Abstract
The manatee family encompasses three extant congeneric species: Trichechus senegalensis (African manatee), T. inunguis (Amazonian manatee), and T. manatus (West Indian manatee). The fossil record for manatees is scant, and few phylogenetic studies have focused on their evolutionary history. We use full mitogenomes of all extant manatee species to infer the divergence dates and biogeographical histories of these species and the effect of natural selection on their mitogenomes. The complete mitochondrial genomes of T. inunguis (16,851 bp), T. senegalensis (16,882 bp), and T. manatus (16,882 bp), comprise 13 protein-coding genes, 2 ribosomal RNA genes (rRNA - 12S and 16S), and 22 transfer RNA genes (tRNA), and (D-loop/CR). Our analyses show that the first split within Trichechus occurred during the Late Miocene (posterior mean 6.56 Ma and 95% HPD 3.81–10.66 Ma), followed by a diversification event in the Plio-Pleistocene (posterior mean 1.34 Ma, 95% HPD 0.1–4.23) in the clade composed by T. inunguis and T. manatus; T. senegalensis is the sister group of this clade with higher support values (pp > 0.90). The branch-site test identified positive selection on T. inunguis in the 181st position of the ND4 amino acid gene (LRT = 6.06, p = 0.0069, BEB posterior probability = 0.96). The ND4 gene encodes one subunit of the NADH dehydrogenase complex, part of the oxidative phosphorylation machinery. In conclusion, our results provide novel insight into the evolutionary history of the Trichechidae during the Late Miocene, which was influenced by geological events, such as Amazon Basin formation.
Similar content being viewed by others
Introduction
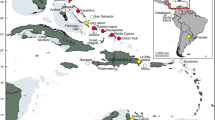
The Order Sirenia comprises a group of Afrotherian aquatic mammals that arose by the Early Paleocene1 and are currently distributed throughout the Indo-Pacific region and from the southeastern part of the United States of America to part of the Brazilian coast, Amazon region, and African coast2,3. Sirenians are classified into two extant families: Trichechidae and Dugongidae. The Family Dugongidae has only one living species (Dugong dugon MÜLLER, 1776), because the Steller’s sea cow (Hydrodamalis gigas ZIMMERMANN, 1780) was driven to extinction in the eighteenth century due to overhunting4. Dugongidae is considered the older of these two families and it was cosmopolitan until its diversity decreased during the Pliocene5,6. The modern dugong is restricted to the Indian Ocean and West Pacific region, specifically in regions of high concentration of seagrass7. The Family Trichechidae is composed of two subfamilies, Miosireninae (extinct species), and Trichechinae, which includes all modern species8. The Subfamily Trichechinae has only one extant genus (Trichechus) and three species: Trichechus senegalensis LINK, 1795 (African manatee) living along the coasts and rivers of western Africa, T. inunguis NATTERER, 1883 (Amazonian manatee) which is endemic to the Amazon Basin, and T. manatus LINNAEUS, 1758 (West Indian manatee) ranging from the southeastern USA and the Caribbean region to the Brazilian northeastern coast9 (Fig. 1).
Distribution of Trichechus species in the world, map made by EMSS with Rstudio v.1.3 using distribution shapefile from IUCN database, script available at author’s Github webpage (https://github.com/souzaems/scripts/blob/master/map_on_R.r).
The fossil record is scant for the Family Trichechidae, which makes it difficult to infer their evolutionary history4. The currently accepted evolutionary scenario for the genus Trichechus is that it originated in South American rivers from where it colonized the marine environment and reached the African continent9,10,11. However, when and how the Trichechus species diverged are still mainly inferred from fossil age and morphological characteristics. Until recently, few genetic studies have attempted to shed light on their evolution, but these studies used single genetic markers or did not include all representatives of the genus9,12,13. One of these studies used a single mitochondrial gene to infer the divergence time between T. manatus and T. inunguis, which was estimated to have been 2–4 million years ago (Ma), in the Plio-Pleistocene3,9.
The mitochondrial genome encodes 13 proteins, which belong to a complex of oxidative phosphorylation pathways (OXPHOS) and have been extensively used to unravel phylogenetic relationships14,15,16. For a long time, the mitochondrial genome was considered to be under neutral or nearly neutral selection17. However, growing evidence has revealed that these genes may be subject to positive directional selection16,18,19. Hence, more recently, attention has been given to the study of molecular adaptation of mitochondrial genes, and many studies have shown that amino acid substitutions on these proteins may improve aerobic capacity and may be related to adaptation to new environments20,21. The aquatic mammals, such as manatees, are textbook examples of lineages that have undergone extreme adaptations related to the transition from land to aquatic environments3,22. Accordingly, positive selection in genes such as cytochrome b23 and ND2, ND4, and ND524 was already detected in killer whales (Orcinus orca, Linnaeus 1758) and dugongs, respectively, but such mitochondrial molecular evolution in manatees has been little studied. In this context, here we sequenced the complete mitogenomes of all extant species of manatees to infer their phylogenetic relationships, to estimate divergence times among the species and to evaluate the effect of natural selection on mitochondrial genes during their evolution.
