Abstract
Respiratory syncytial virus (RSV) is a leading cause of childhood morbidity, however there is no systematic testing in children hospitalised with respiratory symptoms. Therefore, current RSV incidence likely underestimates the true burden. We used probabilistically linked perinatal, hospital, and laboratory records of 321,825 children born in Western Australia (WA), 2000–2012. We generated a predictive model for RSV positivity in hospitalised children aged < 5 years. We applied the model to all hospitalisations in our population-based cohort to determine the true RSV incidence, and under-ascertainment fraction. The model’s predictive performance was determined using cross-validated area under the receiver operating characteristic (AUROC) curve. From 321,825 hospitalisations, 37,784 were tested for RSV (22.8% positive). Predictors of RSV positivity included younger admission age, male sex, non-Aboriginal ethnicity, a diagnosis of bronchiolitis and longer hospital stay. Our model showed good predictive accuracy (AUROC: 0.87). The respective sensitivity, specificity, positive predictive value and negative predictive values were 58.4%, 92.2%, 68.6% and 88.3%. The predicted incidence rates of hospitalised RSV for children aged < 3 months was 43.7/1000 child-years (95% CI 42.1–45.4) compared with 31.7/1000 child-years (95% CI 30.3–33.1) from laboratory-confirmed RSV admissions. Findings from our study suggest that the true burden of RSV may be 30–57% higher than current estimates.
Similar content being viewed by others
Introduction
Respiratory Syncytial Virus (RSV) is a leading cause of morbidity and mortality in young children worldwide, causing 3.2 million detected hospitalisation episodes every year1. The true burden is likely to be much greater, with approximately half of RSV-associated deaths estimated to occur outside of hospital1. In Australia, for every 100,000 hospitalised children aged < 5 years, an estimated 418 have RSV2. A recent population-based study in Western Australia (WA) conducted by our group reported pathogen-specific incidence rates of 247/100,000 child-years for RSV in children aged < 17 years3, with the highest burden among infants in their first 3 months of life (28.1/1000 child-years)4. RSV is most frequently associated with hospitalisations for acute bronchiolitis, but was also identified across other clinical diagnoses including pneumonia, unspecified acute lower respiratory infections, asthma, upper respiratory infections as well as non-specific viral diagnosis codes3,5.
Population-wide in our jurisdiction and elsewhere, there is no systematic approach to RSV testing, nor is it currently a notifiable disease, making estimates of RSV disease burden using microbiological testing datasets alone difficult. From our population-based study, only 10% of children aged < 17 years were ever tested for RSV and variable testing trends were seen between age groups and from year to year. Additionally, 54% of children hospitalised with respiratory infections did not have a corresponding microbiological test3. Improved estimates of RSV burden are required to inform future policy for RSV therapeutics and preventative strategies, as late-stage clinical trials of antivirals, maternal vaccines and monoclonal antibodies progress3,6.
Understanding the demographic and clinical predictors of RSV test positivity in different populations can aid in quantifying the under-ascertainment burden of RSV from standalone datasets. Ideally, a combination of clinical and laboratory data is needed. Such a study has been conducted in England for infants aged < 1 year, using hospitalisation data and RSV positive testing records7.
The aim of our study was to develop a prediction model to estimate the true incidence of RSV associated hospitalisations in children < 5 years of age in WA and to use these findings to determine the under-ascertainment fraction of RSV incidence using laboratory records alone.
Methods
Setting and data sources
WA covers the western third of Australia with a population of approximately 2.6 million people at 31 December 20198. Three quarters of the population reside in the temperate climatic region of metropolitan Perth and its surrounds9. We conducted a population-based cohort study using administrative linked data of all births in WA (1996 to 2012), as previously reported3,10. Data sources used for this study included the Midwives Notifications System, which includes perinatal information on > 99% of births in WA11, Birth and Death Registries, Hospital Morbidity Data Collection and PathWest Laboratory Medicine Database (PathWest). Data were probabilistically linked using best practice protocols through the WA Data Linkage Branch12.
Hospital data
Hospitalisation records, herein referred to as hospital admissions, with an admission and discharge date between 1 January 2000 and 31 December 2012 were included, to match the same time period when routine laboratory data were available. We included all admissions in children aged < 5 years in WA with any diagnosis using International Statistical Classification of Diseases and Related Health Problems,10th Revision, Australian Modification (ICD-10-AM) codes. As per previous analyses of these data, interhospital transfers were collapsed3,10.
