Abstract
WNT ligands can activate several signalling cascades of pivotal importance during development and regenerative processes. Their de-regulation has been associated with the onset of different diseases. Here we investigated the role of the WNT/Calcium Calmodulin Kinase II (CaMKII) pathway in osteoarthritis. We identified Heme Oxygenase I (HMOX1) and Sox-9 as specific markers of the WNT/CaMKII signalling in articular chondrocytes through a microarray analysis. We showed that the expression of the activated form of CaMKII, phospho-CaMKII, was increased in human and murine osteoarthritis and the expression of HMOX1 was accordingly reduced, demonstrating the activation of the pathway during disease progression. To elucidate its function, we administered the CaMKII inhibitor KN93 to mice in which osteoarthritis was induced by resection of the anterior horn of the medial meniscus and of the medial collateral ligament in the knee joint. Pharmacological blockade of CaMKII exacerbated cartilage damage and bone remodelling. Finally, we showed that CaMKII inhibition in articular chondrocytes upregulated the expression of matrix remodelling enzymes alone and in combination with Interleukin 1. These results suggest an important homeostatic role of the WNT/CaMKII signalling in osteoarthritis which could be exploited in the future for therapeutic purposes.
Similar content being viewed by others
Introduction
Osteoarthritis (OA) is a joint disease driven by pathological biomechanics and characterized by cartilage breakdown, subchondral bone sclerosis, osteophytosis, and inconstant low degree inflammation1. OA is a leading cause of chronic disability worldwide: patients suffer from joint pain and impaired motility, which reduces their independence2. No therapeutic treatment other than symptomatic relief is currently available, leaving joint replacement surgery as the ultimate option.
OA progression is driven by an imbalance between cartilage degradation and reparative, homeostatic responses. Pathological biomechanical stimuli activate signalling pathways including interleukin 1 beta (IL1B)3 and the WNT pathway4, which in turn upregulate matrix metalloproteases (MMPs) and A Disintegrin and Metalloproteinase with Thrombospondin motifs (ADAMTS) aggrecanases, which contribute to the degradation of the cartilage extracellular matrix5.
Excessive activation of the WNT signalling pathway has been linked to the onset and progression of cartilage degeneration in OA, both in human and in mice6,7,8,9. WNTs are a family of 19 highly conserved morphogens capable of activating multiple signalling cascades. The most characterised one is the WNT beta-catenin-dependent pathway. The WNT/beta-catenin pathway (also known as canonical WNT pathway) is activated by the interaction of WNT ligands with Frizzled (FZDs) and Lipoprotein-related Protein Receptor 5 and 6 (LRP5/6) which leads to the intracellular accumulation of beta-catenin and its translocation to the nucleus. Within the nucleus, beta-catenin interacts with T-cell factor/lymphoid enhancer-binding factor (TCF/LEF) transcription factors promoting the transcription of target genes such as AXIN210. Other, so called “non-canonical” WNT pathways, are activated by the interaction of WNT ligands with FZDs and other co-receptors such as Receptor Tyrosine Kinase (RYK) and Retinoic Acid-Related Orphan Receptor (RORs), driving the activation of downstream signalling cascades activated by intracellular calcium release and/or associated with intercellular communication to coordinate polarity between adjacent cells11.
Ca2+Calmodulin-dependent Kinase II (CaMKII) is an intracellular kinase that can be activated by several stimuli including WNTs12 and inflammatory stimuli13,14. CaMKII is a Ser/Thr kinase expressed in mammals in 4 isoforms, alpha, beta, gamma and delta15. Binding of Ca2+/Calmodulin to CaMKII transiently activates it. Once activated, CaMKII phosphorylates its substrates, including itself. The autophosphorylation of CaMKII in T287 results in its constitutive activation, even in the absence of further stimulation16. WNT-induced CaMKII activity regulates chondrocyte differentiation12.
We showed that a single WNT ligand could simultaneously activate the beta-catenin-dependent and the CaMKII-dependent WNT pathways with distinct biological outcomes12.
These data suggest an important and still uncharacterised role for the WNT/CaMKII pathway in the maintenance of cartilage homeostasis. To test this hypothesis, the current study investigates the role of the WNT/CaMKII pathway in the context of osteoarthritis.
Methods
Patients
Preserved (Mankin score ≤ 417) human articular cartilage samples were obtained from the femoral condyles removed during total knee replacement for osteoarthritis following informed consent (4 male and 2 female patients aged between 51 and 76). All procedures were approved by the East London and The City Research Ethics Committee 3 (ethics approval REC N. 07/Q0605/29). Full thickness explants were prepared for histology, and chondrocytes were isolated and expanded in-vitro as previously described12. Normal cartilage samples were retrieved from 3 patients (one 58 years-old, and two males aged respectively 14 and 16) undergone lower limb amputation because of malignant bone tumours. The samples were obtained upon favourable opinion of the Ethics Committee of the Ärztekammer Schleswig-Holstein, Bad Segeberg, Germany. We performed all procedures in accordance to relevant guidelines and regulations.