Material and methods
Sampling and DNA extraction
We extracted DNA from one tissue sample of T. senegalensis (Democratic Republic of the Congo12), one tissue sample of Trichechus inunguis (female, Japurá River, Brazil) and one tissue sample from Trichechus manatus (male, Ceará, Brazil). All tissues were collected following the respective environmental regulations. The DNA extraction of T. senegalensis was done following the Phenol:Chloroform:Isoamyl Alcohol 25:24:1 according to manufacturers. The collecting permits were provided by Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio, number 44628-2), and the re-importation tissue for T. senegalensis was provided by CITES number 19BR031212/DF. The activity of access to the Genetic Heritage is registered under the Sistema Nacional De Gestão Do Patrimônio Genético E Do Conhecimento Tradicional Associado (SisGen) number A94D205. This study was carried out according to the Brazilian guidelines for animal care and authorized by the Committee on the Ethics of Animal Use and Care – CEUA of the State University of Campinas – UNICAMP.
The DNA quality and integrity were evaluated on 1% agarose gel. The concentration and purity of samples were verified using the NanoDrop 2000 Spectrophotometer (Thermo Scientific) and subsequently confirmed by fluorimetry on Qubit 2.0 (Invitrogen, Life Technologies). The mitogenome of T. senegalensis was sequenced by an external company (BPI Biotecnologia Pesquisa e Inovação) using the MiSeq Illumina Platform.
mtDNA assembly and annotation
The mitochondrial genomes (mtDNA) from T. inunguis and T. manatus were retrieved from the whole genome sequenced by the HiSeq 2500 platform. We first created two custom BLAST databases on these genomes. Then we used the mtDNA from T. manatus (GenBank access numbers AM904728, and MN105083), as query in BLAST + v. 2.9.0 to search T. inunguis and T. manatus mtDNAs13,25. We subsequently used a custom Python 3 script26 to retrieve BLAST hits and did the assembly with Geneious R9 (https://www.geneious.com). The mtDNA of T. senegalensis was assembled using MitoFinder27.
We used GeSeq—Annotation of Organellar Genomes server28 to annotate mtDNA, and to avoid common annotation errors29, we used reference data (Homo sapiens NC_012920.1, and Mus musculus NC_005089.1) to compare with our annotation. We used OGDRAW—Draw Organelle Genome Maps28 for visualization. Finally, we used ARWEN v1.2.3, ARAGORN v1.2.38 and tRNAscan-SE v2.0.3 as implemented in the MITOS web server30 and Geseq server28 to annotate the tRNA.
Phylogenetic analyses
To infer trichechid phylogeny, we used the following outgroups: Dugong dugon (accession numbers AY075116, and NC003314) as sister group of Trichechidae, Loxodonta africana (NC000934), Loxodonta cyclotis (JN673263), Elephantulus edwardii (NC041486), Echinops telfairi (AB099484), and Dasypus novemcinctus (Y11832). First, we aligned the coding genes with the MAFFT31 algorithm and translated into proteins using Geneious R9 (https://www.geneious.com). The best evolutionary model to be used in phylogenetic analysis was determined by PartitionFinder 2.032 using the Akaike information criterion (AIC); first we tested using all the mitogenome data (tRNAs, rRNA, PCGs), and then only 13 coding genes.
We built a maximum likelihood (ML) tree using RAxML v.8.0, and the GTR + GAMMA model33. Also, the Bayesian inference (BI) tree was inferred by MrBayes v.3.2.634 using the models inferred with PartitionFinder. The Bayesian analysis was conducted using the Markov chain Monte Carlo (MCMC) method with three heated and one cold chain, sampling every 1,000,000 generations from 20,000,000 generations, and discarding the first 500,000 generations as burn-in.
We estimated divergence time in BEAST235, with custom parameters for the calibration nodes that were chosen using fossil data (Table 1). The analyses were performed using the following parameters: for the sites—HKY substitution model, with empirical base frequencies, and gamma site heterogeneity model; for tree—Yule Process Speciation model as the tree prior, and a random starting tree; for the clock—a lognormal relaxed clock uncorrelated prior. After setting the parameters, we performed two independent runs with 100,000,000 generations, sampling every 5000 generations. We used Tracer v.1.6.036 to check for convergence of the chains to stationary distributions, then we summarized the runs using LogCombiner v1.8.237. We built the final tree using the combined results from all trees using TreeAnnotator v1.8.238, and we visualized the tree with FigTree v.1.4.239.