Laboratory data
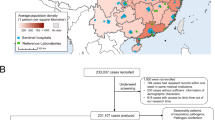
We extracted PathWest testing records of RSV from respiratory specimens with a specimen collection date between 1 January 2000 and 31 December 2012. We then linked these records with hospitalisation records from individuals from the birth cohort when respiratory specimens were collected 48 h before or after the admission date, as per our previous analyses3,10. The laboratory records were linked to the admission closest to the date of specimen collection when the same child had multiple admissions for different reasons within 48 h. During the early study period, RSV was predominantly detected through immunofluorescence antigen detection (65%) and viral culture on respiratory specimens and complement-fixation tests (CFT) on serum while, gradually, polymerase chain reaction (PCR) on respiratory specimens was more frequently used4,10. Using assembled data from hospital and PathWest records, we then identified our source population defined as children in the cohort who had a hospitalisation for any reason in the first 5 years of life with RSV testing records during the study period (n = 37,784 hospitalisations). After exclusion of 3,801 records with missing data for one or more of the variables, we finally included 33,983 hospitalisations with RSV testing records in the final prediction model. Similarly, after excluding observations with missing data, we estimated RSV burden in 321,825 records of hospitalised children under 5 years of age during the study period (Fig. 1).
Statistical analysis
Developing prediction models
A multivariable logistic regression model was fitted to identify predictors of RSV-positivity (binary outcome) amongst children younger than 5 years who were hospitalised and tested for RSV in WA during the study period (2000–2012). We used a robust standard error estimation adjusting for correlated observations due to children having multiple admissions over the study period13. A total of 27 candidate predictors for the prediction models were selected based on a comprehensive literature review, including previous work in our setting14 and clinical plausibility.
Directed acyclic graphs (DAG) were used to inform the choice of these predictors. In modern epidemiology, DAGs are used as a tool in causal inference to identify and demonstrate knowledge, theories and assumptions about causal relationships between variables15,16. Causal inference principles have recently been more widely applied in prediction models17,18. The minimal set of variables identified by DAGs included child age, sex, delivery route (vaginal vs caesarean), season of admission, Aboriginal and/or Torres Strait Islander status, respectfully referred to as (Aboriginal/non-Aboriginal), as identified through a validated algorithm provided by the WA linkage branch19, maternal age (categorised), smoking during pregnancy (yes/no), socioeconomic status (in quantiles), numbers of siblings, gestational age (< 32 weeks, 32–36 weeks and 37 or more weeks), admission year, birth year, length of hospital stay (continuous), admission to intensive care unit (ICU) [binary], mechanical ventilation use during hospital admission (see below), maternal history of asthma (binary), any diagnosis code for acute bronchiolitis, pneumonia, bronchitis, unspecified acute lower respiratory infections (ALRI), asthma, URTI, whooping cough, influenza and diagnosis code for other infections including unspecified viral illness, convulsions, fever and cough, breathing abnormalities (refer to eTable 1). These specific codes were chosen based on our previous work that identified a positive detection of a respiratory virus in hospital admissions with these codes10. Further, we included season of birth and geographical region of residence to account for seasonality and climatic conditions (as a proxy) respectively (eTable 1). Our final prediction model also included a sine and cosine function as per the recommendation by Stolwijk and Edwards to account for the seasonal variation of RSV20,21.
Socioeconomic status (SES) was measured using Socio-Economic Indexes for Areas (SEIFA). Specifically, we used the Index of Relative Advantage and Disadvantage score, calculated at the collection district (CD) level by Australian Bureau of Statistics22.
A child was coded as admitted to an ICU if they spent at least one day in ICU as recorded on the Hospital Morbidity Data Collection. Mechanical ventilation was defined as receipt of at least an hour of continuous ventilatory support or having procedure codes (classified using the 7th edition of the Australian Classification of Health Interventions) for airway management, invasive or non-invasive ventilatory support (eTable 1).
Model specification and estimation
As part of model development, a stepwise backward selection approach using Akaike Information criterion (AIC) was performed. Stepwise backward selection using AIC is a widely used criterion to assess model goodness of fit and parsimony23,24. We assessed the subsequent model improvement through a decrease in the AIC criterion after including all variables in the minimal sufficient adjustment sets plus additional variables not included in our DAG. All variables removed were individually reinserted into the model and reassessed for any model improvement.