Primary cells, cell lines and tissues
Bovine chondrocytes were isolated from metatarsal joints obtained from a local abattoir within 24 h from death. Experiments were performed when primary chondrocytes reached 80% confluence (passage 0, P0) as previously described18. C3H/10T1/2 cells (ATCC) were cultured according to the manufacturer’s instructions.
Murine femoral heads and patellae were explanted from 1 month-old male 129/Sv mice and processed for gene expression analysis (see below).
If not differently indicated, cells were stimulated in complete medium (DMEM/F-12 1:1 plus GlutaMax, 10% FBS, 1 mM sodium pyruvate, and 2% antibiotic antimycotic solution ThermoFisher Scientific) supplemented with human recombinant WNT3A (100 ng/ml; R&D), DKK1 (100 ng/ml, R&D), IL1B (20 ng/ml; InvivoGen), KN93 or its inactive control KN92 (10 μM; EMD Millipore), Autocamtide-2 related inhibitory peptide II, AIP, 5 μM; Calbiochem) or respective vehicles as indicated.
siRNA preparation and transfection
Small interfering RNA for mouse CAMKIIγ was prepared using the Silencer siRNA Construction Kit (Life Technologies) following manufacturer’s instructions. siCaMKIIγ S: AAGGGTTTAAGGGTTTCACTACCTCGTGTCCTGTCTC; AS: AAACACGAAAACACGAGGTAGTGAAACCCCCTGTCTC; scramble siRNA, S: AATTCCGCAATTCCGCTCTGGACTGTAGTCCTGTCTC; AS: AAGAACTTAAGAACTTGATCAACCAGATGCCTGTCTC. The siRNA were used at 20 nM and were transfected in C3H/10T1/2 cells using JetPRIME transfection reagent (Polyplus) as described in18.
RNA isolation and quantitative real time PCR
RNA isolation and PCR were performed as previously described12. For a complete list of primers and conditions refers to Supplementary Table 1.
Microarray analysis
Total RNA was quantified using a Nanodrop ND-1000 Spectrophotometer and assessed for quality (QC) using the Agilent 2100 Bioanalyzer Pico kit. Five nanograms of total RNA with RNA integrity numbers (RINs) higher than 9 were selected for the microarray study. A Poly-A Spiking Control (Life Technologies) was used as amplification control.
Single primer isothermal amplification (SPIA) technology was used to generate cDNA using NuGen Ovation Pico WTA System V2 kit, following manufacturer's instructions. The SPIA cDNA was subjected to QC, fragmented and biotin-labelled using the NuGen Encore Biotin Module according to the manufacturer's instructions. The processed cDNA was subjected to a further round of QC to assess fragmentation size (Agilent 2100 Bioanalyzer Nano kit; fragment size < 200 nt).
Hybridization cocktails were prepared according to Nugen's recommendations for Human Gene 2.0 ST arrays and hybridization took place at 45 °C for 16–20 h in an Affymetrix Genechip Hybridization oven 645.
The arrays were washed and stained using wash protocol FS450_0002 as recommended by Affymetrix on the GeneChip Fluidics station 450. The arrays were scanned using the Affymetrix GeneChip Scanner. CEL files were QC checked using the Expression Console software package (Affymetrix, Thermo Fisher) by using standard metrics and guidelines for the Affymetrix microarray system. Data were normalised together using the Robust Multi-array Average (RMA) sketch algorithm.
The processing of all sample files was performed using R Programming Language (R). In brief, raw data were inputted using the 'read.celfiles' function of the 'oligo' R package19, with data then undergoing Robust Multiarray Average (RMA) normalisation using the 'rma' function. For cross-sample comparisons, the study design model was first created by identifying sample groupings based on KN92, KN92-WNT3a, and KN93-WNT3a status.
Raw and normalized microarray data were submitted to the Gene Expression Omnibus database at the National Center for Biotechnology Information (accession number GSE162076).
Immunohistochemical analysis
CaMKII isoforms and phosphoCaMKII (pCaMKII) were detected by indirect immunofluorescence as described in12. For a list of retrieval methods, antibodies and dilutions please refer to Supplementary Table 2. All images were acquired using identical settings on an Olympus BX61 microscope and consistently modified for best rendering using Adobe Photoshop. The fluorescent and dapi images of the same field were superimposed, or the fluorescent and Nomarski images of the same field were inverted and superimposed (subtraction).
Instability-induced OA in mice
All animal procedures were approved by the UK Home Office (PPL 70-7986). All the experimental procedures were performed in accordance to relevant guidelines and regulations. The study was carried out in compliance with ARRIVE guidelines. Mice were kept in an approved animal care facility, were housed 4 or 5 per cage in standard cages and fed ad libitum. Ten week old, male C57BL/6 mice (Charles River) were subjected to resection of the medial collateral ligament and of the anterior horn of the medial meniscus (menisco-ligament injury—MLI) as previously described20. In each cage, the animals were randomized to receive either PBS or KN93 (see Supplementary Table 3). The contralateral knee underwent sham surgery (arthrotomy but no damage to ligaments or menisci). The joint capsule was sutured with Vicryl 6-0 and the skin with Ethylon 5-0 sutures with atraumatic needles (Ethicon).
