Abstract
Most public health measures to contain the COVID-19 pandemic are based on preventing the pathogen spread, and the use of oral antiseptics has been proposed as a strategy to reduce transmission risk. The aim of this manuscript is to test the efficacy of mouthwashes to reduce salivary viral load in vivo. This is a multi-centre, blinded, parallel-group, placebo-controlled randomised clinical trial that tests the effect of four mouthwashes (cetylpyridinium chloride, chlorhexidine, povidone-iodine and hydrogen peroxide) in SARS-CoV-2 salivary load measured by qPCR at baseline and 30, 60 and 120 min after the mouthrinse. A fifth group of patients used distilled water mouthrinse as a control. Eighty-four participants were recruited and divided into 12–15 per group. There were no statistically significant changes in salivary viral load after the use of the different mouthwashes. Although oral antiseptics have shown virucidal effects in vitro, our data show that salivary viral load in COVID-19 patients was not affected by the tested treatments. This could reflect that those mouthwashes are not effective in vivo, or that viral particles are not infective but viral RNA is still detected by PCR. Viral infectivity studies after the use of mouthwashes are therefore required. (https://clinicaltrials.gov/ct2/show/NCT04707742; Identifier: NCT04707742)
Similar content being viewed by others
Introduction
The coronavirus disease 2019 (COVID-19) outbreak was quickly declared by the World Health Organization (WHO) a public health emergency of international concern and has given rise to one of the most dramatic pandemics in recent human history1. The disease is caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) virus, a single-stranded enveloped RNA virus which belongs to the betacoronavirus genus from the Coronaviridae family2.
Because no effective treatment for COVID-19 is currently available, most public health measures to contain the pandemic are based on preventing the spread of the pathogen. The virus is transmitted by the respiratory route (respiratory droplets and aerosols) and by direct contact with contaminated surfaces and subsequent contact with nasal, oral or ocular mucosa3. Although patients with symptomatic COVID-19 have been the main source of transmission, asymptomatic and pre-symptomatic patients also have the ability to transmit SARS-CoV-24. Higher viral loads are detected after the onset of COVID-19 symptoms, being significantly higher in the nose compared to the throat5.
Angiotensin-converting Enzyme 2 (ACE2) is the main cellular receptor for SARS-CoV-2, which interacts with the spike protein to facilitate its entry. ACE2 receptors are highly expressed in the oral cavity and present at high levels in oral epithelial cells6. The mean expression of ACE2 was higher in the tongue compared to that in other oral tissues and it has been found to be higher in the minor salivary glands than in the lungs6. These findings strongly suggest that the oral cavity and specifically the saliva may be a high-risk route for SARS-CoV-2 infection7. Thus, strategies reducing salivary viral load could contribute to reduce the risk of transmission. Furthermore, studies using macaques as animal models have shown that SARS-CoV-1 (which has an 82% genomic similarity with SARS-CoV-2) persists for 2 days in oral mucous membranes before its diffusion to the lower respiratory tract. This offers an interesting preventive and therapeutic window of opportunity for the control of this disease8. For this reason, the use of mouthwashes with antiseptics that have virucidal activity can be a simple preventive strategy that could easily be applied in the general population.
It is widely known that antimicrobial mouthwashes reduce the levels of oral microorganisms including bacteria and fungi, and there are promising in vitro data and clinical studies showing virucidal effects against influenza9, murine norovirus or herpes simplex virus10,11, among others. In relation to coronaviruses, there was also in vitro evidence that povidone-iodine, a common active ingredient in some oral antiseptics, has efficient virucidal activity against Middle East Respiratory Syndrome Coronavirus (MERS-CoV) and SARS-CoV-112,13. Stimulated by these background, numerous research teams have evaluated the in vitro efficacy of different oral antiseptics against SARS-CoV-2 and other coronaviruses, showing potent virucidal activity of chlorhexidine (CHX), cetylpyridinium chloride (CPC), povidone-iodine (PVP-I), essential oils or hydrogen peroxide (H2O2)14,15,16,17,18. However, there is very limited clinical evidence for the effectiveness of mouth rinses against SARS-CoV-2 in vivo and there is an urgent need to obtain reliable data in placebo-controlled randomized clinical trials where the comparative effect of different oral antiseptics can be experimentally verified19.
Thus, in the current manuscript we present the results of a multi-centre, blind, parallel-group, placebo-controlled randomised clinical trial testing the effect of four different mouthwashes (CPC, CHX, PVP-I and H2O2) in the salivary load of SARS-CoV-2 as measured by quantitative real-time PCR at three different timepoints. Our study aims to test whether any of these standard oral antiseptics appear to diminish viral load in saliva and could therefore be used as a strategy to reduce transmission risk in clinical and social settings.
Material and methods
Study design
To evaluate the effect of several antiseptics to neutralize or reduce the viral load in vivo of SARS-CoV-2 in saliva samples, a multicentre, randomized, double-blind, five-parallel-group, placebo-controlled trial was designed (ClinicalTrials.gov Identifier: NCT04707742). The present study was approved on 2020/05/8th by the Ethical Committee of the Fundación Jiménez Díaz University Hospital with code EO095-20_FJD-HGV-HIE and all research was performed in accordance with relevant guidelines/regulations.
The study was performed in Madrid, Valencia and Murcia regions in Spain, in five different hospitals: Fundación Jiménez Díaz University Hospital (Madrid, Spain), Villalba University General Hospital (Madrid, Spain), Infanta Elena University Hospital (Madrid, Spain), Virgen de la Arrixaca University Hospital (Murcia, Spain) and Clínico de Valencia University Hospital (Valencia, Spain).
Patients
Every patient included was previously diagnosed and hospitalized because of SARS-COV-2 infection, being admitted mainly for respiratory pathology. All were adults (age > 18 years) and provided their voluntary written or oral consent to participation according to the ethics committee recommendations. The inclusion criteria included a positive result for SARS-COV-2 RT-PCR test of a respiratory sample in the previous 7 days and the ability to donate saliva samples and perform a mouthwash. Exclusion criteria included patient participation in a COVID-19 research study using experimental drugs, the use of an antiseptic mouthwash for 48 h before the start of the study and any known hypersensitivity or allergy to components of the mouthwashes.
Randomisation and masking
After approval of consent, the hospital staff responsible for the interventions consecutively assigned each participant a code following the order from a previously randomly generated table provided by other member of the research team. The code consisted of a patient number and a letter corresponding to one of the five study groups (A, B, C, D and E), that were unknown to the personnel who performed RNA extraction and qPCR and to those that analyzed the data. In this way, participants were randomly assigned to one of the five treatment groups and the blind for the patients was achieved by using identical tubes with the same volume for both mouthwashes and placebo. Patients receiving antiviral drugs (n = 6, all taking remdesivir) were randomly assigned to the different groups (three patients to the H2O2 group, one to the PVP-I group and two to the control group).
Procedures
Every included patient was asked not to eat, drink anything but water, chew gum, smoke or brush their teeth for 1 h prior to sample collection. In addition, it was not allowed to drink for half an hour after the mouthwash and eat for the entire test. For standardization, participants were instructed how to perform the mouthwash and how to donate the sample.
Five mouthwashes were randomized: 2% povidone-iodine (group A), 1% hydrogen peroxide (group B), 0.07% cetylpyridinium chloride (group C), 0.12% chlorhexidine (group D), and distilled water (group E), as the control group. Group C (Vitis Xtra Forte©) and D (Clorhexidina Dental PHB©) rinses were ready to use in their commercial formulas. In the case of the mouthwashes A (Betadine® Bucal 100 mg/mL) and B (Oximen©), the concentrations were adjusted to those indicated by the manufacturer, by diluting commercial formulas with distilled water minutes before rinsing (3 mL of povidone-iodine 10% for oral use with 12 mL of distilled water, in group A; and 5 mL of hydrogen peroxide 3%—with 10 mL of distilled water, in group B).