Predictive performance and model validation
The ability of the variables to predict RSV-positivity in the source population was determined by a tenfold cross-validated area under the receiver operating characteristic (AUROC) curve. We constructed Receiver Operator Characteristic (ROC) by plotting the true positive rate (sensitivity) against false positive rate (1-specificity) at various levels of the marker. In k-fold cross validation, the dataset is randomly divided into k approximately equally sized subsamples (or folds)25. At each iteration, one-fold is retained as the validation data for testing the model and estimating the Area Under Curve (AUC), while the remaining k − 1 folds are used as training data for model estimation. This process is repeated k times, with each of the k folds used once as the validation data. The ‘cvauroc’ command in Stata was employed to derive and average the AUCs corresponding to each fold (here k = 10) and bootstrapping the cross-validated AUC was used to obtain statistical inference and bias corrected 95% confidence intervals (CIs). K-fold cross validation avoids the optimistic estimates of predictive performance known to exist when the full dataset is used for assessing model specification and prediction25. To explore diagnostic accuracy of the models, sensitivity, specificity, positive predictive values (PPV) and negative predictive values (NPV) were generated. We determined these characteristics at different false positive rates [eTable 3]. However, we chose a 0.5 probability threshold as the cut-off, which maximises sensitivity and specificity of the model. Furthermore, we used a ‘calibration belt’ to evaluate the calibration of our predictive model. The ‘calibration belt’ is a plot depicting the relationship between the model’s fit probabilities and the observed proportions of the response across all ranges of risk, which reflects the reliability or degree of bias of the model26. In addition, reliability was tested by the Hosmer–Lemeshow goodness-of-fit test27. As a sensitivity analysis, we repeated the same procedure for developing the prediction model by method of testing to see if predictors of RSV-positivity differ between PCR or immunofluorescence (IF) detection methods (eFigs. 8 and 9).
All data were analysed using STATA v.16.028. We used DAGitty v2.3 to produce the DAG29. We conducted a complete case analysis excluding records from the analysis with missing values for one or more of the predictor variables. We calculated incidence rates of predicted RSV-positivity using survival analysis techniques, allowing for multiple hospital admissions per person. We used person-time-at-risk as the denominator (calculated from date of birth until first date of hospital admission). All children were censored at the end of the study period or date of death, whichever was the earlier. Incidence rates were reported per 1000 child-years with associated 95% confidence intervals (CIs) by age group, year of hospital admission, admission season and birth month. Finally, we plotted predicted RSV rates by calendar week throughout the study period.
Estimating burden of RSV
After running each logistic regression model, we estimated a predicted probability of RSV-positivity and applied that to all hospital admissions in children aged less than 5 years during the study period given all non-missing variables in the model. All admissions in children aged less than 5 years during the study period with a predicted probability threshold of 0.5 were classified as an RSV-associated admission (hereafter referred to as predicted RSV). Finally, we estimated the under-ascertainment fraction of RSV rates, computed as rate differences between laboratory-confirmed RSV and predicted RSV.
Transparency of reporting
The Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD) statement was followed for this study (eTable 4)30. The TRIPOD statement provides a framework for the full and clear reporting of a prediction model study, such that risk of bias and potential usefulness can be adequately assessed.
Ethics statement
The authors assert that all methods were carried out in accordance with relevant guidelines and regulations. Ethical approvals were obtained from the WA Department of Health Human Research Ethics Committee and the WA Aboriginal Health Ethics Committee. As the study utilised de-identified linked administrative data, a waiver of informed consent was granted by the WA Department of Health Human Research Ethics Committee.
Results
Cohort description
From the birth cohort of 321,825 hospitalised children under the age of 5 years at the time of hospital admission, 37,784 were tested for RSV (11.7%). Of these 8,471 (22.4%) were infants aged < 3 months and 5,768 (15.3%) were Aboriginal. From all hospitalised children tested for RSV, laboratory confirmation was determined in 22.8% (n = 8,604 episodes). RSV positivity was more common in children that were younger, non-Aboriginal, male, had mothers with average SES, lived in a metropolitan residence and had hospital admission in the Australian winter months between June and August. One in three (2,594 admissions) of RSV positive admissions were among infants aged < 3 months. Approximately three quarters of all laboratory-confirmed RSV-positive children had a discharge diagnosis of bronchiolitis (Table 1).