Pharmacological blockade of CaMKII
Four weeks after surgery the mice were randomized to receive either the water-soluble version of the CaMKII inhibitor KN93 or PBS as control (15 animals per group). For the first 3 days KN93 (10 µmol/kg/day) or PBS were administered by intraperitoneal injection. Subsequently, ALZET osmotic minipumps (Charles River Laboratories) were inserted subcutaneously on the back of the mice allowing a continuous release of KN93 (5 µmol/kg/day) or PBS for additional 28 days. The pumps were changed once during this time. The animals were then killed by cervical dislocation.
Histology and OA scoring
Knee joints were dissected, the majority of the soft tissue was removed, and the joints were fixed in 70% ethanol following which they were decalcified in formic acid, embedded in paraffin and sectioned as described in7. The sections were stained with Safranin O (SO) (0.1%, pH 4). Images were taken using the same settings on an Olympus BX61 microscope and consistently modified for best rendering using GIMP software. The extent of cartilage degradation was scored by two blinded, independent investigators using the Osteoarthritis Research Society International (OARSI) scoring system21. At least 5 sections/knee were scored.
Histomorphometry
Proteoglycan loss, osteophyte size and differentiation, and subchondral bone thickness were measured by histomorphometry using ImageJ22. The mean values from at least 3, but on average 5 sections from each knee (1 section every 50 µm, spanning the entire depth of the joint) were calculated and used for statistical analysis.
Proteoglycan loss was analysed by densitometry of the articular cartilage as previously described7. In brief, images were rotated so the cartilage-bone junction in the middle of the plateau was horizontal. This is important so that non‐cartilage staining in the bone marrow spaces can be eliminated. The tibial plateau was selected to include the growth plate, any osteophytes and up to the end of the cartilage; and selections from every section were placed on one canvas, so they were perfectly aligned horizontally. To isolate the metachromatic Safranin O staining from the background and most of the orthochromatic staining (subchondral bone), the RGB image was transformed into an HSB stack. On the ‘Saturation’ slice, a rectangle 600 microns wide and the full height of the canvas was placed over the first section to include the tibial plateau, making sure to exclude any osteophytes and the “bulgy” part near the intercondylar notch. Ctrl + 1 was pressed, and then the rectangle was moved laterally to the next section and Ctrl + 2 was pressed. This was repeated until all sections were selected and then Ctrl + 3 plotted the density profile of each section. The first peak is the articular cartilage. A horizontal line was drawn to cut residual background if any, and a lateral line to limit the articular cartilage peak and eliminate any staining of bone marrow spaces. Using the wand tool to select the inside of the density profile returned the area. These results were copied from the measurements box to a statistics programme for further analysis.
Osteophytes consistently developed only on the medial tibial compartment of the operated joints. On individual sections, osteophyte size was determined by selecting the osteophyte with the polygon tool, and ensuring a global scale had been set, measuring the area. Osteophyte differentiation was assessed by comparing the intensity of SO staining with the staining of the growth plate, which was set as the threshold for comparison in Image J and defined as the staining for fully differentiated cartilage. The percentage of the thresholded area in each individual section was then normalised for the area of the osteophyte. The points in the graph represent the average of the measurements for multiple sections (at least 3/mouse).
Subchondral bone thickness was determined by selecting the subchondral area underneath the load-bearing area of the articular cartilage of the tibiae as described by Botter et al.23.
Cell density
The number of cells was counted within a defined region of the articular cartilage of the tibia and the femur of the operated joint of 5 mice (1 slide/mouse) either treated with PBS or KN93. Cell density was measured as number of cells/mm2.
Micro-CT scanning
Tibiae were fixed in 70% ethanol and scanned with Skyscan 1174 Micro-CT scanner (Skyscan, Antwerp, Belgium) at 53 kV, 785 µA, 0.7° rotation and 12.57 µM isometric voxel resolution. The individual slices were reconstructed using NRecon software (Skyscan) with an algorithm that included maximum ring artefact correction, 70% beam-hardening correction and misalignment compensation.
Micro-CT analysis, Tibiae epiphyseal volume measurement
The reconstructed scan was rotated and resliced using ImageJ22 in a coronal orientation. Using CTAn software (Skyscan), the tibial epiphysis was reconstructed as a volume of interest (VOI) by manually selecting the epiphysis on every 10th slice for the entire stack of images. A threshold of 90 density units was set to distinguish mineralised from non-mineralised tissue within the trabeculae.
Statistical analysis
Parametric data were compared with the t-test or Anova with Tukey-Post test for multiple comparisons. When possible, data transformation was applied to satisfy the assumptions of parametric tests as described24. Non-parametric data were analysed using the Wilcoxon–Mann–Whitney test. p values < 0.05 were considered significant: *p < 0.05; **p < 0.01; ***p < 0.001. For the analysis of the microarray experiment, individual comparisons within the study design model were conducted by fitting a linear model independently for each probe, with group as the y variable, using 'lmfit' ('limma' R package)25. The linear fit for each comparison was subsequently modified using the empirical Bayes ('eBayes') approach, which aims to bring the probe-wise variances across samples to common values, resulting in modified t-statistics, F-statistic, and log odds differential expression ratios. Finally, for each comparison, log2 fold-change (logFC), P value, and corrected P value (false discovery rate, FDR) was output. A cut-off of 10% FDR was used. Volcano plots were generated using ‘EnhancedVolcano’ (‘EnhancedVolcano’ R package)26.
