Abstract
At present, only a single Rhizopus species, R. microsporus, can be found in fresh tempeh produced in Java, Indonesia. The loss of diversity of Rhizopus in tempeh has been associated with the widespread use of commercial tempeh starter in Indonesia since the 2000s. However, the identities of the previous Rhizopus strains associated with tempeh, which have been preserved in a culture collection in Indonesia, have not been verified. The present study aimed to verify the identities of 22 Rhizopus strains isolated from tempeh produced using the traditional tempeh starters from the 1960s to the 2000s. Phylogenetic analysis based on the ITS regions in the rRNA gene sequence data, revealed that the Rhizopus strains belonged to the species R. arrhizus (five strains); R. delemar (14 strains); and R. microsporus (three strains). Verification of the identities of these Rhizopus strains in the present study confirmed the loss of diversity of Rhizopus species in tempeh produced in Indonesia, particularly in Java. Our findings confirmed that the morphological changes in Rhizopus species isolated from tempeh as a result of centuries of domestication.
Similar content being viewed by others
Introduction
Tempeh is a soybean-based fermented food that is popular worldwide. It is regarded as a good source of protein and is easily digestible food. Tempeh has been a very popular traditional fermented food in Indonesia for many centuries that reported by Nout and Kiers in 20051. It is a very important protein source in the Javanese diet2. The production of traditional tempeh is thought to have started in Indonesia in the early 1600s3. It originated from Central or East Java. English word tempeh comes from Indonesian “tempe”. The word “tempe” originated from Central Java, Indonesia. Serat Centhini is the first known manuscript in Java to mention this word “tempe”3. As written in “The History of Tempeh”3, traditionally since at least 1875, the name for this food in Indonesia was written témpé, with various accents being used. Then in August 1972, when Indonesia modernized its language, the accents were dropped and the word came to be spelled tempe (still pronounced TEM-pay). In English and other European languages, the word has come to be spelled “tempeh,” the final “h” being added to prevent the word from being pronounced “temp”. Hendrik Coenraad Prinsen Geerligs was the first European man who use the spelling tempeh in German article in 18963,4. Van Veen and Schaefer in 19505 were the first scientists used term tempeh in an English language article. Then Steinkrauss et al.6 were the first in the US. Since then, the word has consistently been spelled tempeh in European languages.
The taxonomy of the genus Rhizopus (Ehrenb. 1821) has undergone dramatic changes, especially in the last 40 years. It has been significantly changed from traditional7,8,9 to molecular approaches10,11,12,13,14. Since the description of R. arrhizus by Fischer in 1892 (Fischer 1892), hundreds of species have been described based on discrete morphological and physiological features7,8. In 1965, Inui et al.7 examined 449 Rhizopus species in their monographs. Almost 20 years later, Schipper8 and Schipper and Stalpers9 revised the classification of Rhizopus based on comprehensive morphological characters, temperature tolerance and mating. They separated the genus into three groups—R. microsporus, R. stolonifer, and R. arrhizus (= oryzae), with the re-integration of many species. Schipper8 synonymized 29 species with R. arrhizus (= oryzae). The group classification of Schipper8 and Schipper and Stalpers9 are widely accepted. In 1985, Ellis15 concluded that R. arrhizus, Amylomyces rouxii, and R. delemar are conspecific based on DNA renaturation experiments and proposed to accommodate them in three varieties.
Abe et al.16 established the first molecular phylogeny of Rhizopus based on three molecules of the ribosomal RNA-encoding DNA (rDNA) and confirmed the same taxonomic grouping microsporus-group, stolonifer-group, and R. arrhizus. Liu et al.11 combined analyses of the ITS regions of rRNA and pyrG genes data and only allowed eight species to be distinguished: R. americanus, R. caespitosus, R. homothallicus, R. microsporus, R. reflexus, R. schipperae, R. sexualis, and R. stolonifer. The remaining two morphologically distinct species, R. arrhizus and R. niveus, formed an unresolved cluster. They considered A. rouxii as synonymous of R. arrhizus. In the same year, Zheng et al.12 revised the monograph of Rhizopus and organized the genus into 10 species and seven varieties by reanalyzed the data from Liu et al.11 along with morphological data. Abe et al.17 used rDNA ITS, actin-1, and translation elongation factor 1a (TEF-1a) sequences to confirm the eight-species division of Rhizopus. They showed that the R. microsporus complex consisted of a single species. Dolatabadi18 investigated the species boundaries of R. microsporus using ITS, ACT, and TEF markers in combination with mating tests, morphology, physiology, ecology, geography, and MALDI-TOF MS data, and reduced the six varieties of R. microsporus (vars microsporus, azygosporus, chinensis, oligosporus, rhizopodiformis, and tuberosus; Liu et al.11) to synonyms. The widely held suggestion that the strains with the morphology of R. oligosporus represent a separate species that can be found just in fermented food sources i.e. tempeh should thus be rejected because positive mating results have been found between all varieties of R. microsporus, therefore all strains concluded as a single species, R. microsporus.
