Abstract
To identify circulating proteins predictive of acute cardiovascular disease events in the general population, we performed a proteomic screen in plasma from asymptomatic individuals. A “Discovery cohort” of 25 individuals who subsequently incurred a cardiovascular event within 3 years (median age = 70 years, 80% male) was matched to 25 controls remaining event-free for > 5 years (median age = 72 years, 80% male). Plasma proteins were assessed by data independent acquisition mass spectrometry (DIA-MS). Associations with cardiovascular events were tested using Cox regression, adjusted for the New Zealand Cardiovascular Risk Score. Concentrations of leading protein candidates were subsequently measured with ELISAs in a larger (n = 151) independent subset. In the Discovery cohort, 76 plasma proteins were robustly quantified by DIA-MS, with 8 independently associated with cardiovascular events. These included (HR = hazard ratio [95% confidence interval] above vs below median): fibrinogen alpha chain (HR = 1.84 [1.19–2.84]); alpha-2-HS-glycoprotein (also called fetuin A) (HR = 1.86 [1.19–2.93]); clusterin isoform 2 (HR = 1.59 [1.06–2.38]); fibrinogen beta chain (HR = 1.55 [1.04–2.30]); hemoglobin subunit beta (HR = 1.49 [1.04–2.15]); complement component C9 (HR = 1.62 [1.01–2.59]), fibronectin isoform 3 (HR = 0.60 [0.37–0.99]); and lipopolysaccharide-binding protein (HR = 1.58 [1.00–2.49]). The proteins for which DIA-MS and ELISA data were correlated, fibrinogen and hemoglobin, were analyzed in an Extended cohort, with broader inclusion criteria and longer time to events, in which these two proteins were not associated with incident cardiovascular events. We have identified eight candidate proteins that may independently predict cardiovascular events occurring within three years in asymptomatic, low-to-moderate risk individuals, although these appear not to predict events beyond three years.
Similar content being viewed by others
Introduction
Cardiovascular disease (CVD) is a leading cause of death and disability worldwide1. In 2015 the prevalence of CVD and number of CVD-related deaths was estimated at 423 million and 18 million respectively, with ischemic heart disease (IHD) and stroke contributing the most to loss of age-standardized disability-adjusted life years1. Determining an individual’s risk for developing CVD is commonly based on a set of key variables, including age, gender, ethnicity, blood pressure, diabetes mellitus, smoking and lipid status. These established risk factors for CVD have been incorporated into risk prediction models such as the Framingham risk score (FRS), which is used to predict CVD incidence within 10 years2,3. Subsequent strategies to improve CVD risk prediction, such as the updated 5-year New Zealand (NZ) primary prevention equations, PREDICT-1°, have incorporated additional variables including the NZ Deprivation score (an area-based measure of socioeconomic deprivation), atrial fibrillation confirmed by electrocardiograph (ECG) and use of blood-pressure-lowering, lipid-lowering, and antithrombotic drugs in the 6 months before the index assessment4. The QRISK3 risk prediction model implemented in the UK takes into account 22 risk variables to estimate 10-year CVD risk, with the additional factors being chronic kidney disease, variability of systolic blood pressure, migraine, treatment with corticosteroids, systemic lupus erythematosus, atypical antipsychotic medication, severe mental illness, HIV or AIDS, and erectile dysfunction5.
While risk prediction models such as these work well on the population level, they suffer from poor discrimination at the level of the individual. It remains difficult to predict which individuals within any broad risk stratum will subsequently experience a CVD event6. CVD events still occur frequently in people predicted to be at low-to-moderate risk6. Addition of endogenous biomarkers to CVD risk scores may refine risk stratification for the individual. The only circulating biomarkers routinely incorporated in current CVD risk prediction models are lipids i.e. total cholesterol (TC), high-density lipoprotein (HDL) or their ratio (TC/HDL) and calculated low-density lipoprotein (LDL). Additional biomarker molecules used to diagnose comorbidities in conjunction with CVD risk estimation include glycated hemoglobin (HbA1C) for diabetic status and creatinine for renal status7,8. Plasma concentrations of B-type natriuretic peptide (BNP), its amino-terminal pro-hormone congener (NT-proBNP) and the cardiac troponins T and I (TNT, TNI) are key biomarkers in the clinical diagnosis of heart failure (HF) and myocardial infarction (MI), respectively9,10,11,12. Both markers have also been shown to provide powerful information about CVD events in asymptomatic community populations, with modest elevations in these markers being independently predictive of atherosclerotic CVD, HF, fatal coronary events and total mortality13,14,15.