Predicting RSV positivity
The variables included in the final logistic regression prediction model were child age, gender, delivery route, admission season, Aboriginal status, maternal age, smoking, SES, numbers of siblings, prematurity, admission year, birth year, length of hospital stay, admission to ICU, mechanical ventilation use during admission, maternal history of asthma, any diagnosis code for ALRI and diagnosis code for other infections including unspecified viral illness. To account for seasonality and climatic conditions, we also included season of birth and geographical region of residence respectively (eTable 1). The predictive equation of the final model is presented in (eFig. 2).
In the multivariable analysis, there was a higher odd of RSV-positivity in those aged < 3 months (Adjusted odds ratio (AOR) = 1.91, 95% CI (1.27–2.87)), and children who were not Aboriginal (AOR = 1.44. 95% CI (1.29–1.61)). Children with a primary or secondary diagnosis of acute bronchiolitis had 16-fold increased odds of RSV-positivity (AOR = 16.8, 95% CI (15.3–18.5)). RSV-positivity was also significantly associated with a diagnosis of any pneumonia, unspecified ALRI and bronchitis. Similarly, children born in remote or rural areas, who had a long hospital stay (three or more days) and who required mechanical ventilation during admission had an increased odds of RSV positivity.
Conversely, prematurity, low SES and maternal history of asthma were associated with reduced odds of RSV-positivity (eTable 2).
The tenfold cross-validated model showed accurate and robust performance of the prediction model (AUROC = 0.87, 95% CI 0.86 to 0.88), reflecting excellent ability of the model to predict RSV-positivity (Fig. 2). The sensitivity and specificity of the final model were 58.4% (95% CI 57.3–59.6%), and 92.2% (95% CI 91.8–92.5%) respectively. The model had a PPV of 68.6% (95% CI 67.5–69.7%) and NPV of 88.3% (95% CI 87.9–88.7%). Additionally, the calibration belt demonstrated that our prediction model is well calibrated (eFig. 3). The overall goodness of fit of the model was satisfactory, as indicated by a nonsignificant Hosmer–Lemeshow test (P = 0.66). Our sensitivity analysis suggested that the prediction model performance slightly differed by laboratory detection method (immune-fluorescence vs PCR) (eFigs. 8 and 9).
Laboratory-confirmed and predicted RSV rates by age at admission
The predicted incidence rates of RSV were higher in the younger age groups with the highest admission rates among infants aged less than 3 months. Our model predicted RSV-related admissions for children aged less than 3 months to be 43.7/1000 child-years (95% CI 42.1–45.4) compared with 31.7/1000 child-years (95% CI 30.3–33.1) from laboratory-confirmed RSV admissions. Similarly, the rate of predicted RSV admissions ranged from 5/1000 child-years for 12–24 months to 28/1000 child-years for 3–6 months of age. We estimated that the average annual RSV associated hospitalisation rates were 404 per 1000 children for < 3 months, 499 per 1000 children and 302 per 1000 children for 3–6 months and 6–11 months respectively, which is equivalent to under-ascertainment fractions of 32% for < 3 months, 57% for 3–6 months and 35% for 6–11 months of total admissions respectively (Table 2).
A total of 76% of our predicted RSV-associated admissions were in infants aged less than 12 months, accounting for 39% of the estimated under-ascertainment. For infants aged less than 12 months, the predicted RSV-associated admissions peaked during the winter season (n = 6859, 204/1000 child years [95% CI 199.1, 208.7)]) and in infants born in months of April–June (Table 3). Similarly, for children 12–24 months of age the predicted RSV admissions peaked during the winter season and in children born in March–May (Table 4).
RSV rates by calendar week
Rates of predicted RSV admissions for children aged less than 2 years showed a seasonal pattern. The peak in predicted RSV-associated admissions for this age group were observed during week 26–29 each year between 2000 to 2012, with exception of year 2007 and 2009 with a peak observed during week 34 (66/1000 child-years) and week 32 (55.9/1000 child-years), respectively. We observed a similar pattern to the peak in admissions for laboratory-confirmed RSV admissions during the study period (Fig. 3).