The species boundaries among R. arrhizus and R. delemar was studied by Abe et al.10 and Gryganskyi et al.19, they show clearly that R. arrhizus and R. delemar represent taxonomic entities that either deserves the rank of varieties or species. Dolatabadi et al.13 considered R. arrhizus consisted of two varieties, e.g. var. arrhizus and var. delemar, based on sequence data of multi-locus studies as well as amplified fragment length polymorphism (AFLP) and mating experiment. They found there is still zygospore formation between members of both varieties, although their number is reduced suggesting that the mating barrier is not complete yet. There is also a nomenclatural issue with arrhizus. Rhizopus arrhizus was described first, but R. oryzae has been used by most authors. Schipper8 treated R. arrhizus as a doubtful species. Ellis et al.15 took up the name R. arrhizus again by designating NRRL 1469 as ex-neotype strain of R. arrhizus. Zheng et al.12 in their monograph on Rhizopus preferred R. arrhizus over R. oryzae. Similarly, Dolatabadi et al.13 also use the name R. arrhizus based on the protologue of the first described R. arrhizus. Gryganskyi et al.14, also use the name R. arrhizus in their classification of the genus Rhizopus using phylogenomic approaches based on 192 orthologous genes. They classified Rhizopus strains into four species, e.g. R. microsporus, R. stolonifer, R. arrhizus, and R. delemar. In the present study, we followed this classification system (taxonomy of Rhizopus sensu Gryganskyi et al.14).
Earlier studies on tempeh before the Second World War by Dutch microbiologists3 revealed that tempeh in Java was fermented with R. arrhizus. The first scientific report on tempeh was published in 1896 and was written by the Dutchman H.C. Prinsen Geerligs, who lived in Java3,4. He stated that tempeh was fermented by the mold R. arrhizus. The same species was also mentioned by van Veen and Schaefer5. Some reports around the 1960s20 also found that R. arrhizus, was the dominant species from highly preferred tempeh samples in Java, such as tempeh “Malang” and tempeh “Purwokerto”.
In the 1960s, the cottage-scale tempeh industry spread to all regions in Indonesia by using traditional methods for tempeh production and producing tempeh with various local tastes and flavors. The method for preparing the inoculum (tempeh starter) varied based on locality. In the traditional process, the previous batch of tempeh or the mold grown and dried on Hibiscus tiliaceus leaves (daun Waru) was used as the tempeh inoculum. These leaves are used to carry tempeh inoculum as natural starters (known as usar in Indonesia). Following this, beans were wrapped using banana or other large leaves and finally placed in a warm location to ferment for 1 or 2 days5,21 Tempeh has a pleasant odor and a slight cheese-like flavor6. In the earlier study of tempeh by a group of scientists from Cornell University, USA, around the 1960s, revealed that R. arrhizus to be the essential microorganism isolated from Indonesian tempeh scrapings6.
The interest in tempeh produced in Indonesia rapidly increased among Indonesian scientists after the late 1960s. Several Rhizopus species associated with tempeh produced using the traditional process in Indonesia have been reported by Indonesian mycologists. Dwidjoseputro and Wolf22 reported R. arrhizus, R. microsporus, and R. stolonifer to be associated with tempeh and tempeh starters in Malang, Surakarta, and Jakarta.