However, despite some improvement in risk prediction algorithms over the past decades, traditional risk factor profiling fails to identify many individuals at impending risk of an acute CVD event, highlighting the need for new strategies for risk prediction in the general population. We sought to identify novel circulating protein markers associated with incident CVD events in asymptomatic individuals by performing an unbiased proteomics screen using data independent acquisition mass spectrometry (DIA-MS), followed up in an independent cohort using enzyme-linked immunosorbent assays (ELISA).
Methods
Study participant recruitment and sample collection
Plasma samples for this case–-control study (n = 50 for the Discovery cohort and n = 151 for the Extended cohort) were selected from the Canterbury Healthy Volunteers cohort (HVols, n = 3,358). HVols participants were randomly selected from the Canterbury (New Zealand) electoral rolls. Participants were screened for prior hospital admissions and cardiac diagnoses. CVD risk factors, anthropometric measures, personal health information and family history of cardiovascular events were recorded for each participant. CVD risk was estimated using both FRS and PREDICT-1° equations3,4. Plasma samples were biobanked at − 80 °C and subsequently assayed for high-sensitivity Troponin I (hsTnI) and NT-proBNP. Clinical events of participants have been continuously documented and updated 6-monthly (median follow-up 9.2 years). The study conformed to the Declaration of Helsinki and was approved by the New Zealand Health and Disability Ethics Committee (Reference CTY/01/05/062). All participants gave written, informed consent.
Patient selection for DIA-MS discovery study
For the initial Discovery arm of the study using data independent acquisition mass spectrometry (DIA-MS), a subset of HVols aged < 80 years, BMI < 30 kg/m2, systolic BP < 150 mmHg, without diabetes and non-smokers, who subsequently experienced an acute CVD event within 3 years were identified (n = 25 cases, Fig. 1). Incident ischemic CVD events were defined as non ST-elevation myocardial infarction, ST-elevation myocardial infarction, ischemic stroke, transient ischemic attack, unstable angina, other angina or death due to coronary heart disease. Matched controls (n = 25) meeting the same criteria but without incurring any CVD events for at least 5 years from recruitment were selected using the R package MatchIt16 with “nearest” matching for age, BMI, and systolic blood pressure, and exact matching for gender.
Sample Selection Criteria. The Canterbury Healthy Volunteers cohort (HVols) is a community-based cohort of 3358 individuals of middle to older age with no previously diagnosed cardiovascular disease (CVD). The Discovery cohort included 25 participants who subsequently incurred an acute CVD event within 3 years and 25 who remained event-free for at least 5 years. The Extended cohort was an independent sample of 76 HVols who incurred an acute CVD event within 5 years and 75 controls who remained event-free for at least 7 years. Figure generated using Microsoft PowerPoint 2019.
Patient selection for ELISA extended cohort study
For the Extended arm of the study, a separate subset of healthy volunteers aged < 80 years, BMI < 35 kg/m2, systolic BP < 160 mmHg, without diabetes and non-smokers, who experienced an acute CVD event within 5 years were selected (n = 76 cases, Fig. 1). Criteria for the Extended arm had to be relaxed compared with the Discovery Cohort, particularly the time to first event, due to limited sample numbers. Matched controls (n = 75) were selected as described above.
DIA-MS
To mitigate batch effects a randomized blocked experiment design was used with equal numbers of cases and controls processed at the same time. Plasma was thawed on ice and centrifuged to remove particulate matter. The detailed sample preparation and data analysis methods are outlined in supplementary information. In brief, plasma (2 µL) was denatured (Supplementary Table 1) and hydrolyzed with trypsin. Tryptic peptides were C18 purified and then spiked with retention time calibration peptides to correct for relative retention time differences between runs (Escher et al. 2012). DIA-MS of the 50 individual, trypsinized plasma samples was undertaken using an AB Sciex 5600 + TripleTOF mass spectrometer coupled to an ekspert nanoLC 415 system (eksigent, AB Sciex, Dublin, CA, USA). DIA-MS data were compared to a spectral library generated from pooled samples. Data analysis was carried out with AB Sciex software (SWATH Acquisition MicroApp (version 2.0.0.1) in PeakView (version 2.2) Software) and R (version R-3.6.1) and RStudio (version 1.2.5001)17,18.
Immunoassays
The concentrations of three leading candidate plasma proteins were measured using commercial ELISA kits, including fetuin A (DFTA00 Human Fetuin A Quantikine ELISA Kit, R&D Systems, Minneapolis, MN, USA), hemoglobin (AB157707 Human Hemoglobin ELISA Kit, Abcam, Cambridge, MA, USA) and fibrinogen (AB208036 Human Fibrinogen SimpleStep ELISA Kit, Abcam), with each assay performed according to manufacturer’s instructions. All samples were run in duplicate, using a randomized, blocked experiment design to minimize bias due to inter-plate variability.
Protein concentration was calculated from a calibration curve using StatLIA (Brendan Technologies, Inc, Carlsbad, CA, USA)19. Samples were re-analyzed if the coefficient of variation between duplicates was ≥ 20%.
Ethics approval and consent to participate
The study conformed to the Declaration of Helsinki and was approved by the New Zealand Health and Disability Ethics Committee (Reference CTY/01/05/062). All participants gave written, informed consent.
Results
A flow chart describing participant selection criteria is shown in Fig. 1. The baseline characteristics of participants in the Discovery and Extended arms of the study are summarized in Table 1. There were no significant differences in baseline characteristics or cardiovascular risk factors between cases and controls in either the Discovery or the Extended sub-study, with the exception of hsTNI, which was higher in cases compared with controls in the Extended cohort (p = 0.015).
Plasma mass spectrometric analysis
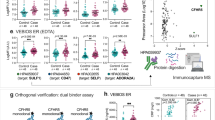
In the Discovery cohort, the 5-year risk PREDICT-1° score did not correspond to time-to-CVD-event (Cox model HR = 1.11 [0.79–1.56], p = 0.53), reflecting that the score alone was not able to distinguish those with subsequent CVD events from those who remained event free in this low-moderate risk sample. In the Discovery cohort, 76 proteins were robustly quantified in human plasma by DIA mass spectrometry. Of these, eight proteins/subunits were associated with CVD events independent of the PREDICT-1° score (Supplementary Table 2). The hazard ratios of these associations were (HR = hazard ratio [95% confidence interval] above vs below median): fibrinogen alpha chain (HR = 1.84 [1.19–2.84]); alpha-2-HS-glycoprotein (fetuin A) (HR = 1.86 [1.19–2.93]); clusterin isoform 2 (HR = 1.59 [1.06–2.38]); fibrinogen beta chain (HR = 1.55 [1.04–2.30]); hemoglobin subunit beta (HR = 1.49 [1.04–2.15]); complement component C9 (HR = 1.62 [1.01–2.59]), fibronectin isoform 3 (HR = 0.60 [0.37–0.99]); and lipopolysaccharide-binding protein (HR = 1.58 [1.00–2.49]), (all p unadjusted < 0.05, Fig. 2). On average, protein levels differed 1.2-fold (range 1.1- to 1.3-fold) between cases who had an event compared with controls who remained event free (Supplementary Table 2). Fibrinogen alpha chain, fetuin A, clusterin isoform 2 and hemoglobin were also independent of BNP, NT-proBNP, and hsTNI in Cox models (Supplementary Tables 3 & 4). Fetuin A and clusterin isoform 2 were highly correlated to one another (Supplementary Fig. 1) leading to the final choice of candidates, fibrinogen, fetuin A and hemoglobin, for further analysis via ELISA assay.
Cox Hazard Ratios of z-score standardized protein abundance in the Discovery cohort, adjusted for the log2-transformed PREDICT-1° 5-year CVD risk score. The whiskers represent 95% confidence intervals. The dashed vertical line corresponds to a Hazard Ratio of 1 (i.e. no difference of CVD risk based on this protein). The dashed horizontal line divides those proteins with p < 0.05 from those proteins with p 0.05. Z-score scaling of protein biomarker data was used instead of using raw protein mass spectrometric intensity to allow easy comparison between proteins. A one unit increase in z-score scaled data represents a one standard deviation increase in protein concentration. Protein IDs are the mnemonic of the uniprot ID (Supplementary Table 5). Figure generated using the ‘survival’ (https://github.com/therneau/survival) and ‘ggplot2′ (https://github.com/tidyverse/ggplot2) packages within R/RStudio17,18,37,38.
ELISA versus DIA-MS assessment
The concentration of fibrinogen, hemoglobin and fetuin A in the same 50 samples from the Discovery arm were also measured using commercially-available ELISAs (Fig. 3). Limits of detection for DIA-MS were applied on each of the detected 9576 ions using IQR guided lower-end cut-offs (see Supplementary Methods). Our stringent quality control also included CV filtering of ions resulting in 1288 high-confidence ions for relative quantification with > 97% of these ions being detectable in each of the 50 people of the Discovery cohort. The coefficient of variation between duplicates measured by ELISA was ≤ 20% with all samples diluted appropriately to lie within the range of the standard curves. DIA mass spectrometry and ELISA methods were strongly correlated for hemoglobin (Spearman correlation r = 0.76 and 0.70, p < 0.05 for the hemoglobin alpha chain and the hemoglobin beta chain, respectively), and moderately correlated for fibrinogen (Pearson correlation r = 0.45, r = 0.45, r = 0.50, p < 0.05 for fibrinogen chains alpha, beta and gamma). However, no correlation was observed between DIA-MS and ELISA quantitation for fetuin A (Pearson correlation r = 0.15, p = 0.30). Therefore, we took both hemoglobin and fibrinogen (but not fetuin A) as candidates to validate our findings in the larger Extended cohort using ELISA assays.
Scatterplots comparing protein concentrations measured via ELISA versus DIA-MS. Panel A1) hemoglobin (Hb) vs Hb alpha chain (HBA) and Hb beta chain (HBB). Panel B) fibrinogen (FIB) vs FIB chains alpha, beta, and gamma (FIBA, FIBB, FIBG) and Panel C) fetuin A (FetA). Correlations were assessed via Pearson for fibrinogen and fetuin A and via Spearman test for hemoglobin due to slight non-normal distribution of the HB ELISA data. Trendlines represent fits using linear regression models. Residual plots for model assessment provided as Supplementary Fig. 2. Figure generated using the ‘ggplot2′ (https://github.com/tidyverse/ggplot2) package within R/RStudio17,18,37.
Extended cohort
There were statistically significant differences between the risk factor profiles of the Extended and the Discovery cohorts, including a longer duration between recruitment and first CVD event (median [IQR] = 1178.0 [791.2, 1432.5] days in the Extended cohort versus 411.0 [244.0, 748.0] days in the Discovery cohort, p < 0.001) and a higher body mass index (BMI) (26.4 ± 3.4 kg/m2 in Extended cohort versus 25.1 ± 2.7 kg/m2 in the Discovery cohort, p = 0.01) (Table 1). As was the case in the Discovery cohort, the PREDICT-1° score alone did not contribute significantly to risk prediction in our Cox models in the Extended cohort (HR = 1.16 [0.86–1.57], p = 0.33). In the Extended cohort, concentrations measured by ELISA of neither hemoglobin nor fibrinogen were significantly different between cases and controls. The measured hemoglobin concentration (median [IQR]) in controls was 141.5 µg/mL [103.3, 317.3] compared with 148.3 µg/mL [78.3, 304.0] in cases, p = 0.537. The fibrinogen concentration was 2.2 mg/mL [1.9, 2.6] in controls compared with 2.1 mg/mL [1.8, 2.4] in cases p = 0.099. When included in Cox-proportional hazard models, adjusted for PREDICT-1° 5-year CVD risk, neither fibrinogen nor hemoglobin showed significant associations with the time-to-event in Cox models (fibrinogen (HR = 0.86 [0.68–1.10]), p = 0.37 and hemoglobin (HR = 0.92 [0.73–1.15]), p = 0.59).
Discussion
Biomarkers to improve prediction
This study provides the first proof of principle that a DIA-MS proteomic approach can be used to identify additional circulating plasma biomarkers to improve current population-based screening approaches to predict future CVD events in asymptomatic individuals. The absence of predictive value of the PREDICT-1° score on its own in this low-to-moderate risk population sample demonstrates the need for improved models to estimate CVD risk. The three top ranked candidate plasma biomarkers from our DIA-MS Discovery screen (hemoglobin, fibrinogen and fetuin A) were independently associated with near-future CVD events (≤ 3 years) in asymptomatic individuals, independent of the PREDICT-1° score. However, in the Extended cohort, where the median time-to-event was considerably longer and individuals with a broader range of risk factors were included, neither hemoglobin nor fibrinogen were independently predictive of future CVD events in Cox-modelling. It is conceivable that these two proteins are associated with more immediate risk as the time to first event was ~ threefold shorter in the Discovery cohort (median of 244 days) compared with 791 days in the Extended Cohort, despite the Discovery cohort being characterised by a generally lower burden of other known risk variables than the Extended cohort (Table 1). Further, it is possible that the moderate correlation between detection techniques, DIA-MS and ELISA, contributes to the fact that the relationship between protein and CVD outcome observed in the Discovery cohort was not demonstrated in the Extended cohort. In general, a high correlation between ELISA and mass spectrometric results can be expected if the peptides used for quantification in DIA-MS and the detection of epitopes by ELISA antibodies allow equivalent detection of the analyte by the respective technology. However, there are many examples of mass spectrometry and ELISA not correlating strongly, oftentimes due to post-translational modification or due to cross-reactivity issues in ELISA20,21,22. Our findings indicate that certain circulating markers may improve risk prediction of impending CVD events in low-to-moderate CVD risk individuals.
Risk prediction models such as the NZ PREDICT-1° score, and the UK QRISK3 work best on the population level but suffer from poor discrimination at the level of the individual. Up to 20% of patients diagnosed with CVD have no traditional risk factors, and up to 40% have only one, suggesting that the accuracy of risk prediction models for individuals is modest and that CVD commonly occurs in people predicted to be at low-to-moderate risk6. Biomarkers may add value for individual risk stratification, especially for those in the low-to-moderate risk “grey zone,” where clinicians often do not have appropriate tools available to guide their decision making, and in those of older age (i.e. > 75 years). In our study of older individuals with a relatively low burden of cardiovascular risk factors, the NZ PREDICT-1° risk score was unable to discriminate between those who subsequently incurred a CVD event and those that remained event-free. Our findings highlight the feasibility of an unbiased DIA-MS approach for discovery of additional circulating plasma biomarkers that may add value to established risk factors and current approaches.
In the Discovery cohort we identified eight proteins (i.e. fibrinogen alpha chain, fetuin A, clusterin isoform 2, fibrinogen beta chain, hemoglobin subunit beta, fibronectin isoform 3, complement component C9, lipopolysaccharide-binding protein) that have potential to improve risk prediction of a near-future CVD event (within 3 years). Of these, fibrinogen is probably the best characterized in terms of its relation to CVD risk prediction. It has been previously proposed that circulating fibrinogen concentration be included in CVD risk stratification23,24. A previous study reported the age- and sex-adjusted hazard ratio per 1 g/L increase in usual fibrinogen concentration was 2.42 (95% CI, 2.24–2.60) for coronary heart disease; 2.06 (95% CI, 1.83–2.33) for stroke; 2.76 (95% CI, 2.28–3.35) for other vascular mortality and 2.03 (95% CI, 1.90–2.18) for nonvascular mortality25. Our data extend these findings by suggesting that fibrinogen remains independently predictive of outcomes even when adjusted for multiple other cardiovascular risk factors.
Hemoglobin circulating freely in the plasma and not contained within the erythrocyte has been implicated in pathological conditions, and has been proposed to contribute to abnormal production of highly reductive and oxidative compounds via the four heme ligands incorporated into each hemoglobin protein26. While not specifically associated with CVD risk, increased concentrations of cell-free hemoglobin have been linked to micro ruptures in the fragile vasculature of unstable atherosclerotic plaques27. In this context, hemoglobin has been reported to function as a “locally active disease modifier” and “intrinsic alarm molecule” (i.e. a biomarker) indicating bleeding and tissue destruction28.
The multifaceted biological role of Fetuin A has been recently reviewed and has been variably reported to be inversely correlated with coronary artery disease and atherosclerotic burden (presumed due to protection against vascular calcification) and positively correlated with coronary artery disease, possibly via its role in diabetes mellitus through the inhibition of insulin receptor tyrosine kinase29.
Proteomics and biomarker discovery approaches
We have used a robust and unbiased analysis protocol for measuring plasma proteins in human plasma. Applying DIA mass spectrometry to biomarker discovery allows the identification and quantifation of > 300 proteins30. All of these proteins tend to be in the high- to medium-abundant concentration range in plasma, and so are readily detectable by a number of clinical assay platforms. However, depletion or further fractionation steps may be required to detect low-abundance proteins using DIA31. Plasma is the ideal, accessible sample reservoir for discovery of potential disease biomarkers, as it captures a considerable proportion of the proteome (~ 2,000 out of ~ 20,000 known proteins detectable using mass spectrometry, not considering isoforms)32. Furthermore, DIA approaches interrogating phosphorylation and glycosylation patterns, important for cell-signalling and protein function, are also being elucidated and likely to contribute further in the identification of novel biomarkers33,34. Continuous improvements in mass spectrometry technology, analysis pipelines and sample throughput are likely to contribute to future precision medicine biomarker discovery35.
The selectivity/specificity of mass spectrometry is superior to that of many ELISAs and also facilitates interrogation of post-translational modifications, which carry additional biological information that may be related to pathophysiology36. However, sensitivity of MS methods is still lacking compared with ELISA, especially for complex mixtures such as plasma where a small fraction of proteins dominates the contribution to the overall protein concentration. Additionally, ELISA is easily scalable with high-throughput easily implemented. DIA-MS methods are especially useful for tissue and cell-line proteomics, where the dynamic range of protein concentrations is smaller compared to that of plasma.
Limitations
Our study has several limitations. First, by design we selected individuals with a relatively low burden of cardiovascular risk considering their older age, which limited the sample size of our Discovery and Extended cohorts. Second, our Discovery and Extended cohorts were not well-matched for time to first event (3 years vs. 5 years) and cardiovascular risk, reducing our ability to replicate our findings. Third, the heterogeneity of outcomes and the different proportions of each outcome in the Discovery and the Extended cohorts may reduce power to detect biomarker associations with specific outcomes. Fourth, despite the good correlation in the Discovery phase, the two techniques (DIA-MS and ELISA) have inherent differences affecting their specificity in terms of which part of the protein is detected. Fifth, the presence of abundant plasma proteins in our samples may have prevented detection of less abundant proteins with similar or stronger associations with CVD events. The MS-detectable plasma proteome is approximately 2000 proteins32, of which 76 were robustly quantified across the 50 DIA samples in this study. This is approximately 28% of the number of proteins observed in other studies (272 robustly quantified proteins) using MS1-level BoxCar quantification. This reflects differences in mass spectrometric data acquisition methods and quality filtering for inclusion/exclusion of peptides into the final data-set32.
Conclusions
DIA-MS methods have great potential in biomarker discovery/hypothesis generation due to measuring many proteins simultaneously. Using DIA-MS we have identified eight protein biomarkers that may be independently associated with near-future CVD events (< 3 years) in asymptomatic individuals of older age with a relatively low cardiovascular risk.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- DIA-MS:
-
Data independent acquisition mass spectrometry
- HR:
-
Hazard ratio
- CVD:
-
Cardiovascular disease
- IHD:
-
Ischemic heart disease
- FRS:
-
Framingham risk score
- NZ:
-
New Zealand
- ECG:
-
Electrocardiograph
- PREDICT-1°:
-
New Zealand primary prevention equations
- QRISK3:
-
United Kingdom prediction algorithm for cardiovascular disease
- UK:
-
United Kingdom
- TC:
-
Total cholesterol
- HDL:
-
High-density lipoprotein
- LDL:
-
Low-density lipoprotein
- HbA1C:
-
Glycated hemoglobin
- BNP:
-
B-type natriuretic peptide
- NT-proBNP:
-
Amino-terminal pro-hormone B-type natriuretic peptide
- TNT:
-
Cardiac troponin T
- TNI:
-
Cardiac troponin I
- HF:
-
Heart failure
- MI:
-
Myocardial infarction
- ELISA:
-
Enzyme-linked immunosorbent assay
- Hvols:
-
Canterbury Healthy Volunteers cohort
- hsTnI:
-
High-sensitivity Troponin I
- SWATH-MS:
-
Sequential windowed acquisition of all theoretical fragment ion spectra mass spectrometry
- IQR:
-
Interquartile range
- BMI:
-
Body mass index
- SD:
-
Standard deviation
- SBP:
-
Systolic blood pressure
- BP:
-
Blood pressure
- eGFR:
-
Estimated glomerular filtration rate
- ACS:
-
Acute coronary syndrome
- CoD:
-
Cause of death
- STEMI:
-
ST-elevation myocardial infarction
- NSTEMI:
-
Non-ST-elevation myocardial infarction
- CHD:
-
Coronary heart disease
- Hb:
-
Hemoglobin
- HBA:
-
Hemoglobin alpha chain
- HBB:
-
Hemoglobin beta chain
- FIB:
-
Fibrinogen
- FIBA:
-
Fibrinogen alpha chain
- FIBB:
-
Fibrinogen beta chain
- FIBG:
-
Fibrinogen gamma chain
References
Roth, G. A. et al. Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J. Am. Coll. Cardiol. 70, 1–25 (2017).
Mahmood, S. S., Levy, D., Vasan, R. S. & Wang, T. J. The framingham heart study and the epidemiology of cardiovascular disease: A historical perspective. The Lancet 383, 999–1008 (2014).
D’Agostino, R. B. Sr. et al. General cardiovascular risk profile for use in primary care: The framingham heart study. Circulation 117, 743–753 (2008).
Pylypchuk, R. et al. Cardiovascular disease risk prediction equations in 400000 primary care patients in new zealand: A derivation and validation study. The Lancet 391, 1897–1907 (2018).
Hippisley-Cox, J., Coupland, C. & Brindle, P. Development and validation of qrisk3 risk prediction algorithms to estimate future risk of cardiovascular disease: Prospective cohort study. BMJ 357, j2099 (2017).
Wang, J. et al. Novel biomarkers for cardiovascular risk prediction. J. Geriatr. Cardiol. JGC 14, 135–150 (2017).
Braatvedt, G. D. et al. Understanding the new hba1c units for the diagnosis of type 2 diabetes. N. Z. Med. J. 125, 70–80 (2012).
Hoste, L., Deiteren, K., Pottel, H., Callewaert, N. & Martens, F. Routine serum creatinine measurements: How well do we perform?. BMC Nephrol. 16, 21 (2015).
Yancy, C. W. et al. 2013 accf/aha guideline for the management of heart failure: A report of the american college of cardiology foundation/american heart association task force on practice guidelines. J. Am. Coll. Cardiol. 62, e147-239 (2013).
Pickering, J. W. et al. Rapid rule-out of acute myocardial infarction with a single high-sensitivity cardiac troponin t measurement below the limit of detection: A collaborative meta-analysis. Ann. Intern. Med. 166, 715–724 (2017).
Richards, A. M. et al. Plasma n-terminal pro-brain natriuretic peptide and adrenomedullin: New neurohormonal predictors of left ventricular function and prognosis after myocardial infarction. Circulation 97, 1921–1929 (1998).
Davis, M. et al. Plasma brain natriuretic peptide in assessment of acute dyspnoea. The Lancet 343, 440–444 (1994).
Saunders, J. T. et al. Cardiac troponin t measured by a highly sensitive assay predicts coronary heart disease, heart failure, and mortality in the atherosclerosis risk in communities study. Circulation 123, 1367–1376 (2011).
Jia, X., Sun, W., Hoogeveen, R. C., Nambi, V., Matsushita, K. & Folsom, A. R. et al. High-sensitivity troponin and incident coronary events, stroke, heart failure hospitalization, and mortality in the aric study. Circulation 139, 2642–2653 (2019).
Everett, B. M., Zeller, T., Glynn, R. J., Ridker, P. M. & Blankenberg, S. High sensitivity cardiac troponin i and b-type natriuretic peptide as predictors of vascular events in primary prevention: Impact of statin therapy. Circulation 131, 1851–1860 (2015).
Ho, D., Imai, K., King, G. & Stuart, E. A. Matchit: Nonparametric preprocessing for parametric causal inference. 42, 28 (2011).
R Core Team. R: A language and environment for statistical computing. (2018).
Team R. Rstudio: Integrated development environment for r (2016). Boston, MA 2020.
Brendan Technologies I. Statlia enterprise. Vol. 3.2 Ed. Carlsbad, CA, USA, (2004).
Parker, C. E. & Borchers, C. H. Mass spectrometry based biomarker discovery, verification, and validation—quality assurance and control of protein biomarker assays. Mol. Oncol. 8, 840–858 (2014).
Denburg, M. R. et al. Comparison of two elisa methods and mass spectrometry for measurement of vitamin d-binding protein: Implications for the assessment of bioavailable vitamin d concentrations across genotypes. J. Bone Miner. Res. 31, 1128–1136 (2016).
Hoofnagle, A. N. & Wener, M. H. The fundamental flaws of immunoassays and potential solutions using tandem mass spectrometry. J. Immunol. Methods 347, 3–11 (2009).
Arbustini, E., Narula, N. & D’Armini, A. M. Fibrinogen. Circulation 128, 1276–1280 (2013).
Kaptoge, S. et al. C-reactive protein, fibrinogen, and cardiovascular disease prediction. N. Engl. J. Med. 367, 1310–1320 (2012).
Danesh, J. et al. Plasma fibrinogen level and the risk of major cardiovascular diseases and nonvascular mortality: An individual participant meta-analysis. JAMA J. Am. Med. Assoc. 294, 1799–1809 (2005).
Olsson, M. G. et al. Pathological conditions involving extracellular hemoglobin: Molecular mechanisms, clinical significance, and novel therapeutic opportunities for alpha(1)-microglobulin. Antioxid. Redox Signal 17, 813–846 (2012).
Matic, L. P. et al. Novel multiomics profiling of human carotid atherosclerotic plaques and plasma reveals biliverdin reductase b as a marker of intraplaque hemorrhage. JACC Basic Transl. Sci. 3, 464–480 (2018).
Schaer, D. J. & Buehler, P. W. Cell-free hemoglobin and its scavenger proteins: New disease models leading the way to targeted therapies. Cold Spring Harb. Perspect. Med. 3, a013433 (2013).
Jirak, P., Stechemesser, L., Moré, E., Franzen, M., Topf, A. & Mirna, M. et al. Chapter three - clinical implications of fetuin-a. In: Makowski GS, editor Advances in clinical chemistry, Vol. 89: Elsevier; 2019. p. 79–130.
Bruderer, R. et al. Analysis of 1508 plasma samples by capillary-flow data-independent acquisition profiles proteomics of weight loss and maintenance. Mol. Cell. Proteom. 18, 1242–1254 (2019).
Geyer, P. E., Holdt, L. M., Teupser, D. & Mann, M. Revisiting biomarker discovery by plasma proteomics. Mol. Syst. Biol. 13, 942 (2017).
Niu, L. et al. Plasma proteome profiling discovers novel proteins associated with non-alcoholic fatty liver disease. Mol. Syst. Biol. 15, e8793 (2019).
Bekker-Jensen, D. B., Bernhardt, O. M., Hogrebe, A., del Val, A. M., Verbeke, L. & Gandhi, T., et al. Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition (dia) without the need for spectral libraries. bioRxiv 2019:657858.
Lin, C.-H., Krisp, C., Packer, N. H. & Molloy, M. P. Development of a data independent acquisition mass spectrometry workflow to enable glycopeptide analysis without predefined glycan compositional knowledge. J. Proteom. 172, 68–75 (2018).
Mann, M. The ever expanding scope of electrospray mass spectrometry—a 30 year journey. Nat. Commun. 10, 3744 (2019).
Fert-Bober, J., Murray, C. I., Parker, S. J. & Eyk, J. E. V. Precision profiling of the cardiovascular post-translationally modified proteome. Circ. Res. 122, 1221–1237 (2018).
Wickham, H. Ggplot2: Elegant graphics for data analysis. springer; 2016.
Therneau, T. M. & Lumley, T. Package ‘survival’. R Top Doc 2015;128.
Yoshida, K., Chipman, J. J., Bohn, J., McGowan, L. D., Barrett, M. & Christensen, R. H. B. Package ‘tableone’. 2019.
Acknowledgements
We thank the study participants and the research staff of the Christchurch Heart Institute. This research was supported by the Freemasons Carrell-Espiner Fellowship, the Heart Foundation of New Zealand (Research Fellowship, Ref# 1693; Small Project Grant, Ref# 1838), the Farina Thompson Charitable Trust, the Health Research Council of New Zealand (Programme Grant, Ref# 11/170; Emerging Researcher Grant, Ref# 17/562), Lotteries Health Research (Ref# R-LHR-2017-49246) the Christchurch Heart Institute Trust and the University of Otago.
Author information
Authors and Affiliations
Contributions
M.L., A.P.P., V.A.C. designed the study; V.A.C. was responsible for recruitment of the study cohort and obtaining ethics approval, M.L., A.P.P., V.A.C., R.W.T., A.M.R. contributed grant funding; M.L., E.A., T.K., A.vZ., L.K.L, T.C.R.P., C.J.P. performed or provided significant input into experimental analyses; ML was responsible for data analysis with significant input from A.P.P., C.M.F., K.P.P., V.A.C.; M.L. M.L. drafted the manuscript and all authors provided input into critical revision of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lassé, M., Pilbrow, A.P., Kleffmann, T. et al. Fibrinogen and hemoglobin predict near future cardiovascular events in asymptomatic individuals. Sci Rep 11, 4605 (2021). https://doi.org/10.1038/s41598-021-84046-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-84046-7
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.