Discussion
RSV vaccine development has gained substantial attention globally, with the WHO identifying global RSV disease burden estimates as a global priority31,32. Our study aimed to improve the estimates of RSV incidence which are needed to help advocate for vaccine programs and provide essential baseline data to evaluate vaccine impact studies. We have developed a population-based prediction model to better estimate the true burden of RSV associated hospitalisations in children younger than 5 years in WA with robust performance. Our prediction model was based on readily available patient characteristics and may be useful for identifying hospitalised children likely to test positive for RSV.
Using population-based hospital and laboratory data, our results corroborated previous findings of the large burden of RSV-associated hospital admissions in children3,4,7, and our prediction model suggests that we underestimate this burden by more than 30%. Our results also indicate that the RSV burden is age-specific, with significant seasonal variation. Over the study period, which spanned more than a decade, the burden of predicted RSV-associated hospital admissions was substantial, with an average annual estimated admission rate of 404 per 1000 children for < 3 months and 338 per 1000 children for under 2 years old respectively. This is consistent with our previous results3,4 and with findings from other studies33,34. The peaks in RSV-associated admissions were observed in the winter season, as well as in children born in April, May and June in both < 12 months and 12–24 months age groups. Given the higher burden of RSV-associated hospitalisation among the young infant age group, future vaccine programmes could target them as priority beneficiaries. There are recent suggestions of targeting infants born around the start of RSV season for a possible seasonal vaccination strategy35.
Our analysis confirmed that children who are younger at admission (< 3 months) are at increased risk of RSV positivity, which is consistent with other studies7,36. In addition to younger age, our study also identified additional important predictors of RSV positivity, including length of hospital stay, primary or secondary diagnosis of acute bronchiolitis, pneumonia, bronchitis and unspecified ALRIs. A primary or secondary diagnosis of acute bronchiolitis was the strongest predictor of RSV positivity. A diagnosis of bronchitis was the second most important predictor, followed by pneumonia. A previous study in England similarly reported that infants with a diagnosis of bronchiolitis, unspecified LRTI or with an RSV-specific code had higher odds of RSV-positivity7.
The risk of hospitalisation was higher for non-Aboriginal children and children from a rural or remote region at birth. Similarly, the highest rates of RSV were found in remote and rural regions compared to metropolitan areas. These difference could be attributed to a combination of socio economic disadvantages (such as household crowding) and access to health care37,38. Interestingly, low SES, prematurity and family history of asthma were associated with lower odds of RSV positivity. Similar findings were reported in previous studies7,39. Nonetheless, findings reported elsewhere have suggested that the majority of cases with RSV do not have any underlying co-morbidity40.
Our evaluation using tenfold cross validation showed an excellent performance and was well calibrated, with AUROC of 0.8727. By comparison, in a recent study predicting RSV associated admissions in England reported AUROC of 0.9, our predictive model has higher specificity and NPV but lower sensitivity and PPV compared to that model which focused on infants under the age of 12 months7. Our prediction study differs from others with respect to either the domain or population studied. Other studies focused on RSV hospitalisation in young children7,41, premature infants42,43, or non-hospitalised RSV among healthy term infants44.
Our prediction model showed a good fit to the seasonality and age distribution of RSV-associated hospitalisation. However, the model slightly underestimated the RSV-associated admissions for older children. This could be due to non-inclusion of additional risk factors, or lifestyle factors beyond the perinatal period, where we had limited data to base our prediction model on. Our prediction model was based on more than a decade of population-based hospitalisation data linked with laboratory and socio-demographic records, which is a key strength of our study. Our model included a comprehensive list of maternal, infant, and perinatal predictors which were not included in previous similar RSV prediction studies7,41,45. We also employed a DAG as an efficient strategy to improve predictor selection in the prediction modelling. We believe the definition of ALRIs, and other infections included as risk factors in our prediction model increases the likelihood of positive detection of a respiratory virus in hospital admissions. We also employed a k-fold cross validation, an internal validation method that takes over-optimism into account far better than conventional data splitting46.
Our study has some limitations. Firstly, our results are based on hospitalisation data linked to laboratory records with specimen collection within 48 h of hospital admission. Therefore, we are limiting our prediction model to more severe RSV cases associated with hospitalisation and therefore our model does not estimate the broader community incidence of RSV. Secondly, our analysis only included linked data in the years between 2000–2012, and more recent trends in hospital admissions are not reflected in our results. However, we are confident our model can be applied to more contemporary data and believe that the majority of the known perinatal and environmental predictors are not likely to change over time. Our study did not include other potential predictors such as breastfeeding, immunodeficiencies, day care attendance, and environmental factors, as these factors are not routinely collected in databases available for linkage. However, we do not expect a major underestimation of our prediction model as these variables are not known to be strong predictors of RSV associated admissions7,44. Additionally, even though the testing detection method would not impact the propensity to get tested, our sensitivity analysis suggested that the prediction model performance slightly different by laboratory detection method.
The population-based linkage of routine laboratory and hospitalisation data allowed us to develop a predictive model with excellent predictive performance to identify RSV associated hospitalisation in WA. Applying the model to all hospitalised children aged less than 5 years (irrespective of respiratory infection diagnosis) during the study period enabled us to estimate the true RSV burden in hospitalised children in the state.
Further research is needed that takes into consideration emergency department and community RSV-associated admissions, preferably including recent data, as well as other potential predictors. A development of a simple risk score, and further external validation of the model in other populations must also be considered for possible future clinical use of the prediction model.
Conclusion
We have successfully developed a prediction model using population-based data to estimate the true burden of RSV in hospitalised children in WA with good predictive performance and internal validation. Findings from our study indicate that the true burden of RSV is up to 30–57% higher than figures based solely on laboratory detection data in young children. These estimates can now be used as input parameters in dynamic transmission models to better predict the impact of prevention measures including maternal vaccination47.
Change history
29 January 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41598-024-52791-0
References
Shi, T. et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. The Lancet 390, 946–958 (2017).
Saravanos, G. L. et al. Respiratory syncytial virus-associated hospitalisations in Australia, 2006–2015. Med. J. Aust. 210, 447–453 (2019).
Lim, F. J., Blyth, C. C., Fathima, P., de Klerk, N. & Moore, H. C. Record linkage study of the pathogen-specific burden of respiratory viruses in children. Influenza Other Respir. Viruses 11, 502–510 (2017).
Moore, H. C. et al. Assessing the burden of laboratory-confirmed respiratory syncytial virus infection in a population cohort of Australian children through record linkage. J. Infect. Dis. 2, 106 (2020).
Müller-Pebody, B., Edmunds, W., Zambon, M., Gay, N. & Crowcroft, N. Contribution of RSV to bronchiolitis and pneumonia-associated hospitalizations in English children, April 1995–March 1998. Epidemiol. Infect. 129, 99–106 (2002).
Griffin, M. P. et al. Single-dose nirsevimab for prevention of RSV in preterm infants. N. Engl. J. Med. 383, 415–425 (2020).
Reeves, R. et al. Burden of hospital admissions caused by respiratory syncytial virus (RSV) in infants in England: a data linkage modelling study. J. Infect. 78, 468–475 (2019).
Australian Bureau of Statistics. Australian Demographic Statistics, https://www.abs.gov.au/Population (2019).
Australian Bureau of Statistics. Regional population by age and sex, Australia, http://www.abs.gov.au (2018).
Lim, F., Blyth, C., Keil, A., De Klerk, N. & Moore, H. Using record linkage to examine testing patterns for respiratory viruses among children born in Western Australia. Epidemiol. Infect. 145, 1688–1698 (2017).
Downey, F. A validation study of the Western Australian Midwives’ Notification System. Perth Dep. Health Western Australia 38, 1 (2007).
Holman, C. D. A. J., Bass, A. J., Rouse, I. L. & Hobbs, M. S. Population-based linkage of health records in Western Australia: development of a health services research linked database. Aust. N. Z. J. Public Health 23, 453–459 (1999).
Mansournia, M. A., Nazemipour, M., Naimi, A. I., Collins, G. S. & Campbell, M. J. Reflection on modern methods: demystifying robust standard errors for epidemiologists. Int. J. Epidemiol. 50, 346–351 (2021).
Moore, H. C., De Klerk, N., Richmond, P. & Lehmann, D. A retrospective population-based cohort study identifying target areas for prevention of acute lower respiratory infections in children. BMC Public Health 10, 757 (2010).
Tennant, P. W. et al. Use of directed acyclic graphs (DAGs) in applied health research: review and recommendations. medRxiv 2, 1069 (2019).
Greenland, S., Pearl, J. & Robins, J. M. Causal diagrams for epidemiologic research. Epidemiology 296, 37–48 (1999).
Piccininni, M., Konigorski, S., Rohmann, J. L. & Kurth, T. Directed Acyclic Graphs and causal thinking in clinical risk prediction modeling. arXiv preprint arXiv:2002.09414 (2020).
Blakely, T., Lynch, J., Simons, K., Bentley, R. & Rose, S. Reflection on modern methods: when worlds collide—prediction, machine learning and causal inference. Int. J. Epidemiol. 2, 690 (2019).
Christensen, D. et al. Evidence for the use of an algorithm in resolving inconsistent and missing Indigenous status in administrative data collections. Aust. J. Soc. Issues 49, 423–443 (2014).
Stolwijk, A., Straatman, H. & Zielhuis, G. Studying seasonality by using sine and cosine functions in regression analysis. J. Epidemiol. Community Health 53, 235–238 (1999).
Edwards, J. The recognition and estimation of cyclic trends. Ann. Hum. Genet. 25, 83–87 (1961).
Australian Bureau of Statistics. Socio-Economic Indexes for Areas (SEIFA) 2016 Technical Paper. (Australian Bureau of Statistics Canberra, Australia, 2018).
Sauerbrei, W., Royston, P. & Binder, H. Selection of important variables and determination of functional form for continuous predictors in multivariable model building. Stat. Med. 26, 5512–5528 (2007).
Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control 19, 716–723 (1974).
Luque-Fernandez, M. A., Redondo-Sánchez, D. & Maringe, C. cvauroc: Command to compute cross-validated area under the curve for ROC analysis after predictive modeling for binary outcomes. Stand. Genomic Sci. 19, 615–625 (2019).
Nattino, G., Lemeshow, S., Phillips, G., Finazzi, S. & Bertolini, G. Assessing the calibration of dichotomous outcome models with the calibration belt. Stand. Genomic Sci. 17, 1003–1014 (2017).
Hosmer, D. W., Lemeshow, S. & Sturdivant, R. X. Applied Logistic Regression Vol. 398 (Wiley, 2013).
StataCorp, L. Stata Statistical Software: Release 16. (College Station, 2019).
Textor, J., Hardt, J. & Knüppel, S. DAGitty: a graphical tool for analyzing causal diagrams. Epidemiology 22, 745 (2011).
Collins, G. S., Reitsma, J. B., Altman, D. G. & Moons, K. G. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) the TRIPOD statement. Circulation 131, 211–219 (2015).
Organization, W. H. RSV Vaccine Research and Development Technology Roadmap: Priority Activities for Development, Testing, Licensure and Global Use of RSV Vaccines, with a Specific Focus on the Medical Need for Young Children in Low-and Middle-Income Countries. (World Health Organization, 2017).
Madhi, S. A. et al. Respiratory syncytial virus vaccination during pregnancy and effects in infants. N. Engl. J. Med. 383, 426–439 (2020).
Manuel, B. et al. RSVpredict: an online tool to calculate the likelihood of respiratory syncytial virus infection in children hospitalized with acute respiratory disease. Pediatr. Infect. Dis. J. 38, 678–681 (2019).
Simoes, E. A. Environmental and demographic risk factors for respiratory syncytial virus lower respiratory tract disease. J. Pediatr. 143, 118–126 (2003).
Korsten, K. & Bont, L. Seasonal immunisation against respiratory syncytial virus disease. The Lancet Public Health 2, e344–e345 (2017).
Bont, L. et al. Defining the epidemiology and burden of severe respiratory syncytial virus infection among infants and children in western countries. Infect. Dis. Ther. 5, 271–298 (2016).
Holman, R. C. et al. Respiratory syncytial virus hospitalizations among American Indian and Alaska Native infants and the general United States infant population. Pediatrics 114, e437–e444 (2004).
Brew, B. et al. Identifying preventable risk factors for hospitalised asthma in young Aboriginal children: a whole-population cohort study. Thorax.
Zar, H. J. et al. Early-life respiratory syncytial virus lower respiratory tract infection in a South African birth cohort: epidemiology and effect on lung health. Lancet Glob. Health 8, e1316–e1325 (2020).