Extensive research on tempeh was also conducted in the USA since the 1960s by groups of microbiologists and food scientists2,5,6,23. An Indonesian microbiologist, Ko Swan Djien from the Bandung Institute of Technology, West Java, brought tempeh samples from Java to the laboratory of Dr. Hesseltine at NRRL, USA, in 1961 in order to study tempeh fermentation23. Forty Rhizopus strains were isolated from these tempeh samples. These strains belonged to species: R. achlamydosporus, R. arrhizus, R. formosaensis, R. microsporus (= R. oligosporus), and R. stolonifer23. Hesseltine23 stated that only R. arrhizus and R. microsporus (= R. oligosporus) were commonly used to produce tempeh. Wang and Hesseltine24 reported the best strain for producing tempeh from wheat and soybeans was R. microsporus (= R. oligosporus) NRRL 2710. Since they claimed that R. microsporus (= R. oligosporus) as the best tempeh mould, this species was then used by many Indonesian microbiologists for their study on tempeh (Gandjar and Santoso)20.
Large-scale commercial tempeh production began in the 1980s with the aim of guaranteeing a good tempeh product. The first commercial inoculum for tempeh, which consisted of mixed cultures of R. arrhizus and R. microsporus, was developed by the Chemistry Institute-Indonesian Institute of Sciences (LKN-LIPI) and the Cooperative of Tempeh and Tofu Producers of Indonesia (KOPTI) in 1985; they then distributed it to tempeh producers20. The next generation of commercial tempeh starter developed by LIPI was Raprima, containing only a single species, R. microsporus. Raprima has been produced by PT. Aneka Fermentasi Industri, Bandung, Indonesia, since 2001, and is widely used in tempeh fermentation in Indonesia and abroad.
Taxonomy of Rhizopus strains obtained from tempeh in Indonesia has been well studied by many scientists in abroad and those strains are well maintained at Centraalbureau voor Schimmelcultures KNAW (currently hosted by Westerdijk Institute) (The Netherlands), others in Mycothèque de l'Université catholique de Louvain (MUCL) (Belgium) and Northern Regional Research Laboratory (NRRL) (USA). On the other side, it is difficult to trace the genetic diversity of Rhizopus spp. previously used for tempeh production that preserved in culture collections in Indonesia, because Rhizopus cultures were rarely collected or were never preserved properly in culture collections in Indonesia. Their representation within sequence database is lacking and their molecular study has never been reported.
One of the authors (I. G.), collected Rhizopus strains and accumulated hundreds of strains from almost all regions in Indonesia since the 1960s. These Rhizopus strains have been preserved in the Universitas Indonesia Culture Collection (UICC), Depok, Indonesia. It is the only culture collection in Indonesia that maintains the Rhizopus strains isolated from tempeh produced using the traditional tempeh starters. Because of a lack of budget, this collection of Rhizopus strains was originally maintained only as living cultures; therefore, many strains have been lost. Since 2012, the strains have been maintained using a long-term preservation method, the liquid drying (L-drying) method, after financial support was obtained from the Society for Applied Microbiology of the United Kingdom (SfAM UK) Endangered Collection Grant.
At present, only 127 Rhizopus strains available from those isolated from tempeh produced using traditional starters (1960s–2000s) that are preserved in UICC. The molecular identification of these strains was not performed until 2017, when we sequenced 15 strains of Rhizopus from UICC based on the ITS regions of ribosomal RNA (rRNA) gene25,26,27. The present study aimed to sequence another 22 strains the Rhizopus strains from UICC based on the ITS regions of ribosomal RNA (rRNA) gene, to provide the accurate taxonomic identity of Rhizopus strains that were isolated from tempeh produced using traditional tempeh starters (1960s–2000s).
Materials and methods
Fungal strains and preservation methods
All Rhizopus strains were obtained from UICC, Center of Excellence for Indigenous Biological Resources-Genome Studies, FMIPA Universitas Indonesia, Depok, Indonesia. UICC maintains 127 Rhizopus spp. strains that originated from various types of tempeh (e.g., tempe kedelai, tempe gembus, tempe kopra, tempe kedelai hitam, tempe koro, tempe koro wedus, tempe benguk, tempe kapok, and tempe lamtoro) and traditional tempeh starters (e.g. laru daun waru and laru daun pisang) and were isolated from the 1960s to the 2000s. The origin of the 22 strains used in the present study and their year of isolation are provided in Table 1. Tempeh and tempeh starter samples were obtained from many regions in Indonesia, particularly those in Java, Kalimantan, Nusa Tenggara, Papua, Sulawesi, and Sumatera. The regions were as follows: Java (Jakarta, Cilacap, Magelang, Malang, Pacitan, Salatiga, Semarang, Solo, Surabaya, Tegal, Wonogiri, Wonosari, and Yogyakarta), Kalimantan (Balikpapan and Palangkaraya), Nusa Tenggara (Mataram), Papua (Wamena), Sulawesi (Manado), and Sumatera (Banda Aceh). Three samples per place of tempeh were collected. Long-term preservation of the cultures was performed using Liquid-drying method in lyophilized tubes and in glycerol solution (at − 80 °C).