Adaptive molecular evolution analysis
To identify codon sites with positive selection in the Family Trichechidae, we estimated the ω (dN/dS) rate, where dN is the non-synonymous substitution rate (the rate at which changes in nucleotide sites lead to changes into new amino acids) and dS is the synonymous substitution rate (the rate at which changes in nucleotide sites do not lead to changes in the amino acid chain). A ω > 1 indicates positive selection, i.e. when natural selection is favoring amino acid changes. We used the branch-site test for positive selection40 implemented in Godon software41, which incorporate codon rate variation approaches, and gamma variation between codons41,42,43. We applied this test on the thirteen coding genes of Trichechus species and the outgroups. We ran five tests for each gene, labelling the following lineages: (I) T. manatus only, (II) T. inunguis only, (III) T. senegalensis only, (IV) the lineage of T. manatus and T. inunguis and (V) the entire Genus Trichechus (Supplementary Figure 1).
To estimate whether substitutions with signs of positive selection will affect the structure and function of mitochondrial proteins, we aligned the Trichechus sp. and got the X-ray crystal structures using SWISS-MODEL44. We also estimated domain position using the InterPro web tool45.
Results
Assembly and annotation
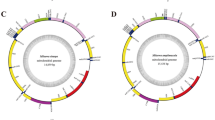
Here we present the complete mitochondrial genomes of the three contemporaneous Trichechinae species: Trichechus inunguis 16,851 base pairs (bp), T. senegalensis 16,882 bp, and T. manatus 16,924 bp, deposited under GenBank accession numbers MW073826, MW073827 and MW073828, respectively. The mitochondrial genome structure of all Trichechus species consists of 13 protein-coding genes, 2 ribosomal RNA genes (rRNA - 12S and 16S), and 22 transfer RNA genes (tRNA). Most of these elements were encoded on the H-strand, except for two protein-coding genes (ATP8 and ND6), and eight tRNA (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, Pro) that were encoded in the L-strand; for all species the tRNA length ranges between 59 and 75 bp (Fig. 2, and Supplementary Table 1).
Phylogenetic analyses
We generated maximum likelihood and Bayesian trees using two types of dataset: one using all mitogenome data, and the second using only the 13 protein-coding genes from the mitogenome. All phylogenetic trees resulted in the same topology, with the highest posterior probability and bootstrap values (Supplementary Figure 2). Our mitochondrial phylogeny depicts Dugong dugon as a sister group of all Trichechus species, with high posterior probability (pp) > 0.99, and bootstrap value of 100. Within the Trichechidae, T. senegalensis is the sister group of a clade composed by T. manatus and T. inunguis, both relationships with strong statistical support in Bayesian analysis (pp > 0.95), and bootstrap of 65 in maximum likelihood analysis.
We estimated the ages of the divergence events within the Trichechidae using nodes of calibration based on the fossil record (Table 2). Our results suggest that the genus Trichechus originated during the Late Miocene, 6.56 Ma (95% HPD 3.81–10.66), and the divergence of T. manatus and T. inunguis may have occurred in the Pleistocene, 1.34 Ma (95% HPD 0.1–4.23) (Fig. 3).
Adaptive molecular evolution
We used the branch-site test to identify positive selection in Trichechus species. We detected evidence for positive selection only in the T. inunguis lineage, at the 181st position of the ND4 amino acid gene (LRT = 6.06, p = 0.0069, Bayes Empirical Bayes posterior probability = 0.96).
Protein structure
We mapped the positively selected site identified by branch-site test onto the mitochondrial protein three-dimensional (3D) crystal structures and searched for the proximity of this site to the functional domains of the protein. The alignment of homologous structures for ND4 and crystal structures revealed that the site with a significant signal of positive selection (site 181) is inside a highly conserved core in the main domain of the ND4 protein. This region corresponds to the discontinuous region of the transmembrane helices 7 (Fig. 4), which enables flexibility for the ND4 subunit.
Discussion
Mitochondrial DNA annotation issues
For the mtDNA final annotation we used primarily the GeSeq information. We found some disagreements regarding codon initiation and termination among the three softwares, which were corrected in the final consensus based on human (Homo sapiens NC_012920.1) and mouse (Mus musculus NC_005089.1) mitogenome alignments. In general, the annotation errors were the identification of the ATP8 gene, which was identified as D-loop, and the ND1 gene was reduced in size and was annotated in the wrong region. These disagreements highlight the importance of using more than one annotation software, to manually inspect and fix mistakes during the gene annotation procedure.