Hall, C. B. et al. The burden of respiratory syncytial virus infection in young children. N. Engl. J. Med. 360, 588–598 (2009).
Rietveld, E. et al. Hospitalization for respiratory syncytial virus infection in young children: development of a clinical prediction rule. Pediatr. Infect. Dis. J. 25, 201–207 (2006).
Sheridan-Pereira, M. et al. Respiratory syncytial virus preterm (32–36 completed weeks of gestation) risk estimation measure for RSV hospitalization in Ireland: a prospective study. Pediatr. Infect. Dis. J. 35, 19–24 (2016).
Ambrose, C. S. et al. Respiratory syncytial virus disease in preterm infants in the US born at 32–35 weeks gestation not receiving immunoprophylaxis. Pediatr. Infect. Dis. J. 33, 576 (2014).
Houben, M. L. et al. Clinical prediction rule for RSV bronchiolitis in healthy newborns: prognostic birth cohort study. Pediatrics 127, 35–41 (2011).
Straňák, Z. et al. Predictors of RSV LRTI hospitalization in infants born at 33 to 35 weeks gestational age: a large multinational study (PONI). PLoS One 11, e0157446 (2016).
Steyerberg, E. W. et al. Internal validation of predictive models: efficiency of some procedures for logistic regression analysis. J. Clin. Epidemiol. 54, 774–781 (2001).
Hogan, A. B. et al. Potential impact of a maternal vaccine for RSV: a mathematical modelling study. Vaccine 35, 6172–6179 (2017).
Acknowledgements
The authors would like to thank the Linkage and Client Services Teams at the Western Australia (WA) Data Linkage Branch, as well as custodians of the datasets used (Birth and Death Register, Midwives Notification System, Hospital Morbidity Data Collection and PathWest Laboratory Medicine Database), for their assistance and support in collating the data.
Funding
This work was funded by Australian Respiratory Council (ARC) Research Support Grant funding 2020. ATG and HCM receive salary support from Wesfarmers Centre of Vaccines and Infectious Diseases. Data used in this study was supported by National Health and Medical Research Council Project Grant (1045668). ABH is supported by an Imperial College Research Fellowship. ABH also acknowledges funding support for the MRC Centre for Global Infectious Disease Analysis (reference MR/R015600/1), jointly funded by the UK MRC and the UK FCDO, under the MRC/FCDO Concordat agreement, also part of the EDCTP2 programme supported by the European Union. CCB receives support from the National Health and Medical Research Council. The funding bodies had no role in either the study design, conduct, analysis or interpretation of the study or the decision to submit for publication.
Author information
Authors and Affiliations
Contributions
A.T.G. cleaned and analysed the data, wrote the first draft of the manuscript, and critically reviewed the manuscript for intellectual content. A.B.H. and K.G. contributed to the design of the study, data analysis, and critically reviewed the manuscript for intellectual content. C.C.B. contributed to the design of the study, provided clinical input on interpretation of the results, and critically reviewed the manuscript for intellectual content. H.C.M. conceptualised and secured funding for the study, contributed to the design of the study, data analysis, and critically reviewed the manuscript for intellectual content. All authors finally approved the final manuscript as submitted.
Corresponding author
Ethics declarations
Competing interests
ABH declares personal fees from the World Health Organization, outside the submitted work. ABH was previously engaged by Pfizer Inc to advise on modelling RSV vaccination strategies for which she received no financial compensation. The remaining authors have no conflicts of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained errors in Table 1, where the values for “RSV-positive N (%)” under the Gestational age variable were interchanged. As a result, the values for “Total admissions N (%)” and “RSV positivity rate (%)” were incorrect.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gebremedhin, A.T., Hogan, A.B., Blyth, C.C. et al. Developing a prediction model to estimate the true burden of respiratory syncytial virus (RSV) in hospitalised children in Western Australia. Sci Rep 12, 332 (2022). https://doi.org/10.1038/s41598-021-04080-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-04080-3
This article is cited by
-
Respiratory syncytial virus disease morbidity in Australian infants aged 0 to 6 months: a systematic review with narrative synthesis
BMC Public Health (2023)
-
Assessing the transportability of clinical prediction models for cognitive impairment using causal models
BMC Medical Research Methodology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.