Fungal growth medium
Potato dextrose agar (PDA, Difco) was used as the growth medium for stock cultures and working cultures, for purifying the cultures, and for preparing DNA isolation, while 4% malt extract (Acumedia) agar (Difco) (MEA 4%) was used as the growth medium for morphological characterization. Macroscopic and microscopic observations of colonies, size and shape of spores were performed using a microscope [ZEISS Primostar Axio-Cam]. Monographs of Rhizopus were used as references for comparing the morphological data8,9,12.
Fungal identification
The extraction of fungal genomic DNA was performed using the PrepMan™ Ultra Kit (Applied Biosystems, Foster City, CA) as described previously25,26,27. The ITS regions in the fungal rRNA gene were amplified using ITS universal fungal primers, namely ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′)28. PCR was performed under the following conditions: 95 °C for 1 min; 40 cycles at 94 °C for 1 min; 60 °C for 1 min; and 72 °C for 1 min; and a final extension cycle at 72 °C for 5 min. The PCR product was purified with a QIAquick Purification Kit (Qiagen). For sequencing of the ITS regions, the primers ITS5 and ITS4 were used)28. Sequencing reactions were conducted using a BigDye Terminator v3.1 Cycle Sequencing Ready Reaction Kit (Applied Biosystems) Foster City, CA, USA) following the manufacturer’s instructions. The gel electrophoresis and data collection were performed on ABI Prism 310 Genetic Analyzer (Applied Biosystems), or the PCR products of the ITS regions of rDNA were sent to 1st BASE (Malaysia) for sequencing. The fungal strains were identified according to their sequence homology with fungal sequences obtained from the GenBank DNA database hosted by NCBI (http://blast.ncbi.nlm.nih.gov) using the BLAST search tool29.
Phylogenetic analyses
Sequence assembly and editing were performed using ChromasPro ver.1.7.7, while sequence alignment and phylogenetic tree construction were performed using ClustalX and MEGA 7, respectively30,31. Phylogenetic trees were constructed using the neighbor-joining (NJ)32, minimum evolution (ME)33, and maximum likelihood (ML)34 methods with 1000 bootstrap replications35. Evolutionary distances in the NJ method were computed using the Kimura 2-parameter method36. Phycomyces blakesleeanus NBRC 5823 was used as an outgroup. The identity of each fungal strain to the species level was verified according to the currently described species concept of the genus Rhizopus28. The ITS rRNA gene sequence accession numbers of the Rhizopus strains identified in the present study (LC514296–LC514335) have been deposited in the DNA Database of Japan (DDBJ, https://www.ddbj.nig.ac.jp) (Table 1).
Results
Re-identification of Rhizopus strains isolated from tempeh
A homology search was performed with the BLAST tool in DDBJ using the ITS regions in the rRNA gene sequence data of the fungal strains as a query; the results indicated that all the 22 strains in this study had very high homology (99–100%) to their closest species (Table 1).
Phylogenetic analyses
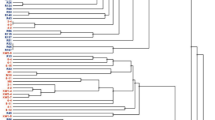
Phylogenetic analyses using the NJ method and two other methods (ME and ML methods; data not shown) based on 70 OTUs revealed that all members of the genus Rhizopus were grouped into three major clades: R. arrhizus–R. delemar, R. microsporus, and R. stolonifer clades (Fig. 1). Analysis of the phylogenetic tree consisting of 22 strains determined in this study and 15 strains from our previous studies25,26,27, revealed that the strains isolated in the different regions in Indonesia before the commercial tempeh starters have been widely used in Indonesia belonged to three species: R. arrhizus (five strains), R. delemar (14 strains), and R. microsporus (three strains). All the strains isolated from Java in the 1960s–1970s belonged to R. arrhizus and R. delemar. The other strains isolated outside Java in 1996–2003 (Aceh, Balikpapan, Manado, Mataram, Palangkaraya, and Wamena) belonged to R. arrhizus, R. delemar, and R. microsporus.