Phylogenetic relationships within the genus Trichechus
One of the first studies that discussed trichechid diversification compared tooth and skeletal characteristics from extant Trichechus species with fossil taxa, and concluded that T. manatus and T. senegalensis have similar morphological characteristics—which could be the consequence of a close relationship, and that T. inunguis has more derived characteristics3. Other studies using molecular data found divergent results: a study using only the cyt-b sequence suggested a sister relationship between T. manatus and T. senegalensis9, while other studies using mitochondrial D-loop as a genetic marker found T. senegalensis as the sister group of T. manatus and T. inunguis with high support values13,46, similar to our data.
The divergence time estimates based on our mitogenomic data in the Family Trichechidae (approximately 7 Ma) are consistent with the fossil record for the genus in South America—register from Early Miocene and Late Miocene3,47. Notably, the history of Trichechus is difficult to explain looking only at living species, especially due the fact that its geographic distribution is very broad with species in the African continent, North America and South America. Also, we have few studies about its evolution using molecular data. Another genetic study9, that used D-loop as a marker, found a split between T. inunguis and T. manatus around 3.1–0.65 Ma, similar to our results.
Evolutionary history of the genus Trichechus
The river courses on the South American continent underwent many modifications during the Neogene (23.03–2.5 Ma)48,49,50, which undoubtedly influenced the evolutionary history of the genus Trichechus. Based on our phylogenetic tree and divergence time estimates, we hypothesize a scenario for the evolutionary history of Trichechus, similar to a scenario suggested in the 1980s3,10.
During the formation of the Amazon Basin, the distribution and connections of wetlands had major influences on the evolution and diversity of many groups such as mammals, birds, reptiles and amphibians51,52,53,54,55. In the Early and Middle Miocene (20.4–10 Ma) there was an extensive wetland known as the Pebas Lake (Western Amazon) which was connected with the Caribbean Sea and associated with the higher sea-levels of the Mid-Miocene Climatic Optimum48,56 (Fig. 5a). It is possible that part of an ancestral lineage that later gave rise to Trichechus (Potamosiren REINHART, 1951, known from the Magdalena Basin of Colombia) inhabited both the Pebas Lake and the Caribbean Sea along the South American coast, which were connected51.
Schematic figure to show the evolution of the Amazon drainage pattern, based on Albert et al. (2018). (a) Early and Middle Miocene (20.4–9.0 Ma): Potamosiren (P) present in Magdalena basin based on Domning (1982), and fossil described during this period66; (b) Late Miocene (9.0–5.3 Ma): Trichechus-like sirenians (T) presumed to be present in the Amazon basin and South America Coast as suggested by Domning (1982), and fossil described by Beatty et al. (2012); (c) Middle Pliocene—Recent (c. 4.5–0 Ma): the possible occurrence of modern Trichechus species T, and a new species of Trichechus described recently by Perini et al. (2020).
After this period, in the Late Miocene (9.0–5.3 Ma), the Pebas Lake changed due to tectonic movements (Andean Orogeny), received a huge amount of Andean-derived sediments, and is known as the Acre Lake, not connected to the Caribbean Sea48,56 (Fig. 5b). The manatees (Trichechus-like) that lived in the Acre Lake were at least semi-isolated in Western Amazonia from the other trichechids living along the South American Atlantic coast. Some authors48 indicate some overflow across the Purús Arch, providing a possible route for coastal manatees to swim upstream to the Acre Lake. These manatees likely became more adapted to the riverine environment, as evidenced by Ribodon AMEGHINO, 1883, considered the immediate ancestor of Trichechus and the first manatee to have had horizontally-replaced supernumerary teeth adapted to an abrasive diet. Ribodon is known only from the Rio Paraná basin in Argentina, and from North America, but it might have dispersed along the Atlantic coast and up the incipiently transcontinental Amazon River. Or, perhaps more likely, Ribodon could have evolved in the Acre Lake and dispersed downstream through the Rio Amazonas to the coast.
During the Plio-Pleistocene (5.3–0.012 Ma) the Andean-derived waters in the Acre Lake finally broke completely through the drainage divide into the Solimões and Amazonas sedimentary basins and discharged into the Atlantic Ocean, the modern situation48,51 (Fig. 5c). These nutrient-rich waters would have nourished abundant aquatic vegetation in these regions, including the abrasive true grasses (Gramineae or Poaceae) for which the supernumerary teeth of the manatees are adapted6,10. It is during this stage that we envision the evolution of the genus Trichechus and its diversification into T. manatus and T. inunguis that we know today, although these modern species maintain the ability to hybridize46,48,51,57. Other taxa now extinct may even have evolved in semi-isolated parts of the Amazon Basin; indeed, a new Late Pleistocene fossil, Trichechus hesperamazonicus PERINI et al., 2020, was recently discovered in a gold mine on the Madeira River (Brazil)58. It suggests that the phylogeny of manatees may prove to be more complex than is portrayed in this outline.
From among the Trichechus that at various times inhabited the Atlantic coast, it seems that the ancestors of modern African manatees reached West Africa by way of transoceanic currents from the Caribbean and South America as already suggested before3,8,10 and in accordance with our phylogenetic tree. From our analyses, we cannot specify when this dispersal might have happened, and as no trichechid fossils are known from Africa, this remains an open question. The discovery of more Trichechus fossils in different locations, like the Caribbean region, the Amazon Basin, and Africa, seems indispensable to refine and eventually confirm this evolutionary scenario10,58,59.
Finally, it has been hypothesized that changes in climate and in the types of Caribbean seagrasses dominant during the Pliocene played a role in the extinction of the dugongs from the Caribbean area, which made possible the manatee expansion throughout that area; in fact, competition from manatees with more durable dentitions may have driven the last Caribbean dugongids to extinction6,10.
Mitochondrial molecular adaptation in the genus Trichechus
Although most mitochondrial genes are conserved and evolved under purifying selection, several studies have reported that the action of positive selection in these genes is more common than previously thought19,20,23. Furthermore, mutations in mitochondrial genes can influence the production of reactive oxygen species in mammals24 and different implications for adaptive evolution of mitogenomes have been suggested in several studies. For example, selection in few branches (episodic selection) was related to niche change when a significantly higher ω ratio was found in mitochondrial genes of subterranean mammals18,60, freshwater dolphins61 and high-altitude alpacas24, and this might also be the case for T. inunguis.
Moreover, studies indicated that amino acid variations in mitogenome proteins may be related to functional implications such as adaptation to low-energy diet versus large body size and adaptation to extremely lowered O2 requirements in different mammal species24. Adaptation to temperature change was also related to mitogenome positive selection for hares62. In most studies, different subunits of the OXPHOS system I complex showed signs of positive selection, including ND418. The ND4 gene encodes one subunit of NADH dehydrogenase complex that is part of the oxidative phosphorylation machinery. This complex initiates the electrochemical proton gradient that leads to ATP synthesis. Hence, it has been suggested that mutations in these subunits may influence with the efficiency of the proton-pumping process63,64,65. We found one positively selected site located inside a critical protein region of the ND4 OXPHOS subunit (site 181), suggesting that substitutions at this site may be adaptive, a finding that hints at its possible functional relevance. As a first step in the study of molecular evolutionary adaptations of these diving mammals, this result suggests the importance of developing a more in-depth and comparative study on the functionality of this subunit within the genus of Trichechus, which may confirm whether this variability represents an adaptive change related to T. inunguis. It is important to note that here we only investigated the mitochondrial genes related to the OXPHOS system; the nuclear genes that are part of this system remain to be tested.
Conclusions
In summary, this first mitogenome phylogeny that includes all living Trichechus species provides a new framework for trichechid evolution, which was influenced by geological events in the formation of the Amazon Basin and by transoceanic currents. Probably the landscape change in the Amazon provided conditions for a population increase of Trichechus ancestors, which spread when the basin connected with the sea. We also showed evidence for positive selection acting in the ND4 Trichechus inunguis mtDNA gene. We suggest that this particular site might have functional implications related to metabolic efficiency in this species, but further experimental studies are needed to test this.
References
Buffrenil, V. de. Mammifères. In Stratotype danien. Muséum national d’histoire naturelle (ed. Montenat, C., Merle, D., de Wever, P., & Cornée, A.) 337–339 (2018).
Berta, A., Sumich, J. L. & Kovacs, K. M. Marine Mammals: Evolutionary Biology Most (Elsevier, Amsterdam, 2005).
Domning, D. P. Evolution of manatees: a speculative history. J. Paleontol. 56, 599–619 (1982).
Shoshani, J. Order hyracoidea. Mammal Species World Taxon. Geogr. Ref. 1, 87–89 (2005).
Berta, A. Return to the Sea: The Life and Evolutionary Times of Marine Mammals (University of California Press, Berkeley, 2020).
Vélez-Juarbe, J. Ghost of seagrasses past: using sirenians as a proxy for historical distribution of seagrasses. Palaeogeogr. Palaeoclimatol. Palaeoecol. 400, 41–49 (2014).
Chilvers, B. L. et al. Diving behaviour of dugongs, Dugong dugon. J. Exp. Mar. Bio. Ecol. 304, 203–224 (2004).
Domning, D. R. A phylogenetic analysis of the Sirenia. Proc. San Diego Soc. Nat. Hist. 29, 177–189 (1994).
Cantanhede, A. M. A. M. et al. Phylogeography and population genetics of the endangered Amazonian manatee, Trichechus inunguis Natterer, 1883 (Mammalia, Sirenia). Mol. Ecol. 14, 401–413 (2005).
Domning, D. P. Sirenians, seagrasses, and Cenozoic ecological change in the Caribbean. Palaeogeogr. Palaeoclimatol. Palaeoecol. 166, 27–50 (2001).
Diagne, L. W. K. Phylogenetics and feeding ecology of the African manatee, Trichechus senegalensis (2014).
Laudisoit, A. et al. West African Manatee Trichechus senegalensis (LINK, 1795) in the Estuary of the Congo River (Democratic Republic of the Congo): review and update. J. Biodivers. Endanger. Species 5, 181 (2017).
Vilaça, S. T. & Santos, F. R. Complete mitochondrial genome of the Florida manatee (Trichechus manatus latirotris, Sirenia). Genet. Mol. Biol. 42, e20190210 (2019).
Li, Y. et al. The molecular evolutionary dynamics of oxidative phosphorylation (OXPHOS) genes in Hymenoptera. BMC Evol. Biol. 17, 269 (2017).
Havird, J. C., Trapp, P., Miller, C. M., Bazos, I. & Sloan, D. B. Causes and consequences of rapidly evolving mtDNA in a plant lineage. Genome Biol. Evol. 9, 323 (2017).
Havird, J. C., Whitehill, N. S., Snow, C. D. & Sloan, D. B. Conservative and compensatory evolution in oxidative phosphorylation complexes of angiosperms with highly divergent rates of mitochondrial genome evolution. Evolution (N.Y.) 69, 3069–3081 (2015).
Ballard, J. W. O. & Kreitman, M. Is mitochondrial DNA a strictly neutral marker?. Trends Ecol. Evol. 10, 485–488 (1995).
Tomasco, I. H. & Lessa, E. P. The evolution of mitochondrial genomes in subterranean caviomorph rodents: adaptation against a background of purifying selection. Mol. Phylogenet. Evol. 61, 64–70 (2011).
Consuegra, S., John, E., Verspoor, E. & de Leaniz, C. G. Patterns of natural selection acting on the mitochondrial genome of a locally adapted fish species. Genet. Sel. Evol. 47, 58 (2015).
Romero, P. E., Weigand, A. M. & Pfenninger, M. Positive selection on panpulmonate mitogenomes provide new clues on adaptations to terrestrial life. BMC Evol. Biol. 16, 164 (2016).
Tian, R. et al. Adaptive evolution of energy metabolism-related genes in hypoxia-tolerant mammals. Front. Genet. 8, 205 (2017).
Uhen, M. D. Evolution of marine mammals: Back to the sea after 300 million years. Anat. Rec. 290, 514–522 (2007).
Foote, A. D. et al. Positive selection on the killer whale mitogenome. Biol. Lett. 7, 116–118 (2011).
Da Fonseca, R. R., Johnson, W. E., O’Brien, S. J., Ramos, M. J. & Antunes, A. The adaptive evolution of the mammalian mitochondrial genome. BMC Genom. 9, 119 (2008).
Arnason, U. et al. Mitogenomic relationships of placental mammals and molecular estimates of their divergences. Gene 421, 37–51 (2008).
Souza, E. M. S. de & Freitas, L. Extract_frag_scaffolds. 1 (2019).
Allio, R. et al. MitoFinder: efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. bioRxiv 685412 (2019).
Tillich, M. et al. GeSeq–versatile and accurate annotation of organelle genomes. Nucl. Acids Res. 45, W6–W11 (2017).
Prada, C. F. & Boore, J. L. Gene annotation errors are common in the mammalian mitochondrial genomes database. BMC Genom. 20, 73 (2019).
Bernt, M. et al. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69, 313–319 (2013).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T. & Calcott, B. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773 (2017).
Stamatakis, A. Phylogenetic Models of Rate Heterogeneity : A High Performance Computing Perspective (2006).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003).
Bouckaert, R. et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 (2014).
Rambaut, A., Suchard, M. A., Xie, D. & Drummond, A. J. Tracer v1. 6. 2014. (2015).
Rambaut, A. & Drummond, A. J. LogCombiner v1. 8.2. (2015).
Rambaut, A. & Drummond, A. J. TreeAnnotator v1. 7.0. Available as part BEAST Packag. http//beast.bio.ed.ac.uk (2013).
Rambaut, A. & Drummond, A. J. FigTree version 1.4. 0. available tree. bio. ed. ac. uk/software/figtree (2012).
Zhang, J., Nielsen, R. & Yang, Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22, 2472–2479 (2005).
Davydov, I. I., Salamin, N. & Robinson-Rechavi, M. Large-scale comparative analysis of codon models accounting for protein and nucleotide selection. Mol. Biol. Evol. 36, 1316–1332 (2019).
Scheffler, K., Martin, D. P. & Seoighe, C. Robust inference of positive selection from recombining coding sequences. Bioinformatics 22, 2493–2499 (2006).
Rubinstein, N. D., Doron-Faigenboim, A., Mayrose, I. & Pupko, T. Evolutionary models accounting for layers of selection in protein-coding genes and their impact on the inference of positive selection. Mol. Biol. Evol. 28, 3297–3308 (2011).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucl. Acids Res. 46, W296–W303 (2018).
Mitchell, A. L. et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucl. Acids Res. 47, D351–D360 (2019).
Vianna, J. A. et al. Phylogeography, phylogeny and hybridization in trichechid sirenians: implications for manatee conservation. Mol. Ecol. 15, 433–447 (2006).
Beatty, B. L., Vitkovski, T., Lambert, O. & Macrini, T. E. Osteological associations with unique tooth development in manatees (Trichechidae, Sirenia): a detailed look at modern Trichechus and a review of the fossil record. Anat. Rec. Adv. Integr. Anat. Evol. Biol. 295, 1504–1512 (2012).
Albert, J. S., Val, P. & Hoorn, C. The changing course of the Amazon River in the Neogene: center stage for Neotropical diversification. Neotrop. Ichthyol. 16, e180033 (2018).
Hoorn, C. et al. Amazonia through time: andean. Science 330, 927–931 (2010).
Figueiredo, J., Hoorn, C., Van der Ven, P. & Soares, E. Late Miocene onset of the Amazon River and the Amazon deep-sea fan: evidence from the Foz do Amazonas Basin. Geology 37, 619–622 (2009).
Hoorn, C. et al. Amazonia through time: Andean uplift, climate change, landscape evolution, and biodiversity. Science 330, 927–931 (2010).
Ribas, C. C., Aleixo, A., Nogueira, A. C. R., Miyaki, C. Y. & Cracraft, J. A palaeobiogeographic model for biotic diversification within Amazonia over the past three million years. Proc. R. Soc. B Biol. Sci. 279, 681–689 (2012).
Ribas, C. C. et al. Biogeography and diversification of Rhegmatorhina (Aves: Thamnophilidae): implications for the evolution of Amazonian landscapes during the Quaternary. J. Biogeogr. 45, 917–928 (2018).
Alfaro, J. W. L. et al. Biogeography of squirrel monkeys (genus Saimiri): South-central Amazon origin and rapid pan-Amazonian diversification of a lowland primate. Mol. Phylogenet. Evol. 82, 436–454 (2015).
Alfaro, M. E. et al. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc. Natl. Acad. Sci. 106, 13410–13414 (2009).
Jaramillo-Giraldo, C., Soares Filho, B., Ribeiro, S. M. C. & Gonçalves, R. C. Is it possible to make rubber extraction ecologically and economically viable in the Amazon? The Southern Acre and Chico Mendes Reserve case study. Ecol. Econ. 134, 186–197 (2017).
Hoorn, B. C. & Flantua, S. An early start for the Panama land bridge. Science https://doi.org/10.1126/science.aab0099 (2015).
Perini, F. A., Nascimento, E. R. & Cozzuol, M. A. A new species of Trichechus Linnaeus, 1758 (Sirenia, Trichechidae), from the upper Pleistocene of southwestern Amazonia, and the evolution of Amazonian manatees. J. Vertebr. Paleontol. 39, e1697882 (2020).
Benoit, J. et al. Cranial remain from Tunisia provides new clues for the origin and evolution of Sirenia (Mammalia, Afrotheria) in Africa. PLoS ONE 8, e54307 (2013).
Tomasco, I. H. & Lessa, E. P. Two mitochondrial genes under episodic positive selection in subterranean octodontoid rodents. Gene 534, 371–378 (2014).
Caballero, S., Duchene, S., Garavito, M. F., Slikas, B. & Baker, C. S. Initial evidence for adaptive selection on the NADH subunit two of freshwater dolphins by analyses of mitochondrial genomes. PLoS ONE 10, e0123543 (2015).
Slimen, H. B., Schaschl, H., Knauer, F. & Suchentrunk, F. Selection on the mitochondrial ATP synthase 6 and the NADH dehydrogenase 2 genes in hares (Lepus capensis L., 1758) from a steep ecological gradient in North Africa. BMC Evol. Biol. 17, 46 (2017).
Walker, J. E. The NADH: ubiquinone oxidoreductase (complex I) of respiratory chains. Q. Rev. Biophys. 25, 253–324 (1992).
Efremov, R. G. & Sazanov, L. A. Structure of the membrane domain of respiratory complex I. Nature 476, 414–420 (2011).
Sazanov, L. A. A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat. Rev. Mol. Cell Biol. 16, 375–388 (2015).
Antoine, P.-O. et al. A 60-million-year Cenozoic history of western Amazonian ecosystems in Contamana, Eastern Peru. Gondwana Res. 31, 30–59 (2016).
Gheerbrant, E., Amaghzaz, M., Bouya, B., Goussard, F. & Letenneur, C. Ocepeia (Middle Paleocene of Morocco): the oldest skull of an afrotherian mammal. PLoS ONE 9, e89739 (2014).
Benoit, J., Orliac, M. & Tabuce, R. The petrosal of the earliest elephant-shrew Chambius (Macroscelidea: Afrotheria) from the Eocene of Djebel Chambi (Tunisia) and the evolution of middle and inner ear of elephant-shrews. J. Syst. Palaeontol. 11, 907–923 (2013).
Gheerbrant, E., Sudre, J. & Cappetta, H. A Palaeocene proboscidean from Morocco. Nature 383, 68–70 (1996).
Savage, R. J. G., Domning, D. P. & Thewissen, J. G. M. Fossil Sirenia of the West Atlantic and Caribbean region. V. The most primitive known sirenian, Prorastomus sirenoides Owen, 1855. J. Vertebr. Paleontol. 14, 427–449 (1994).
Domning, D. P. The earliest known fully quadrupedal sirenian. Nature 413, 625–627 (2001).
Shoshani, J. et al. A proboscidean from the late Oligocene of Eritrea, a “missing link” between early Elephantiformes and Elephantimorpha, and biogeographic implications. Proc. Natl. Acad. Sci. 103, 17296–17301 (2006).
de Paula Couto, C. Mamíferos fósseis de cenozóico de Amazônia (Boletim do Conselho Nacional de Pesquisa, 1956).
Acknowledgements
This study is part of EMSS’s PhD research at the Universidade Estadual de Campinas (UNICAMP). This work was supported by the São Paulo Research Foundation (FAPESP 2015/18269-1), and a scholarship granted to EMSS by CAPES (PROEX-IS/88882.329516/2019-01). AL and EV thank the ICCN for providing CITES certificate 6543 export of the T. senegalensis sample we used, originating from the Park Marin de Mangroves of the DR Congo. Also, we want to thank the Congolese Institute for Nature Conservation (ICCN), and Mr Marcel Collet, Mangrove Marine Park Director, for their administrative, scientific, logistical and financial support in the DRCongo.
Author information
Authors and Affiliations
Contributions
É.M.S.S. and E.K.S.R. extracted the genomic DNA; É.M.S.S., M.F.N., E.V., A.L., D.P.D. and M.M. wrote the manuscript; É.M.S.S., E.K.S.R, L.F., G.S.V., M.C.R.R., F.A.S. participated in data analyses; M.M.; F.S., A.L., E.V. collected and shared the samples; É.M.S.S. and M.F.N. designed the project; All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
de Souza, É.M.S., Freitas, L., da Silva Ramos, E.K. et al. The evolutionary history of manatees told by their mitogenomes. Sci Rep 11, 3564 (2021). https://doi.org/10.1038/s41598-021-82390-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-82390-2
This article is cited by
-
Evolutionary Dynamics of American Manatee Species on the Northern Coast of South America: Origins and Maintenance of an Interspecific Hybrid Zone
Evolutionary Biology (2024)
-
Ancestral chromosomal signatures of Paenungulata (Afroteria) reveal the karyotype of Amazonian manatee (Trichechus inunguis, Sirenia: Trichechidae) as the oldest among American manatees
BMC Genomics (2023)
-
Molecular Footprints on Osmoregulation-Related Genes Associated with Freshwater Colonization by Cetaceans and Sirenians
Journal of Molecular Evolution (2023)
-
Signatures of positive selection in the mitochondrial genome of neotropical freshwater stingrays provide clues about the transition from saltwater to freshwater environment
Molecular Genetics and Genomics (2023)
-
Positive selection over the mitochondrial genome and its role in the diversification of gentoo penguins in response to adaptation in isolation
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.