Results
Transcriptional signature of the WNT3A/CaMKII pathway in the articular chondrocytes
We reported that WNT3A can activate both the WNT/beta-catenin and the WNT/CaMKII pathways in articular chondrocytes with distinct, dose-dependent biological outcomes12. This implies that the modulation of some transcriptional targets of WNT3A will be due to the activation of the WNT/beta-catenin-dependent pathway and others due to the activation of the CaMKII pathway. To elucidate the specific transcriptional signature of the CaMKII pathway activation by WNT3A, we performed an expression microarray analysis of human articular chondrocytes treated with WNT3A in presence of KN93 or of its inactive control compound KN92 (Fig. 1a–c). We proposed that genes that are activated in a WNT/CaMKII-dependent manner will be upregulated by WNT3A, but only in the absence of the CaMKII inhibitor KN93.
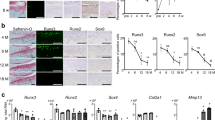
HMOX1 is a selective target of the WNT/CaMKII pathway in the articular chondrocytes. (a–d) Human articular chondrocytes isolated from OA patients were treated with human recombinant WNT3A in presence of the CaMKII inhibitor KN93 or its inactive control molecule KN92. (a) Volcano plot showing genes regulated by WNT3A (KN92 + WNT3A vs KN92), (b) genes modulated by CaMKII inhibition in the presence of WNT3A (KN93 + WNT3A vs KN92 + WNT3A) and (c) genes activated by WNT3A independently of CaMKII (KN93 + WNT3A vs KN92). (d) Venn diagram showing the number of significantly modulated genes in each group. (e) qPCR confirming downregulation of HMOX1 in response to WNT3A but not in the presence of the CaMKII inhibitor KN93. Microarray data were obtained by testing samples from 4 human donors. For 2 donors all conditions were tested in two independent cartilage explants; for one donor the condition KN92 + WNT3A was tested in a single explant; for the remaining donor all the conditions were tested in a single explant per condition. (f) qPCR showing that HMOX1 is a selective target of the WNT/CaMKII branch and it is not modulated via activation of the WNT/beta-catenin pathway. n = 3 human donors, 2 technical replicates per donor. (a–d) data were analysed with a linear models with 10% FDR p value correction. (e,f) Data were analysed with one-way ANOVA followed by TukeyHSD post-hoc test.
This analysis revealed that 44 genes were modulated by WNT3A in the absence of CaMKII inhibitor (Fig. 1d), but only 3 were modulated by WNT3A in a CaMKII-dependent manner (Fig. 1d). The 3 genes modulated by WNT3A in a CaMKII-dependent manner included SRY-Box Transcription Factor 9 (SOX9) (upregulated), the uncharacterized transcript RP11-474G23.3 (downregulated) and Heme-Oxygenase 1 (HMOX1) (downregulated) mRNA. The upregulation of SOX9 confirmed our previous results12. The downregulation of HMOX1 mRNA in response to WNT/CaMKII activation was confirmed at qPCR level in human articular chondrocytes (Fig. 1e). Consistent with the requirement for CaMKII activation, KN93 effectively rescued the downregulation of HMOX1 induced by WNT-3A and significantly upregulated its expression of this gene without exogenous WNT3A.
We previously showed that the WNT/beta-catenin pathway antagonizes the WNT/CaMKII pathway in chondrocytes and that, therefore, its suppression using DKK1 results in hyper-activation of the CaMKII-dependent targets12. In keeping with this model, treatment of chondrocytes with DKK1 alone decreased the expression of HMOX1 (Fig. 1f) and did not rescue the downregulation of HMOX1 induced by WNT3A, confirming that HMOX1 is a negative target of the WNT3A/CaMKII pathway but not of the beta-catenin-dependent WNT pathway (Fig. 1f).
Activated CaMKII and HMOX1 expression are inversely correlated in osteoarthritis
We next sought to confirm our in vitro findings in human cartilage and human OA pathology. We mined transcriptomics data in publicly available OA cartilage databases27. CaMKIIγ and –δ RNA were the most expressed CaMKII isoforms at mRNA levels in normal as well as in osteoarthritic cartilage (Fig. 2c). A similar expression pattern was detected in a previously reported expression microarray in human cartilage explants6 (Fig. 2a) and in cultured human chondrocytes through in vitro culture (Fig. 2b). Confirming a previous report28, CaMKII phosphorylation was increased in osteoarthritic cartilage as assessed with immunostaining with an antibody directed to the T287 residue (Fig. 2d). In keeping with our in vitro data showing that HMOX1 is a negative CaMKII target, HMOX1 expression decreased in cartilage isolated from OA patients (Fig. 2e). These results therefore suggest that the WNT/CaMKII pathway is activated in osteoarthritic cartilage. Whilst our data show a correlation between CaMKII activation and HMOX1 downregulation, they do not establish causality in vivo and other factors may contribute to HMOX1 suppression in OA.
CaMKII and HMOX1 expression are oppositely modulated in osteoarthritis. (a) Relative expression of the four CaMKII mRNA isoforms as assessed by expression microarrays in human cartilage explants6. (b) Gene expression analysis by semiquantitative PCR for CaMKIIα, -β, -γ, -δ, in P0 human articular chondrocytes (upper row). A pool of cDNA from brain, placenta and lymph node was used as positive control for the PCR (lower row) (n cycles = 40). All the samples were loaded and run in the same gel whose image was cut for best rendering. A picture of the full original gel can be found in Supplementary Figure 3. (c) RNA expression of CaMKII isoforms in human cartilage retrieved from normal and OA human cartilage as assessed by Soul and colleagues in a previously performed RNAseq27. (d) Immunodetection of phospho-CaMKII in normal and OA articular cartilage. Image representative of n = 3. (e) RNA deep sequencing analysis of HMOX1 expression in normal and OA articular cartilage.
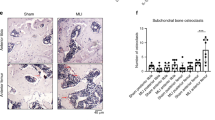
CaMKIIγ and –δ were the most expressed isoforms in cartilage isolated from murine femoral heads and patellae (Fig. 3a,b), as previously seen in human cartilage. However, the different CaMKII isoforms displayed a tissue specific-expression pattern in the murine joint. CaMKIIγ was expressed in the most superficial layer of the articular cartilage, with some limited expression in the menisci (Fig. 3h–j). CaMKIIβ and –δ were also detectable in the surrounding soft tissues and in the bone marrow (Fig. 3c–g and k–o). To further validate a direct dependence of HMOX1 downstream the activation of CaMKII, we silenced CaMKIIγ in the murine fibroblast cell line C3H/10T1/2 by siRNA (Fig. 3p,q). Silencing CaMKIIγ did not result in compensation from the other isoforms (Fig. 3p) and was sufficient to significantly upregulate HMOX1 mRNA in these cells (Fig. 3q).
Expression of CaMKII isoforms in murine cartilage. (a,b) qPCR analysis of murine femoral heads and patellae removed from 1 month-old 129/Sv male mice (n = 4). Gene expression was normalized for the housekeeping gene beta-actin. (c–o) Immunofluorescence for CaMKII-β, -γ and δ in the murine joint. (p) mRNA expression of the CaMKII isoforms after CaMKIIγ silencing by siRNA in C3H/10T1/2 cells. n = 4. (q) Downregulation of CaMKII by siRNA was sufficient to upregulate HMOX1 in C3H/10T1/2 cells n = 4. Data analysed by t-test. F = femur; T = tibia; M = meniscus; BM = bone marrow; CL = cruciate ligament; SM = synovial membrane.
Pharmacological inhibition of CaMKII promotes cartilage degeneration and subchondral bone remodelling in a murine model of OA
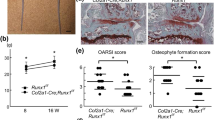
To investigate if CaMKII inhibition affects the outcome of OA, we induced OA in adult mice by meniscus-ligament injury (MLI)20,29 and 4 weeks later we inhibited CaMKII by systemic administration of the CaMKII inhibitor KN93 or PBS as control for additional 4 weeks (Fig. 4a). The weight and general health status of the animals were monitored throughout the entire length of the stimulation and no significant difference between vehicle-treated vs inhibitor-treated animals was noted (Supplementary Table 3). CaMKII phosphorylation was effectively inhibited in KN93-treated animals (Supplementary Figure 1). Animals receiving KN93 developed more severe OA, as quantified using the OARSI scoring system (Fig. 4b,c) and a corresponding decrease in proteoglycan content (Fig. 4d,e), measured by histomorphometry. A statistically significant decrease in cell density was detected in the articular cartilage of KN93-treated animals (Supplementary Figure 2).
Effect of CaMKII inhibition in vivo in a murine model of OA. (a) Osteoarthritis was induced by removal of the anterior horn of the medial meniscus and transection of the medial collateral ligament (MLI model). Four weeks after surgery, mice were administered KN93 or PBS vehicle subcutaneously via a minipump for additional 4 weeks (the initial minipump was replaced after 2 weeks). The total volume administered over 4 weeks was equal to 400 µl. n = 15/treatment. (b) Safranin O staining of representative sections of the medial compartment of the knee of mice treated with PBS or KN93. (c) OARSI score of the medial and the lateral compartments of the MLI knees n = 15/treatment. (d) Histomorphometry analysis of the MLI knees. (e) Correlation of the OARSI score and histomorphometry analysis. MF: medial femur; MT: medial tibia; LF: lateral femur; LT: lateral tibia. OARSI scores were analysed with Mann–Whitney test with Wilcoxon post-test. Histomorphometry data were analysed by t-test. Linear regression was used to compare the OARSI scores and histomorphometry data.
Bone changes were assessed by microCt. As expected, BV/TV of the entire tibial epiphysis increased in the MLI-operated compared to the sham-operated knee, but no difference was observed between treatment groups (Fig. 5a,b). Subchondral bone thickness, however, was significantly less increased in KN93-treated mice (Fig. 5c). Osteophyte size and bone vs cartilage ratio was not affected by KN93 treatment (Fig. 5c).
CaMKII inhibition decreases subchondral thickness. (a,b) MicroCt analysis of operated and contralateral sham-operated knee joints of mice treated with KN93 or PBS vehicle upon surgical destabilization of the knee joint. (a) Epiphyseal Bone volume/tissue volume of the entire tibial epiphysis of the operated knees. (b) Comparison of the percent change of the BV/TV between the MLI and the sham operated knee in the two treatment groups. (c) Histomorphometrical assessment of osteophyte area, % of cartilage area and histomorphometrical quantification of subchondral bone thickness. Data were analysed by t-test. n = 15/treatment.
CaMKII inhibition enhanced the capacity of IL1B to upregulate catabolic enzymes in articular chondrocytes
To gain insight into the mechanisms leading to increased cartilage degeneration in mice treated with KN93, we treated monolayer cultures of primary bovine articular chondrocytes with IL1B, a potent driver of extracellular matrix degradation5. CaMKII inhibition with KN93 (Fig. 6a,c,e,g) or AIP (Fig. 6b,d,f,h) induced the upregulation of the matrix-degrading enzymes MMP3 and ADAMTS5 alone and in synergy with IL1B.
CaMKII inhibition enhanced the capacity of IL1B to upregulate catabolic enzymes in the articular chondrocytes. (a–h) mRNA expression of ADAMTS4/5 and MMP3/13 measured by qPCR of bovine articular chondrocytes stimulated for 24 h with IL1B (20 ng/ml) in presence or absence of KN93 or its inactive analogue KN92 (both 10 µM) (a,c,e,g) or in the presence or absence of the CaMKII inhibitor AIP (b,d,f,h) (5 µM). Data were analysed with 2 ways ANOVA and Tukey post-test. n = 3.
Discussion
Deregulation of Wnt signalling is associated with OA in in patients30 and in animal models8,9,31.
We previously showed that WNT3A could simultaneously activate both the beta-catenin dependent and the CaMKII-dependent pathways in chondrocytes, resulting in distinct transcriptional and biological outcomes12.
In this study we established the transcriptional signature of WNT/CaMKII signalling and identified HMOX1 as a specific transcriptional target of this branch of the Wnt pathway (see schematic representation in Fig. 7). We showed that CaMKII phosphorylation was increased in human and murine OA, while the expression of HMOX1 was decreased. CaMKII blockade increased cartilage breakdown in an instability-induced OA model in mice. CaMKII inhibition increased the capacity of IL1B to upregulate catabolic enzymes in chondrocytes.
The microarray analysis also confirmed SOX9 mRNA upregulation in response to CaMKII blockade, as we previously showed by qPCR12. The mechanism by which CaMKII blockade results in SOX9 upregulation and, simultaneously, in upregulation of cartilage-degrading enzymes is unknown. It could be speculated that SOX9 upregulation occurs as a homeostatic response to the increased levels of extracellular matrix degradation. HMOX1 is a cytoprotective enzyme exerting antioxidant effects and chondroprotective effects in vitro32,33,34.
The three CAMKII isoforms expressed in the joint had distinct expression patterns. CaMKIIγ was selectively expressed at the articular interface in the cartilage and in the menisci, while -CaMKIIβ and CaMKIIδ had a broader expression pattern. Unfortunately, because of their high similarity across the phosphorylation site, no antibody specifically detecting the phosphorylated form of the different isoforms is available, making impossible to determine if only one isoform or all of them are activated in OA. Interestingly, however, we demonstrated that downregulation of CaMKIIγ alone is sufficient to mimic the effect of KN93 in inducing the upregulation of HMOX1, suggesting that this isoform might be downstream the activation of the WNT/CaMKII in the adult articular cartilage. The understanding of the differences in the activity of the four isoforms is going to be important for targeting CaMKII for therapeutic purposes.
In our in vivo experiments, CaMKII inhibition affected subchondral bone thickness. During embryonic development CaMKII is a modulator of chondrocyte hypertrophic differentiation during development35,36 and therefore we cannot exclude that altered skeletal morphogenesis may have also contributed to the worsening of the chondropathy.
CaMKII appears to modulate the final outcomes of major homeostatic effects in cartilage homeostasis in a highly context-dependent manner both embryonic development and in adulthood12,28,35,36. Saitta et al. also recently reported that CaMKII blockade reduced the capacity of bone morphogenetic proteins (BMPs) to upregulate cartilage phenotypic markers37. Calcification and PI turnover play a crucial role in OA progression38. It has been reported that the calcium channel TRPM7 links Ca2+ fluctuations to CaMKII activation and downstream signalling in growth plate chondrocytes during skeletal development39. It is possible that a similar mechanism may be responsible for CaMKII activation in OA articular cartilage. The identification of the calcium channels responsible for CaMKII activation in OA may offer a therapeutic opportunity. Therefore, understanding the biology of CaMKII activation is going to be key to successfully harness such pathways in musculoskeletal medicine.
References
Goldring, S. R. & Goldring, M. B. Clinical aspects, pathology and pathophysiology of osteoarthritis. J. Musculoskelet. Neuronal Interact. 6, 376–378 (2006).
Fernandes, G. S. & Valdes, A. M. Cardiovascular disease and osteoarthritis: Common pathways and patient outcomes. Eur. J. Clin. Investig. 45, 405–414 (2015).
van den Bosch, M. H. J., van Lent, P. L. E. M. & van der Kraan, P. M. Identifying effector molecules, cells, and cytokines of innate immunity in OA. Osteoarthr. Cartil. https://doi.org/10.1016/j.joca.2020.01.016 (2020).
Lories, R. J., Corr, M. & Lane, N. E. To Wnt or not to Wnt: The bone and joint health dilemma. Nat. Rev. Rheumatol. 9, 328–339 (2013).
Fernandes, J. C., Martel-Pelletier, J. & Pelletier, J.-P. The role of cytokines in osteoarthritis pathophysiology. Biorheology 39, 237–246 (2002).
Dell’accio, F., De Bari, C., Eltawil, N. M., Vanhummelen, P. & Pitzalis, C. Identification of the molecular response of articular cartilage to injury, by microarray screening: Wnt-16 expression and signaling after injury and in osteoarthritis. Arthritis Rheum. 58, 1410–1421 (2008).
Nalesso, G. et al. WNT16 antagonises excessive canonical WNT activation and protects cartilage in osteoarthritis. Ann. Rheum. Dis. https://doi.org/10.1136/annrheumdis-2015-208577 (2016).
Zhu, M. et al. Activation of beta-catenin signaling in articular chondrocytes leads to osteoarthritis-like phenotype in adult beta-catenin conditional activation mice. J. Bone Miner. Res. 24, 12–21 (2009).
Zhu, M. et al. Inhibition of beta-catenin signaling in articular chondrocytes results in articular cartilage destruction. Arthritis Rheum. 58, 2053–2064 (2008).
Wiese, K. E., Nusse, R. & van Amerongen, R. Wnt signalling: Conquering complexity. Development 145, dev165902 (2018).
van Amerongen, R. Alternative Wnt pathways and receptors. Cold Spring Harb. Perspect. Biol. 4, a007914–a007914 (2012).
Nalesso, G. et al. WNT-3A modulates articular chondrocyte phenotype by activating both canonical and noncanonical pathways. J. Cell Biol. 193, 551–564 (2011).
Wu, C. Y., Hsieh, H. L., Sun, C. C. & Yang, C. M. IL-1β induces MMP-9 expression via a Ca2+-dependent CaMKII/JNK/c-Jun cascade in rat brain astrocytes. Glia 57, 1775–1789 (2009).
Monnerat, G. et al. Macrophage-dependent IL-1β production induces cardiac arrhythmias in diabetic mice. Nat. Commun. 7, 1–5 (2016).
Griffith, L. C. Calcium/calmodulin-dependent protein kinase II: An unforgettable kinase. J. Neurosci. 24, 8391–8393 (2004).
Bangaru, M. L. Y. et al. Differential expression of CaMKII isoforms and overall kinase activity in rat dorsal root ganglia after injury. Neuroscience 300, 116–127 (2015).
Mankin, H. J., Dorfman, H., Lippiello, L. & Zarins, A. Biochemical and metabolic abnormalities in articular cartilage from osteo-arthritic human hips: II. Correlation of morphology with biochemical and metabolic data. J. Bone 53, 523–537 (1971).
Eldridge, S. et al. Agrin mediates chondrocyte homeostasis and requires both LRP4 and α-dystroglycan to enhance cartilage formation in vitro and in vivo. Ann. Rheum. Dis. https://doi.org/10.1136/annrheumdis-2015-207316 (2015).
Carvalho, B. S. & Irizarry, R. A. A framework for oligonucleotide microarray preprocessing. Bioinformatics 26, 2363–2367 (2010).
Hamada, D., Sampson, E. R., Maynard, R. D. & Zuscik, M. J. Surgical induction of posttraumatic osteoarthritis in the mouse. Methods Mol. Biol. 1130, 61–72 (2014).
Glasson, S. S., Chambers, M. G., van Den Berg, W. B. & Little, C. B. The OARSI histopathology initiative—Recommendations for histological assessments of osteoarthritis in the mouse. Osteoarthr. Cartil. 18(Suppl 3), S17–S23 (2010).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Botter, S. M. et al. ADAMTS5-/- mice have less subchondral bone changes after induction of osteoarthritis through surgical instability: Implications for a link between cartilage and subchondral bone changes. Osteoarthr. Cartil. 17, 636–645 (2009).
Chambers, J. M. Graphical Methods for Data Analysis (Wadsworth International Group, Belmont, 1983).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Blighe, K., Rana, S., Lewis, M. Bioconductor-EnhancedVolcano. https://bioconductor.org/packages/release/bioc/html/EnhancedVolcano.html.
Soul, J. et al. Stratification of knee osteoarthritis: Two major patient subgroups identified by genome-wide expression analysis of articular cartilage. Ann. Rheum. Dis. 77, 423–423 (2018).
Sugita, S. et al. Transcription factor Hes1 modulates osteoarthritis development in cooperation with calcium/calmodulin-dependent protein kinase 2. Proc. Natl. Acad. Sci. 112, 3080–3085 (2015).
Sampson, E. R. et al. Teriparatide as a chondroregenerative therapy for injury-induced osteoarthritis. Sci. Transl. Med. 3, 101ra93 (2011).
Loughlin, J. et al. Functional variants within the secreted frizzled-related protein 3 gene are associated with hip osteoarthritis in females. Proc. Natl. Acad. Sci USA. 101, 9757–9762 (2004).
Lories, R. J. et al. Articular cartilage and biomechanical properties of the long bones in Frzb-knockout mice 1. Arthritis Rheum. 56, 4095–4103 (2007).
Forootan, S. S. et al. Real-time in vivo imaging reveals localised Nrf2 stress responses associated with direct and metabolism-dependent drug toxicity. Sci. Rep. 7, 16084 (2017).
de An, Q. et al. Inhibition of bromodomain-containing protein 4 ameliorates oxidative stress–mediated apoptosis and cartilage matrix degeneration through activation of NF-E2–related factor 2-heme oxygenase-1 signaling in rat chondrocytes. J. Cell. Biochem. 119, 7719–7728 (2018).
Hu, B. et al. Heme oxygenase-1 attenuates IL-1β induced alteration of anabolic and catabolic activities in intervertebral disc degeneration. Sci. Rep. 6, 21190 (2016).
Taschner, M. J., Rafigh, M., Lampert, F., Schnaiter, S. & Hartmann, C. Ca2+/Calmodulin-dependent kinase II signaling causes skeletal overgrowth and premature chondrocyte maturation. Dev. Biol. 317, 132–146 (2008).
Li, Y., Ahrens, M. J., Wu, A., Liu, J. & Dudley, A. T. Calcium/calmodulin-dependent protein kinase II activity regulates the proliferative potential of growth plate chondrocytes. Development 138, 359–370 (2011).
Saitta, B., Elphingstone, J., Limfat, S., Shkhyan, R. & Evseenko, D. CaMKII inhibition in human primary and pluripotent stem cell-derived chondrocytes modulates effects of TGFβ and BMP through SMAD signaling. Osteoarthr. Cartil. https://doi.org/10.1016/j.joca.2018.08.017 (2018).
Bertrand, J. et al. Decreased levels of nucleotide pyrophosphatase phosphodiesterase 1 are associated with cartilage calcification in osteoarthritis and trigger osteoarthritic changes in mice. Ann. Rheum. Dis. 71, 1249–1253 (2012).
Qian, N. et al. TRPM7 channels mediate spontaneous Ca2+ fluctuations in growth plate chondrocytes that promote bone development. Sci. Sign. 12(576), eaaw4847 (2019).
Acknowledgements
We wish to thank Mr Pramod Achan, Barts and the London NHS Trust, for the provision of the human cartilage samples used to perform this study. This work has been supported by the following grants: Medical Research Council MR/S008608/1 and MR/K013076/1; FOREUM grant 1016807, Versus Arthritis grant 21621, Versus Arthritis grant 21515; William Harvey Research Foundation Grant dd060116.
Author information
Authors and Affiliations
Contributions
G.N. and F.D.A., conceived and designed the project, collected, analysed and interpret the data, obtained funds and wrote the manuscript; A.D.P., A.K., and K.P. collected and analysed the data. A.S.T., S.E.E. and B.L.T., helped interpreting the data and in revising the manuscript; K.B. and S.R. analysed the microarray data, BS prepared the murine samples for histological examination, J.B. and J.S. provided human and murine materials for the study, T.L.V., A.F. and C.P. provided logistic support and revised the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nalesso, G., Thorup, AS., Eldridge, S.E. et al. Calcium calmodulin kinase II activity is required for cartilage homeostasis in osteoarthritis. Sci Rep 11, 5682 (2021). https://doi.org/10.1038/s41598-021-82067-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-82067-w
This article is cited by
-
Re-thinking osteoarthritis pathogenesis: what can we learn (and what do we need to unlearn) from mouse models about the mechanisms involved in disease development
Arthritis Research & Therapy (2023)
-
CircRNA/lncRNA–miRNA–mRNA network and gene landscape in calcific aortic valve disease
BMC Genomics (2023)
-
Cartilage calcification in osteoarthritis: mechanisms and clinical relevance
Nature Reviews Rheumatology (2023)
-
Molecular Insights to the Wnt Signaling During Alzheimer’s Disorder: a Potential Target for Therapeutic Interventions
Journal of Molecular Neuroscience (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.