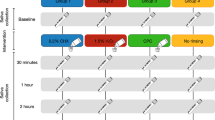
A total of 4 non stimulated saliva samples were taken for each patient in the morning: one basal and three after a 1-min mouthwash, specifically at 30, 60 min and 120 min. Figure 1 shows a scheme of the study design. The patients were asked to provide each unstimulated saliva sample into a sterile plastic container (at least 0.5 mL of sample) by drooling, avoiding spilling secretions of bronchial origin. Immediately after each sampling, 0.5 mL of saliva was transferred to a sterile Eppendorf tube with 1.5 mL of virus inactivating buffer (60% w/v Guanidine thiocyanate, 50% (v/v) 0.1 M Tris HCl, 11% v/v 0.2 M EDTA pH 8 and 1.3% v/v Triton X-100) (Crick Institute Standard Operation Procedures, accessed May 16th 2020) labeled with the patient code and sample time and kept at 4 °C. The four Eppendorf tubes per patient were introduced in an airtight bag, inside which there was protecting absorbent material in case of undesired opening or breaking. The secondary containers were introduced in a rigid box according to UN3733 standards and sent to a centralized laboratory (FISABIO) by courier service for analysis.
Diagram of the experimental study protocol. The upper left panel illustrates the multicentre study with five hospitals involved, the number of enrolled patients diagnosed and hospitalized because of SARS-COV-2 infection and the record of clinical variables. The sampling of the four non stimulated saliva samples for each patient is represented by the numbers 1, 2, 3 and 4 (corresponding to one basal and three post-mouthwash samples, at 30 min, at 60 min and at 120 min, respectively), as well as the rinsing time for 1 min. In this blind study, participants were randomly assigned to one of the five parallel group: PVP-I (group A), H2O2 (group B), CPC (group C), CHX (group D) and distilled water (group E, control group). After each sampling, 0.5 mL of saliva was transferred to a sterile Eppendorf tube with 1.5 mL of virus inactivating buffer, illustrated in the figure with a syringe. All samples were taken to the laboratory where RNA extraction and RT-qPCR (reverse transcription—quantitative polymerase chain reaction) was performed.
Nucleic acid extraction and real-time RT-PCR
An aliquot of 1 mL of each saliva sample diluted in the virus inactivating buffer in proportion 1:3, was used for RNA extraction using a standard TRIzol™ reagent (Invitrogen) method20.
An assay for SARS-CoV-2 was developed based in the WHO-Charité and U.S. CDC assays, to detect the SARS-CoV-2 E-gene in a multiplex assay together with the human cellular control RNAse-P to ensure reliability of sample collection and nucleic acid extraction before RT-PCR assays21,22. SARS-CoV-2 E-gene (FAM) and the housekeeping human RNP gene (Cy5) primer and probe sets were mixed with the qScript XLT One-Step RT-qPCR ToughMix (Quanta BioSciences, USA) and 8 μL of extracted RNA, as per manufacturer instructions. Real-time RT-PCR was performed in a Lightcycler 480II apparatus (Roche Diagnostics, Spain). Only those samples with RNAse-P Ct values < 35 were considered valid. The limit of detection of the assay was established to 35 copies using the SARS-CoV-2 E-gene as reference material, either as an insert in a pCDNA plasmid (a gift from Dr. Luis Enjuanes laboratory, CNB-CSIC, Spain) or as encapsulated RNA (a gift from Charité Medical Center, Berlin, though the European Virus Archive program). All samples were run in two replicates, together with previously known positive and negative controls. Virus copies were quantified using a tenfold dilution series standard curve of synthetic RNA (SARS-CoV-2 Standard, Exact Diagnostics, USA). Virus copies were normalized by mL of saliva.
Statistical analysis
Considering that each volunteer act as their own control, when comparing the viral load values in saliva at every time with respect to the levels prior to rinsing with the mouthwash, a sample size of 15 patients per branch was considered sufficient to identify significant differences between groups of more than 20%, assuming a 10% loss of patients due to abandonment or low viral load and adopting an alpha of 0.05 and a power of 0.8. As 5 branches were programmed in the trial (CPC, CHX, PVP-I, H2O2 and the control), 75 patients were the minimum number of individuals to be recruited, who were distributed among the different hospitals.
A Wilcoxon signed-rank test was used to test average differences, both for the paired case, when comparing observations in different times for the same individuals, and for the un-paired case, when comparing different treatments based on relative increments of viral load. Also, some tests for association between paired samples using Spearman's correlation coefficient were performed to assess relationship between viral load and other clinical continuous variables. Finally, to evaluate associations between clinical categorical variables and the frequency of responders/non-responders to the treatments, we carried out chi-squared contingency table tests. We define as responder an individual exhibiting an improvement equal to or greater than 90% of basal viral load. All computations and tests were performed using R environment for statistical computing version 3.6.3 and its 'stats' package23.
Results
From the 98 individuals screened, 14 did not fulfil inclusion criteria due to COVID-19 diagnostic other than PCR or to a period of more than 7 days since the PCR. The 84 individuals enrolled were randomly assigned to one of the 5 study groups (Fig. 2). After RNA extraction, a control qPCR for the human ribonuclease P gene was performed for all 336 samples, from which 17 produced no result and were discarded. A sample size of 12–15 individuals per group were analysed with 3–4 valid samples (total number of patients: 67), and for paired statistical comparisons, a sample size of 9–14 individuals per group (total number: 58) was used for which all four samples produced valid results.
Trial profile. Boxes display the number of individuals recruited for the clinical study and those assigned to each treatment, indicating the number of samples discarded due to undetectable viral load. aNo viral load detected at all four times. bNo viral load detected in any one of the four sampling times. *Unpaired statistical analysis (three samples per patient available, including basal values). **Paired statistical analysis (all four samples available).
The mean age of recruited patients was between 54–55 for the five groups with a range of 19–87 years (Table 1). There was a higher proportion of males than females in all five groups. Before the mouthwash, basal viral loads for most patients ranged between 102–106 copies/mL of saliva, and a maximum load of 1010 copies/mL was detected. There was no statistically significant correlation in basal viral load with the number of days elapsed since the original diagnostic RT-PCR (Spearman’s correlation coefficient between viral load and days since PCR was − 0.014) or since the day of first symptoms (Spearman’s correlation coefficient − 0.064). There were no statistically significant differences in basal viral load between sexes, nor between patients receiving antiviral drugs (n = 6, all of them taking Remdesivir) and those that did not (n = 61). A trend towards higher basal viral load values in older patients was detected (Spearman’s correlation coefficient 0.24, p-value = 0.067; Supplementary Fig. 1).
Viral loads in saliva for all five groups through time are shown in Fig. 3 (individual data for each patient are provided in Supplementary Table 1). None of the tested mouthwashes significantly reduced viral load at any timepoint compared with baseline. However, individual responses were highly divergent, with clear decreases in viral load in some individuals or at some time points and increases at other times. When looking at the relative changes compared to the values before the mouthwash, the maximum effects on viral load were observed 2 h after treatment in PVP-I and CPC groups, with mean viral load reductions around 30% (Fig. 4). The number of individuals that underwent a 50% decrease in viral load increased with the time elapsed since the mouthwash in the PVP-I and CPC groups, reaching 65% and 59% of participants, respectively (Fig. 5a). In the H2O2 group, the largest effect was seen 1 h after treatment, whereas in the CHX group the effect was seen already at 30 min and was maintained with time. A similar pattern was observed for individuals undergoing a 90% decrease in viral load (Fig. 5b). The advantage of using an antiseptic mouthwash was limited when compared with the distilled water control, where a significant number of patients also underwent a decrease in viral load.
Salivary viral loads for all five treatment groups through time. Box plots show viral load levels, as measured by qPCR, for each of the patients in the group A—PVP-I (povidone-iodine), B—Hydrogen peroxide (H2O2), C—CPC (cetylpyridinium chloride), D—CHX (chlorhexidine) and E—Distilled water (placebo group) at the four different timepoints (1 for basal and 2, 3 and 4 for the three times after the mouthwash, corresponding to 30, 60 and 120 min after the mouthwash, respectively). Lines joining the samples from the same patient are also shown.
Relative changes in salivary viral load for all five treatment groups after the mouthwash. Box plots show changes of viral load (%) for each of the patients in the group A—PVP-I (povidone-iodine), B—H2O2, C—CPC (cetylpyridinium chloride), D—CHX (chlorhexidine) and E (placebo group) at the three different times after the oral rinse treatment, relative to the baseline viral load prior to the mouthwash (changes at 30 min: t2–t1; changes at 60 min: t3–t1; changes at 120 min: t4–t1). Lines joining the points from the same patient through time are also shown.
Patients with improvement in viral load values. Bar plots show the percentage of individuals lowering salivary viral load by 50% or higher (a), or 90% or higher (b) relative to baseline values prior to the oral rinsing treatment, at different times after the mouthwash (t2: 30 min, t3: 60 min and t4: 120 min) in each study group: PVP-I (povidone-iodine), H2O2, CPC (cetylpyridinium chloride), CHX (chlorhexidine) and control group, respectively.
Given the high inter-individual variability in the response, the potential effect of clinical features on treatment efficacy was evaluated. Only a trend was observed for viral load decrease (higher than one order of magnitude) in patients in the PVP-I group 2 h post-treatment when considering individuals for which saliva was collected less than 6 days after first symptoms appeared (p = 0.06, Wilcox test). The effect of oral mouthwashes on salivary viral load remained non-significant when considering only samples with a high or low basal viral load.
Discussion
The data obtained in the current clinical study indicate that SARS-CoV-2 salivary load, as measured by real-time RT-PCR, was not significantly affected by the use of four widely used mouthwashes. This contrasts dramatically with previous in vitro studies where short exposure times (15–60 s.) of SARS-CoV-2 or other coronaviruses to different oral antiseptics resulted in strong virucidal effects ranging from one to four orders of magnitude15,16,24,25. These studies were performed using direct contact of the virus with antiseptics prior to adsorption and infection essays using Vero E6 or Huh7 cell lines. In some cases, addition of compounds like mucin, BSA, fetal calf serum or yeast extract was performed to mimic oral secretions or the naso/oropharynx. However, our data suggest that artificial conditions used to test mouthwashes in vitro may not be extrapolated to the actual effect of those antiseptics in the human oral cavity. For example, salivary glycoproteins are likely to interfere with the antiseptics’ antimicrobial activity, and compounds introduced in the oral cavity have been found to undergo a 1:4 dilution due to the presence of saliva26, potentially reducing their effect. In addition, all oral antiseptics tested in the current study have been previously shown to have activity against other microorganisms, including bacteria and/or fungi. Thus, a lower product availability in vivo is expected as a consequence of the antiseptic binding to other microorganisms in the oral cavity.
When present, the virucidal effects observed did not always match the temporal expected pattern based on their substantivity (i.e. their ability to persist in the oral cavity for a period of time). Hydrogen peroxide, for example, is expected to have an immediate effect but its activity may not remain over time due to chemical decomposition. However, the proportion of individuals with a large (> 1 log) reduction in viral load in this group at 1 and 2 h doubled that observed after 30 min of the mouthwash (Fig. 5). CPC, on the other hand, has a high expected substantivity (between 4–6 h), but a 30% of patients with a decreased viral load relative to the control group was already observed 30 min after the mouthwash. Results with CHX did match its expected persistence in the oral cavity, as it increased its efficacy through time. It must be considered that oral clearance of food or drinks due to saliva swallowing can take up to half an hour for a person with a healthy salivary flow. Thus, a potential increase in viral load after the mouth rinse could be expected if viral particles are released from oral mucosa and tongue tissues, but our data did not show this potential increase at the 30-min sample. Future studies should therefore also focus on determining SARS-CoV-2 levels during the first 30 min after the mouthwash, in order to determine the initial dynamics of viral load after the use of oral antiseptics. In the current manuscript, a higher proportion of patients appeared to show a decrease in viral load with time, especially in the CHX group, suggesting an initial virucidal effect in those individuals, after which viral particles would be swallowed and not replaced by new viruses.
The data presented in the current study reveal a large inter-individual variation in the observed response. This high variability can partly be due to age differences among subjects, as a trend was found in higher basal viral load in older individuals. The use of anti-viral treatments could of course affect the response, but this effect is probably limited in our dataset because only six out of 84 individuals were under antiviral drug treatment. In addition, although specific instructions were given to participants on how to perform the mouthwash, we cannot discard that the strength or intensity of the procedure was different for different individuals and that this mouthwash performance influenced inter-individual variability in treatment response. Apart from methodological effects, the temporal variability in viral load, which was also observed in the control group, could be influenced by naturally occurring changes in the shedding of viruses from other body niches like the nasopharynx27.
The randomized assignment to the different treatments produced groups that did not vary in age or sex, which are factors known to affect disease severity, thereby facilitating inter-group comparisons. Interestingly, a considerable number of patients underwent reduced viral loads through time in the control group, highlighting the importance of a placebo branch to extract valid conclusions. In three of the four previous clinical studies, for example, no control group was included when testing the effect of chlorhexidine (n = 228), H2O2 (n = 1229) or povidone-iodine (n = 430) and the effect observed could have therefore been taking place also in the absence of a mouthwash. In a fourth clinical study to test the effect of three oral mouthwashes (n = 6 on each treatment group), a control branch was included with only two individuals, which happened to increase viral load after the mouth rinse31. Thus, when the changes in viral load after the mouthwashes were compared to those observed in the control group, a small relative benefit could be deduced, but the viral load in all individuals when compared to their corresponding basal values did not change after the mouthwash. In our dataset, up to half of the patients in the control group decreased their viral load by 50% after 2 h (Fig. 5), underlining the importance of a control branch with an equivalent sample size to the other branches. In addition, the lack of efficient antiviral drugs that could serve as a positive control in our study limits the evaluation of mouthwashes. This may change in the near future as a continuous, unprecedented effort is being performed aiming to develop efficient treatments against SARS-CoV-2, for example by drug repositioning32. Thus, although these drugs are mostly envisaged for systemic treatment, some may be suitable for topic application through mouthwashes.
Although our data reveal that oral mouthwashes have no significant effect on SARS-CoV-2 viral load as measured by qPCR, the possibility remains that those viral particles detected after the treatment could be non-viable. This was proposed by Gottsauner and collaborators in their study with a H2O2 mouthwash but unfortunately, viral cultures in their samples could only be obtained from one baseline sample in one of the patients, and this hypothesis could not therefore be tested29. The possibility of a mouthwash effect on viral infectivity would explain why the virucidal activity appears to increase with time even in the H2O2 treatment, which is a compound without a prolonged effect, as in the absence of viral colonization from the nasopharynx, oral viral replication would be diminished. Thus, there is an urgent need of doing viral culture experiments with saliva samples collected after mouthwash use to confirm viral viability. Although RNA is a highly unstable molecule and therefore its integrity in saliva is expected to be quickly compromised, the possibility that the qPCR protocol could be detecting RNA from dead viral particles is supported by the fact that persistent viral amplification by PCR has been documented in upper respiratory or fecal samples weeks after the COVID-19 symptoms have disappeared33,34.
The potent virucidal effects against different coronavirus detected in vitro resulted in oral mouthwashes being recommended during the pandemic and that they are routinely used around the world in different clinical situations and before dental procedures to reduce infection risk by SARS-CoV-21,19. However, if the action of oral antiseptics in vivo is limited, their use could provide a false sense of security. Thus, we recommend that strict preventive measures should stand in place in clinical settings until viral infectivity studies in cell lines with post-mouthwash saliva are performed. These viral culture studies with oral samples after the use of oral mouthwashes are warranted.
Data availability
Clinical data has been previously published in the clinical trial database (https://clinicaltrials.gov/ct2/show/NCT04707742; Identifier: NCT04707742). Additional data are available from the authors upon request.
References
Meng, L., Hua, F. & Bian, Z. Coronavirus Disease 2019 (COVID-19): Emerging and future challenges for dental and oral medicine. J. Dent. Res. 99, 481–487 (2020).
Mohan, S. V., Hemalatha, M., Kopperi, H., Ranjith, I. & Kumar, A. K. SARS-CoV-2 in environmental perspective: Occurrence, persistence, surveillance, inactivation and challenges. Chem. Eng. J. 405, 126893 (2021).
Whitworth, J. COVID-19: A fast evolving pandemic. Trans. R. Soc. Trop. Med. Hyg. 114, 227–228 (2020).
Chan, J. F. et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 395(10223), 514–523 (2020).
Wang, W. et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA J. Am. Med. Assoc. https://doi.org/10.1001/jama.2020.3786 (2020).
Xu, H. et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 12(1), 1–5 (2020).
Xu, J., Li, Y., Gan, F., Du, Y. & Yao, Y. Salivary glands: Potential reservoirs for COVID-19 asymptomatic infection. J. Dent. Res. 99(8), 989 (2020).
Sabino-Silva, R., Jardim, A. C. G. & Siqueira, W. L. Coronavirus COVID-19 impacts to dentistry and potential salivary diagnosis. Clin. Oral Investig. 24(4), 1619–1621 (2020).
Popkin, D. L. et al. Cetylpyridinium chloride (CPC) exhibits potent, rapid activity against influenza viruses in vitro and in vivo. Pathog. Immun. 2(2), 252–269 (2017).
Eggers, M., Koburger-Janssen, T., Ward, L. S., Newby, C. & Müller, S. Bactericidal and virucidal activity of povidone-iodine and chlorhexidine gluconate cleansers in an in vivo hand hygiene clinical simulation study. Infect. Dis. Ther. 7(2), 235–247 (2018).
Wood, A. & Payne, D. The action of three antiseptics/disinfectants against enveloped and non-enveloped viruses. J. Hosp. Infect. 38, 283–295 (1998).
Eggers, M., Koburger-Janssen, T., Eickmann, M. & Zorn, J. In vitro bactericidal and virucidal efficacy of povidone-iodine gargle/mouthwash against respiratory and oral tract pathogens. Infect. Dis. Ther. 7(2), 249–259 (2018).
Eggers, M., Eickmann, M. & Zorn, J. Rapid and effective virucidal activity of povidone-iodine products against Middle East Respiratory Syndrome Coronavirus (MERS-CoV) and Modified Vaccinia Virus Ankara (MVA). Infect. Dis. Ther. 4(4), 491–501 (2015).
Kampf, G., Todt, D., Pfaender, S. & Steinmann, E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 104(3), 246–251 (2020).
Meyers, C. et al. Lowering the transmission and spread of human coronavirus. J. Med. Virol. https://doi.org/10.1002/jmv.26514 (2020).
Steinhauer, K. et al. Comparison of the in vitro-efficacy of different mouthwash solutions targeting SARS-CoV-2 based on the European Standard EN 14476. bioRxiv. https://doi.org/10.1101/2020.10.25.354571 (2020).
Meister, T. L. et al. Virucidal efficacy of different oral rinses against severe acute respiratory syndrome coronavirus 2. J. Infect. Dis. 222(8), 1289–1292 (2020).
Anderson, D. E. et al. Povidone-iodine demonstrates rapid in vitro virucidal activity against SARS-CoV-2, the virus causing COVID-19 disease. Infect. Dis. Ther. 9(3), 669–675 (2020).
O’Donnell, V. B. et al. Potential role of oral rinses targeting the viral lipid envelope in SARS-CoV-2 infection. Function (Oxf) 1(1), zqaa002 (2020).
Paz, S., Mauer, C., Ritchie, A., Robishaw, J. D. & Caputi, M. A simplified SARS-CoV-2 detection protocol for research laboratories. PLoS One 15(12), e0244271. https://doi.org/10.1371/journal.pone.0244271.PMID:33338082;PMCID:PMC7748277 (2020).
WHO. Molecular assays to diagnose COVID-19: Summary table of available protocols. 24 January 2020. https://www.who.int/publications/m/item/molecular-assays-to-diagnose-covid-19-summary-table-of-available-protocols
Corman, V. M. et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 25(3), 2000045 (2020).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2020).
Statkute, E., Rubina, A., O’Donnell, V. B. & Stanton, D. W. T. R. J. The virucidal efficacy of oral rinse components against SARS-CoV-2. In Vitro https://doi.org/10.1101/2020.11.13.381079 (2020).
Kariwa, H., Fujii, N. & Takashima, I. Inactivation of SARS coronavirus by means of povidone-iodine, physical conditions and chemical reagents. Dermatology 212(Suppl 1), 119–123 (2006).
Duke, S. & Forward, G. The conditions occurring in vivo when brushing with toothpastes. Br. Dent. J. 152, 52–54 (1982).
Sun, J. et al. The kinetics of viral load and antibodies to SARS-CoV-2. Clin. Microbiol. Infect. 26(12), 1690.e1-1690.e4 (2020).
Yoon, J. G. et al. Clinical significance of a high SARS-CoV-2 viral load in the saliva. J. Korean Med. Sci. 35(20), e195 (2020).
Gottsauner, M. J. et al. A prospective clinical pilot study on the effects of a hydrogen peroxide mouthrinse on the intraoral viral load of SARS-CoV-2. Clin. Oral Investig. 24(10), 3707–3713 (2020).
Martínez, L. L. et al. Is povidone iodine mouthwash effective against SARS-CoV-2? First in vivo tests. Oral Dis. https://doi.org/10.1111/odi.13526 (2020).
Seneviratne, C. J. et al. Efficacy of commercial mouth-rinses on SARS-CoV-2 viral load in saliva: Randomized control trial in Singapore. Infection 14, 1–7 (2020).
Riva, L. et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature 586(7827), 113–119 (2020).
Xu, Y. et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 26(4), 502–505 (2020).
Herrera, D., Serrano, J., Roldán, S. & Sanz, M. Is the oral cavity relevant in SARS-CoV-2 pandemic?. Clin. Oral Investig. 24(8), 2925–2930. https://doi.org/10.1007/s00784-020-03413-2 (2020).
Acknowledgements
We thank Gonzalo Díaz Tapia, Raúl Rubio Yangüas, Eduard Teixeira de Freitas, Christian Ruminot Lehmann, Gabriel Álvarez Curro, Fernando González Galán, Hander Guillermo Acosta Díaz, Mireya Bonet Loscertales, Daniella Laguado Bulgheroni, María José Hernández García and Jeanette Saenz Piñones for help and assistance during patient recruitment and sample collection. We also thank Elena Buetas for help and assistance during RNA extraction. The authors are indebted to the FISABIO Foundation for providing reagents and plastic materials for performing sample collection and analysis. We also thank Drs Vanessa Blanc and Rubén León for critically revising an earlier version of the manuscript. We want to thank DentAid SA for providing CHX and CPC mouthwashes free of charge.
Author information
Authors and Affiliations
Contributions
A.M., M.V.M., V.A. and M.D.F. conceived and planned the study. A.S.B., E.G.V., M.P.T. and M.J.F.G. coordinated the clinical study in the different hospitals. M.D.F. coordinated logistics and arranged materials and reagents. Y.M.B., V.A., A.C.G., J.M.S.C., I.A.R., J.M.V.A., C.C.E., A.L.V. and D.S.A. informed patients and collected samples. M.D.F. and S.G.E. extracted RNA. A.A. and M.D.F. did the statistical analysis. M.D.F. made a first version of the figures with input from AM. X.L.L. coordinated RT-PCR work. S.G.E. performed RT-PCR work. AM wrote the first draft of the report with input from M.D.F., X.L.L., Y.M.B., A.S.B. and M.V.M. M.V.M. reviewed the literature. M.D.F., S.G.E. and A.M. have accessed and verified the data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ferrer, M.D., Barrueco, Á.S., Martinez-Beneyto, Y. et al. Clinical evaluation of antiseptic mouth rinses to reduce salivary load of SARS-CoV-2. Sci Rep 11, 24392 (2021). https://doi.org/10.1038/s41598-021-03461-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-03461-y
This article is cited by
-
Efficacy of mouthwash on reducing salivary SARS-CoV-2 viral load and clinical symptoms: a systematic review and meta-analysis
BMC Infectious Diseases (2023)
-
Efficacy of three antimicrobial mouthwashes in reducing SARS-CoV-2 viral load in the saliva of hospitalized patients: a randomized controlled pilot study
Scientific Reports (2023)
-
Preprocedural mouthwashes for infection control in dentistry—an update
Clinical Oral Investigations (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.