Phylogenetic tree of 37 Rhizopus strains from tempeh based on ITS rRNA gene sequence data: 22 strains determined in this study (indicated in bold face) and 15 strains from our previous works25,26,27. Asterisks (*) indicated Rhizopus strains from tempeh isolated in Java in 1960s–1980s. The tree was constructed using the NJ method32. Bootstrap values less than 50% are not shown.
Morphological characteristics
Morphological characterization was performed after 3 days incubation on 4% MEA to confirm the identities of the strains based on molecular identification. Light microscopic examination showed that sporangiospores of R. arrhizus (UICC 10, UICC 36, UICC 85, UICC 116, UICC 119, UICC 135) are angular, globose, subglobose, and irregular, striated, up to 8 µm in length; R. delemar (UICC 121, UICC 67, UICC 27, UICC 26, UICC 40, UICC 39) are angular, globose, subglobose, and irregular, striated, up to 8 µm in length; and R. microsporus (UICC 500, UICC 531) are globose to subglobose, some are large and irregular, smooth, up to 9 µm maximum diameter (Fig. 2).
Sporangiospores of Rhizopus from tempeh as seen under light microscope. (A–F) R. arrhizus UICC 10, UICC 36, UICC 85, UICC 116, UICC 119, UICC 135; sporangiospores are angular, globose, subglobose, and irregular, striated. (G–L) R. delemar UICC 121, UICC 67, UICC 27, UICC 26, UICC 40, UICC 39, sporangiospores are angular, globose, subglobose, and irregular, striated. (M–R) R. microsporus UICC 500, UICC 531, sporangiospores are globose to subglobose, some are large and irregular, smooth. Three days on 4% MEA. Bars = 10 μm.
Light microscopic examination revealed a peculiar form of sporangiophores in Rhizopus strains. Rhizopus arrhizus, R. delemar, and R. microsporus strains possessed sporangiophores with unique morphological characteristics. These sporangiophores showed swelling and branching or were sometimes forked at the middle, upper-half, or apical position. The number of sporangia in a single sporangiophore varied between two and six, and they were arranged in a verticillate pattern (Fig. 3).
Branching sporangiophores with multi-sporangia of Rhizopus from tempeh: (A,B) R. arrhizus UICC 36, UICC 120, (C,D) UICC 10, (E) UICC 119; (F,G R. delemar UICC 40, UICC 26; (H) R. microsporus UICC 539. Seven days on 4% MEA. (G) photo credit to Vebliza47. Scale bar = 10 μm.
Discussion
In the present study, we accurately identified the Rhizopus strains from tempeh isolated from traditional starters, before the widespread used of commercial tempeh starter in Indonesia. Based on the current taxonomy of Rhizopus14, the 22 Rhizopus strains determined in this study belonged to three species: R. microsporus (three strains), R. delemar (14 strains), and R. arrhizus (five strains) (Table 1). Based on ITS rRNA sequence data, the identities of many Rhizopus strains isolated from tempeh were rectified (Table 1). For example, R. arrhizus was changed to R. delemar, R. oryzae to R. arrhizus, R. cohnii to R. arrhizus, R. microsporus to R. delemar, R. oryzae to R. delemar, and R. oligosporus to R. delemar.
All Rhizopus strains isolated from tempeh belonged to three major groups in the phylogenetic tree: R. arrhizus, R. delemar, and the R. microsporus complex (Fig. 1). The resolution of sequences from the ITS regions was sufficient for identifying these Rhizopus species. The tree topology as seen in Fig. 1 was in agreement with the molecular taxonomic studies of Rhizopus13,17,37 and generally congruent with the tree topology based on phylogenomic approaches as inferred from a dataset of 192 orthologous genes14. The tree in Fig. 1 clearly indicates that R. delemar is a sibling or cryptic species of R. arrhizus and that they are very closely related, as evidenced by a 95% bootstrap value. The results from morphological characterizations (Fig. 2) also confirmed the results from molecular identification that all Rhizopus strains isolated from tempeh belong to R. arrhizus, R. delemar, and the R. microsporus.
Our group member (I.G.), isolated at least five species, namely R. arrhizus, R. cohnii, R. microsporus, and R. stolonifer from various tempeh products in Indonesia (mainly Java) in the 1960s–2000s. Previous identification of 127 Rhizopus strains isolated from tempeh based on morphological and physiological data revealed that R. arrhizus and R. microsporus were the most common species isolated from various tempeh products in Java in the 1960s–1970s (data not shown). These species identification results are in agreement with the findings of Hesseltine23, who reported Rhizopus species that were commonly used to produce tempeh: R. arrhizus and R. microsporus.
The results of molecular identification (Table 1) and the phylogenetic tree based on the ITS regions in the rRNA gene sequence data of Rhizopus strains isolated from tempeh before the use of commercialized tempeh starter revealed that the strains belonged to three species: R. delemar, R. arrhizus, and R. microsporus (Fig. 1). It is clear that both R. delemar and R. arrhizus are the most commonly isolated Rhizopus species from tempeh in Java. The identity of tempeh molds verified in the present is in accordance with the findings of Arbianto38, who reported that two Rhizopus species are involved in the traditional process for tempeh production. In this process, R. arrhizus are a strong amylase, protease, and pectinase producers. After the temperature increases, R. microsporus, which can better withstand higher temperatures (37–40 °C), completes the process. Another study, Samson et al.39 also reported that R. arrhizus and R. microsporus were the most commonly isolated Rhizopus species from 110 commercial tempeh products in the Netherlands. Tempeh was introduced to the Netherlands by immigrants from Indonesia3.
As shown in Fig. 1, two species, R. arrhizus and R. delemar, were most commonly species found in tempeh in Java in the 1960s–1970s and in other regions in Indonesia before the 2000s. These species were originated from the leaves of Hibiscus tiliaceus, the leaves that were used as traditional tempeh starter (usar) in that period in Indonesia. Nout et al.40 found that R. arrhizus and R. microsporus were the predominant epiphytic molds on Hibiscus tiliaceus leaves (daun Waru) in Indonesia.
In the 1960s, I. G. isolated many Rhizopus strains from tempeh “Malang” and tempeh “Purwokerto” and found that R. arrhizus to be dominant tempeh molds from tempeh “Malang” and tempeh “Purwokerto”. However, 30 years later (in the 1990s), on isolating Rhizopus species from tempeh “Malang” and tempeh “Purwokerto”, she found R. microsporus to be dominant20.
These days, Rhizopus species other than R. microsporus are rarely found in tempeh in Indonesia. Hartanti et al.41 reported that 35 out of 36 Rhizopus strains isolated from fresh tempeh from 26 locations (mainly in Java) in Indonesia in 2012 and 2013 belonged to R. microsporus. Only one isolate from Sulawesi belonged to R. delemar. In a recent study on the genetic diversity of Rhizopus species isolated from the traditional inoculum of tempeh (daun Waru) all isolates were found to belong to a single species, R. microsporus42. In other surveys43,44, tempeh producers in Indonesia generally do not use their own traditional starters anymore. They use commercial starters, such as Raprima which can be purchased online and sold abroad. The use of commercial of tempeh starters is not limited to Java; it has spread to other regions in Indonesia since the 2000s.
Interestingly, Anggriawan42 performed RAPD typing of 471 pure Rhizopus isolates obtained from 247 samples of fresh tempeh and its inoculum from 16 provinces in Indonesia in 2013–2015 and found that R. arrhizus, R. delemar, and R. stolonifer were present in the samples collected outside Java, while the R. microsporus complex, was present in the samples collected within Java. These findings indicated that some tempeh producers outside Java still use the traditional process for tempeh production. Therefore, other Rhizopus species could be detected.
Sukardi et al.45 reported that the use of commercial tempeh starters containing R. microsporus is not suitable for tempeh production in Malang, East Java (which is located on a cool plateau). The R. microsporus inoculum results in the production of a less compact tempeh cake, which sometimes has an alcoholic smell. Moreover, the optimum growth temperature of R. arrhizus is lower than that of R. microsporus; therefore, Rhizopus arrhizus is more suitable for tempeh production in Malang.
Based on molecular evidence from ITS rRNA gene data, many strains validated in the present study had been misidentified (Table 1). The identification of Rhizopus species from tempeh based on morphological observation is not easy, especially within the R. arrhizus complex and R. microsporus complex. The high similarity in their morphological characteristics often leads to misidentification. Rhizopus arrhizus and R. delemar are sibling species and morphologically identical10. Despite the close genetic relationships between members of the R. arrhizus sensu lato and among the members of the R. microsporus complex, Zheng et al.12 mentioned in their monograph that these species have already undergone marked changes in their morphology while adapting themselves to their artificial environment, as fermentative agents over many generations. These morphological changes make the identification of these species very difficult based on their descriptions provided in a previous monograph by Schipper8. Therefore, Zheng et al.12 produced a monograph of Rhizopus based on the sporangial morphology, making it one of the most important references for the classification of the genus Rhizopus. Specifically, in the synoptic key to the classification of Rhizopus, sporangiophores exhibit simple, sometimes forked, branching at the upper portion and at the base and are rarely verticillate. Swelling is common, mostly at the middle or apical portion. As shown in Fig. 3, some R. arrhizus, R. delemar, and R. microsporus strains isolated from tempeh exhibited the branching and swelling of sporangiophores at the middle and upper portions. Multiple sporangia were observed at the upper or apical portion of sporangiophores and were verticillate.
In the present study, using a light microscope, we identified unique characteristics in some Rhizopus strains isolated from tempeh; these included the presence of sporangiophores with more than two branches or a single sporangiophore with more than two sporangia (multiple sporangia). Multiple sporangia were observed in R. arrhizus, R. delemar, and R. microsporus (Fig. 3). Similarly, Jennessen et al.46 reported double sporangia in R. microsporus CBS 112.586; however, a case of multiple sporangia has never been reported in this species. Our findings confirmed that the morphological changes in Rhizopus species isolated from tempeh as a result of centuries of domestication. Zheng et al.12 reported morphological changes in other Rhizopus species of economic importance, such as R. microsporus and R. stolonifer.
The present study verified the identity of Rhizopus strains used 40–50 years ago to produce tempeh using the traditional process in Indonesia. Phylogenetic analyses revealed that R. arrhizus and R. delemar were commonly found in various locations in Java 40–50 years ago (Table 1). However, neither species is found today because of the widespread use of the commercial tempeh starter Ragi Raprima® containing only R. microsporus in Indonesia.
The loss of genetic diversity of Rhizopus species in tempeh has changed the taste and flavor of tempeh. We do feel the impact of using commercialized inoculum in Java. Good–quality tempeh “Malang” and tempeh “Purwokerto”, which contain R. arrhizus complex, cannot be found anymore. The white wooly appearance and pleasant aroma of the famous tempeh Malang and tempeh Purwokerto have been replaced by plain white tempeh, because the aforementioned species have been replaced by R. microsporus, which is present in the commercial inoculum. Fortunately, the precious Rhizopus strains that were isolated from tempeh Malang in the 1960s–1970s are still preserved in UICC.
In summary, the ITS regions of the rRNA gene sequence data and phylogenetic analyses confirmed that the Rhizopus strains associated with tempeh fermentation using traditional inocula in Indonesia belong to three species: R. arrhizus, R. delemar, and R. microsporus. The wide use of commercial tempeh starters containing a single species, R. microsporus in Java has decreased the genetic diversity of Rhizopus species in tempeh and reduced the quality of tempeh Malang. The heavy commercialization of these tempeh starters has thus resulted in a change in the diversity of Rhizopus species associated with tempeh in Java in the last 30 years (since the 1990s).
Our findings confirmed the loss of Rhizopus diversity in tempeh currently produced in Indonesia, particularly in Java, where tempeh originated. We concluded that R. delemar and R. arrhizus have been lost from tempeh in Java. The present study makes an important contribution to validating the diversity of Rhizopus species, which were commonly used for tempeh production in Indonesia in the past (before the use of commercial tempeh starters). These strains have been securely deposited in a culture collection in Indonesia, and their sequence data have been deposited in a sequence database. The present findings emphasize the importance of conserving the Rhizopus strains isolated from tempeh produced using the traditional process in the past in culture collections in order to preserve and restore our precious genetic resources for conservation and sustainable use.
References
Nout, M. J. R. & