions may provide valuable insights for identifying downstream genes that affect PCa progression. Genetic markers, such as a panel of SNPs, are excellent predictive biomarkers as they remain unchanged during patients’ lifetime. Thus, the SNP-based scores can be a useful tool used for early prediction of future PCa progression, not just early detection when PCa progression already occurs. In addition, SNPs are tissue-independent, and can be measured non-invasively, so our SNP interaction-based PRS of PCa aggressiveness may be used clinically for disease classification and treatment guidance. Further investigation of the biological functions of the identified genes and additional validation of this prediction model is needed.
Methods
Study population
This study included 20 270 PCa cases (22.6% of which were aggressive PCa) with European ancestry from 21 studies within the Collaborative Oncological Gene-Environment Study (COGS) in the Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL) Consortium. Details of the PRACTICAL Consortium study have been previously reported20. PCa aggressiveness was defined as Gleason score ≥ 8, PSA level > 100 ng/mL, distant disease stage at diagnosis, or PCa-related death. Ethnic groups were defined based on ~ 37,000 uncorrelated markers that passed quality control, including ~ 1000 that were selected as ancestry informative markers. Half of the cases from each study site were randomly assigned to discovery and validation sets (10,135 cases in each set).
Selection of genes and SNPs
We evaluated 8587 SNPs from 2122 genes within 4 pathways: the angiogenesis-, mitochondria-, miRNA-, and androgen metabolism-related pathways. These pathways were selected based on our previous studies13,35, the literature, and PCa GWAS. These SNPs were genotyped on a custom iCOGS Illumina array (Illumina, San Diego, CA, USA) using blood DNA samples. SNPs were excluded if they had a call rate < 95%, a call rate < 99% with minor allele frequency (MAF) < 5%, MAF < 1%, or if their genotype frequencies departed from Hardy–Weinberg equilibrium at P < 1 × 10−12.
Statistical analyses
The effects of individual SNPs and 2-way SNP–SNP interactions on the outcome of PCa aggressiveness were evaluated. The analyses were conducted separately for the discovery set and validation set. To control for population substructure, the principal component analysis was performed. All models were adjusted for the study site and the first 6 principal components of population stratification as suggested by the PRACTICAL study20. For evaluating individual SNP effects, we considered three different inheritance modes: dominant [D], recessive [R], and additive [A] modes, which were assigned based on the minor allele. Logistic models were applied, and the best mode with the lowest P value was selected for each SNP.
We assessed 2-way SNP–SNP interactions. LD among all testing SNPs was examined based on r2. In order to avoid multicollinearity in modeling, we tested correlations of neighborhood SNPs within 100 kilobases using PLINK. SNPs with a strong LD of r2 > 0.8 were excluded from interaction analyses. A total of 5345 SNPs with a MAF > 0.05 and no strong LD (r2 < 0.8) were included for SNP–SNP interaction analyses.
For SNP–SNP interactions, we applied the conventional AA_Full approach using PLINK in the discovery, validation and combined sets. In addition, we applied a powerful 2-stage AA9int + SIPI approach to search intensively for SNP–SNP interactions associated with PCa aggressiveness. The AA9int approach, which treats all SNPs as having an additive inheritance mode (additive–additive patterns [AA_]), tested 9 interaction patterns of pairwise SNP–SNP interactions associated with an outcome10. The SIPI approach is an extended version of AA9int and tests 45 biologically meaningful SNP–SNP interaction patterns by considering 2 more inheritance modes (dominant and recessive)9. Thus, SIPI’s test patterns include additive–additive (AA_), dominant–dominant (DD_), dominant–recessive (DR_), recessive–dominant (RD_), and recessive–recessive (RR_) models. The AA9int and SIPI models are composed of the full-interaction model (both main effects plus interaction), the models with 1 main effect and 1 interaction, and models with interaction only. The corresponding patterns’ names are Full, M1_int, M2_int, or Int_, which represent various types of interactions with 3, 2, 2, and 1 term(s) in the statistical model equation, respectively (see the equations below). For easy interpretation, these SIPI interaction patterns can be shown as the 3 × 3 table with a heat table format using the plot3by3 function in the SIPI R package9,10.
The SNP pairs with binary modes (such as DD and RR) categorize the subjects into 2, 3, and 4 distinct risk groups for interaction-only, main + interaction, and full-interaction models, respectively. For AA models, the coding for an SNP with the additive mode is 0, 1, or 2 for the number of the target alleles, so the coding of the interaction term of 2 additive SNPs (SNP1 × SNP2) is 0, 1, 2, or 4. The details of these 2 methods were described previously9,10. We considered the original (0, 1, and 2 for the number of minor alleles) and reverse coding directions. For the binary outcome of PCa aggressiveness, the logistic-based AA9int was applied, and the best model was selected based on the lowest BIC value for each SNP pair. For a large-scale study, the 2-stage AA9int + SIPI approach is suggested. For performing AA9int and SIPI analyses, we used the ‘parAA9int’ and ‘parSIPI’ functions in the SIPI R package in this study. There are four related functions in the SIPI R package: SIPI, parSIPI, AA9int, and parAA9int. The parSIPI and parAA9int functions are parallel computing versions of SIPI and AA9int, which can be used for large-scale studies. We applied logistic-based AA9int and SIPI. The details of SIPI and AA9int parameter settings are listed in the SIPI R package manual. The SIPI R package and its manual are freely available for download at GitHub (https://github.com/LinHuiyi/SIPI).
As shown in Fig. 1, the 2-stage AA9int + SIPI approach was applied in the discovery set to evaluate 1.4 × 107 candidate SNP pairs associated with PCa aggressiveness. Promising SNP pairs with P < 0.001 in the screening stage were further evaluated using the SIPI approach. The SNP pairs in the discovery set with P < 0.001 were further validated in the validation set. We defined significant results using P < 0.001 in both the discovery and validation sets and a P value less than the significance level of Bonferroni correction (P < 5.8 × 10–6 [= 0.05/8587 SNPs] for the individual SNP effects and P < 3.5 × 10–9 [= 0.05/1.4 × 107 pairs] for interactions in the combined set). As it is well-known that Bonferroni correction is too stringent, we also used a P < 1 × 10–5 as the cutoff to select promising SNP–SNP interaction pairs. For testing whether an SNP interaction pair performed better than their composite individual SNP effects, we also compared the P-value of an SNP pair with P values of their composite individual SNP effects for SNP pairs with an interaction-only pattern. For SNP pairs with > 1 terms, the stepwise selection within a pair was applied. The methods used for SNPint-PRS, comparison of model performance, eQTL analyses, and the gene–gene interaction networks are listed in Supplementary Methods. All analyses were based on 2-sided tests.
References
Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 71, 7–33. https://doi.org/10.3322/caac.21654 (2021).
Damber, J. E. & Aus, G. Prostate cancer. Lancet 371, 1710–1721 (2008).
Albertsen, P. C. PSA and the conservative treatment of early prostate cancer. Archivio italiano di urologia, andrologia : organo ufficiale [di] Societa italiana di ecografia urologica e nefrologica 78, 152–153 (2006).
MacArthur, J. et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45, D896–D901. https://doi.org/10.1093/nar/gkw1133 (2017).
Conti, D. V. et al. Two novel susceptibility loci for prostate cancer in men of african ancestry. J. Natl. Cancer Inst. https://doi.org/10.1093/jnci/djx084 (2017).
Schumacher, F. R. et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat. Genet. 50, 928–936. https://doi.org/10.1038/s41588-018-0142-8 (2018).
Karunamuni, R. A. et al. Additional SNPs improve risk stratification of a polygenic hazard score for prostate cancer. Prostate Cancer Prostat. Dis. https://doi.org/10.1038/s41391-020-00311-2 (2021).
Cordell, H. J. Detecting gene–gene interactions that underlie human diseases. Nat. Rev. Genet. 10, 392–404 (2009).
Lin, H. Y. et al. SNP interaction pattern identifier (SIPI): An intensive search for SNP–SNP interaction patterns. Bioinformatics 33, 822–833. https://doi.org/10.1093/bioinformatics/btw762 (2017).
Lin, H. Y. et al. AA9int: SNP interaction pattern search using non-hierarchical additive model set. Bioinformatics 34, 4141–4150. https://doi.org/10.1093/bioinformatics/bty461 (2018).
Fukumori, T. et al. Galectin-3 regulates mitochondrial stability and antiapoptotic function in response to anticancer drug in prostate cancer. Cancer Res. 66, 3114–3119. https://doi.org/10.1158/0008-5472.CAN-05-3750 (2006).
Poliseno, L. et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood 108, 3068–3071. https://doi.org/10.1182/blood-2006-01-012369 (2006).
Lin, H. Y. et al. SNP–SNP interaction network in angiogenesis genes associated with prostate cancer aggressiveness. PLoS ONE 8, e59688. https://doi.org/10.1371/journal.pone.0059688 (2013).
Sarkar, C., Goswami, S., Basu, S. & Chakroborty, D. Angiogenesis inhibition in prostate cancer: An update. Cancers (Basel). https://doi.org/10.3390/cancers12092382 (2020).
Boddy, J. L. et al. The androgen receptor is significantly associated with vascular endothelial growth factor and hypoxia sensing via hypoxia-inducible factors HIF-1a, HIF-2a, and the prolyl hydroxylases in human prostate cancer. Clin. Cancer Res. 11, 7658–7663. https://doi.org/10.1158/1078-0432.CCR-05-0460 (2005).
Eisermann, K. & Fraizer, G. The androgen receptor and VEGF: Mechanisms of androgen-regulated angiogenesis in prostate cancer. Cancers (Basel). https://doi.org/10.3390/cancers9040032 (2017).
McKay, R. R. et al. A randomized phase II trial of short-course androgen deprivation therapy with or without bevacizumab for patients with recurrent prostate cancer after definitive local therapy. J. Clin. Oncol. 34, 1913–1920. https://doi.org/10.1200/JCO.2015.65.3154 (2016).
Audet-Walsh, E. et al. Androgen-dependent repression of ERRgamma reprograms metabolism in prostate cancer. Cancer Res. 77, 378–389. https://doi.org/10.1158/0008-5472.CAN-16-1204 (2017).
Amankwah, E. K. et al. miR-21, miR-221 and miR-222 expression and prostate cancer recurrence among obese and non-obese cases. Asian J. Androl. 15, 226–230. https://doi.org/10.1038/aja.2012.160 (2013).
Eeles, R. A. et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat. Genet. 45, 385–391. https://doi.org/10.1038/ng.2560 (2013).
Szklarczyk, D. et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368. https://doi.org/10.1093/nar/gkw937 (2017).
Sullivan, J. et al. An analysis of the association between prostate cancer risk loci, PSA levels, disease aggressiveness and disease-specific mortality. Br. J. Cancer 113, 166–172. https://doi.org/10.1038/bjc.2015.199 (2015).
Batra, J., O’Mara, T., Patnala, R., Lose, F. & Clements, J. A. Genetic polymorphisms in the human tissue kallikrein (KLK) locus and their implication in various malignant and non-malignant diseases. Biol. Chem. 393, 1365–1390. https://doi.org/10.1515/hsz-2012-0211 (2012).
He, Y. et al. The prostate cancer susceptibility variant rs2735839 near KLK3 gene is associated with aggressive prostate cancer and can stratify gleason score 7 patients. Clin. Cancer Res. 20, 5133–5139. https://doi.org/10.1158/1078-0432.CCR-14-0661 (2014).
Kotarac, N., Dobrijevic, Z., Matijasevic, S., Savic-Pavicevic, D. & Brajuskovic, G. Association of KLK3, VAMP8 and MDM4 genetic variants within microRNA binding sites with prostate cancer: Evidence from Serbian population. Pathol. Oncol. Res. 26, 2409–2423. https://doi.org/10.1007/s12253-020-00839-7 (2020).
Sutherland, G. R. et al. Human prostate-specific antigen (APS) is a member of the glandular kallikrein gene family at 19q13. Cytogenet. Cell Genet. 48, 205–207. https://doi.org/10.1159/000132629 (1988).
Lawrence, M. G., Lai, J. & Clements, J. A. Kallikreins on steroids: Structure, function, and hormonal regulation of prostate-specific antigen and the extended kallikrein locus. Endocr. Rev. 31, 407–446. https://doi.org/10.1210/er.2009-0034 (2010).
Katnik-Prastowska, I., Przybysz, M. & Chelmonska-Soyta, A. Fibronectin fragments in human seminal plasma. Acta Biochim. Pol. 52, 557–560 (2005).
Xiao, K. et al. Use of two gene panels for prostate cancer diagnosis and patient risk stratification. Tumour Biol. 37, 10115–10122. https://doi.org/10.1007/s13277-015-4619-0 (2016).
Gan, L. et al. Inhibition of the androgen receptor as a novel mechanism of taxol chemotherapy in prostate cancer. Cancer Res. 69, 8386–8394. https://doi.org/10.1158/0008-5472.CAN-09-1504 (2009).
Das, D. K. & Ogunwobi, O. O. A novel microRNA-1207-3p/FNDC1/FN1/AR regulatory pathway in prostate cancer. RNA Dis. 4 (2017).
Stegeman, S. et al. A large-scale analysis of genetic variants within putative miRNA binding sites in prostate cancer. Cancer Discov. 5, 368–379. https://doi.org/10.1158/2159-8290.CD-14-1057 (2015).
Mostafavi, S., Ray, D., Warde-Farley, D., Grouios, C. & Morris, Q. GeneMANIA: A real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 9(Suppl 1), S4. https://doi.org/10.1186/gb-2008-9-s1-s4 (2008).
Fortier, A. H. et al. Recombinant prostate specific antigen inhibits angiogenesis in vitro and in vivo. Prostate 56, 212–219. https://doi.org/10.1002/pros.10256 (2003).
Amankwah, E. K., Sellers, T. A. & Park, J. Y. Gene variants in the angiogenesis pathway and prostate cancer. Carcinogenesis 33, 1259–1269. https://doi.org/10.1093/carcin/bgs150 (2012).
Jedroszka, D., Orzechowska, M., Hamouz, R., Gorniak, K. & Bednarek, A. K. Markers of epithelial-to-mesenchymal transition reflect tumor biology according to patient age and Gleason score in prostate cancer. PLoS ONE 12, e0188842. https://doi.org/10.1371/journal.pone.0188842 (2017).
Giri, A., Edwards, T. L., Motley, S. S., Byerly, S. H. & Fowke, J. H. Genetic determinants of metabolism and benign prostate enlargement: Associations with prostate volume. PLoS ONE 10, e0132028. https://doi.org/10.1371/journal.pone.0132028 (2015).
Andreeva, A. V. & Kutuzov, M. A. Cadherin 13 in cancer. Genes Chromosomes Cancer 49, 775–790. https://doi.org/10.1002/gcc.20787 (2010).
Dai, Y. et al. The TGF-beta signalling negative regulator PICK1 represses prostate cancer metastasis to bone. Br. J. Cancer 117, 685–694. https://doi.org/10.1038/bjc.2017.212 (2017).
Sharifi, N., Hurt, E. M., Kawasaki, B. T. & Farrar, W. L. TGFBR3 loss and consequences in prostate cancer. Prostate 67, 301–311. https://doi.org/10.1002/pros.20526 (2007).
Wu, Y. P. et al. Identification of prostate cancer-related circular RNA through bioinformatics analysis. Front. Genet. 11, 892. https://doi.org/10.3389/fgene.2020.00892 (2020).
Fortier, A. H., Nelson, B. J., Grella, D. K. & Holaday, J. W. Antiangiogenic activity of prostate-specific antigen. J. Natl. Cancer Inst. 91, 1635–1640 (1999).
Baker, S. A., Lombardi, L. M. & Zoghbi, H. Y. Karyopherin alpha 3 and karyopherin alpha 4 proteins mediate the nuclear import of methyl-CpG binding protein 2. J. Biol. Chem. 290, 22485–22493. https://doi.org/10.1074/jbc.M115.658104 (2015).
Wang, X. et al. The prostaglandin E2-EP3 receptor axis regulates anaplasma phagocytophilum-mediated NLRC4 inflammasome activation. PLoS Pathog. 12, e1005803. https://doi.org/10.1371/journal.ppat.1005803 (2016).
Sundaram, M. K. et al. Phytochemicals induce apoptosis by modulation of nitric oxide signaling pathway in cervical cancer cells. Eur. Rev. Med. Pharmacol. Sci. 24, 11827–11844. https://doi.org/10.26355/eurrev_202011_23840 (2020).
Augsten, M. et al. Cancer-associated fibroblasts expressing CXCL14 rely upon NOS1-derived nitric oxide signaling for their tumor-supporting properties. Cancer Res. 74, 2999–3010. https://doi.org/10.1158/0008-5472.CAN-13-2740 (2014).
Ota, M. et al. Association between receptor interacting serine/threonine kinase 2 polymorphisms and gastric cancer susceptibility. Oncol. Lett. 15, 3772–3778. https://doi.org/10.3892/ol.2018.7785 (2018).
Mertins, P. et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 55–62. https://doi.org/10.1038/nature18003 (2016).
Reid-Lombardo, K. M. et al. Inflammation-related gene variants as risk factors for pancreatic cancer. Cancer Epidemiol. Biomark. Prev. 20, 1251–1254. https://doi.org/10.1158/1055-9965.EPI-11-0264 (2011).
Backes, D. M. et al. Single-nucleotide polymorphisms of allergy-related genes and risk of adult glioma. J. Neurooncol. 113, 229–238. https://doi.org/10.1007/s11060-013-1122-6 (2013).
Ibarrola-Villava, M. et al. Genetic polymorphisms in DNA repair and oxidative stress pathways associated with malignant melanoma susceptibility. Eur. J. Cancer 47, 2618–2625. https://doi.org/10.1016/j.ejca.2011.05.011 (2011).
Nassar, Z. D. et al. PTRF/Cavin-1 decreases prostate cancer angiogenesis and lymphangiogenesis. Oncotarget 4, 1844–1855. https://doi.org/10.18632/oncotarget.1300 (2013).
Cunningham, J. M. et al. Evaluation of genetic variations in the androgen and estrogen metabolic pathways as risk factors for sporadic and familial prostate cancer. Cancer Epidemiol. Biomark. Prev. 16, 969–978. https://doi.org/10.1158/1055-9965.EPI-06-0767 (2007).
Zambon, C. F. et al. Effectiveness of the combined evaluation of KLK3 genetics and free-to-total prostate specific antigen ratio for prostate cancer diagnosis. J. Urol. 188, 1124–1130. https://doi.org/10.1016/j.juro.2012.06.030 (2012).
Wako, K. et al. Expression of androgen receptor through androgen-converting enzymes is associated with biological aggressiveness in prostate cancer. J. Clin. Pathol. 61, 448–454. https://doi.org/10.1136/jcp.2007.050906 (2008).
Hamid, A. et al. Early upregulation of AR and steroidogenesis enzyme expression after 3 months of androgen-deprivation therapy. BMC Urol. 20, 71. https://doi.org/10.1186/s12894-020-00627-0 (2020).
Davidson, B. et al. High levels of MMP-2, MMP-9, MT1-MMP and TIMP-2 mRNA correlate with poor survival in ovarian carcinoma. Clin. Exp. Metastasis 17, 799–808. https://doi.org/10.1023/a:1006723011835 (1999).
Wong, N. & Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 43, D146-152. https://doi.org/10.1093/nar/gku1104 (2015).
Aslan, G. et al. Vascular endothelial growth factor expression in untreated and androgen-deprived patients with prostate cancer. Pathol. Res. Pract. 201, 593–598. https://doi.org/10.1016/j.prp.2005.07.003 (2005).
Tennakoon, J. B. et al. Androgens regulate prostate cancer cell growth via an AMPK-PGC-1alpha-mediated metabolic switch. Oncogene 33, 5251–5261. https://doi.org/10.1038/onc.2013.463 (2014).
Acknowledgements
This study was supported by the National Cancer Institute (R21CA202417, PI: Lin, HY; R01CA128813, PI: Park, JY). Some parts of this research were conducted using the high-performance computational resources provided by the Louisiana Optical Network Infrastructure (LONI, http://www.loni.org). Editorial assistance was provided by the Moffitt Cancer Center’s Scientific Editing Department. Additional members from the PRACTICAL consortium http://practical.icr.ac.uk/) are provided in the Supplementary Information.
Author information
Authors and Affiliations
Consortia
Contributions
H.L., P.H., C.C., and H. T. conducted data analyses. H.L. and J.Y.P. were mainly responsible for the study design and drafted the initial version of the paper. H.L., J.P. A.E.B., Z.F., A.C., J.F., D.H., J.P.S., K.Y., J.L.C., S.A., R.R., T.G., and J.D. were part of the writing group and were mainly responsible for the interpretation of the data and critical commenting on the initial draft versions of the paper. All other co-authors were responsible for cohort-level data collection, cohort-level data analysis, and critical reviews of the draft paper. All authors approved the final version of the paper that was submitted to the journal.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lin, HY., Huang, PY., Cheng, CH. et al. KLK3 SNP–SNP interactions for prediction of prostate cancer aggressiveness. Sci Rep 11, 9264 (2021). https://doi.org/10.1038/s41598-021-85169-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-85169-7
This article is cited by
-
SNPxE: SNP-environment interaction pattern identifier
BMC Bioinformatics (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.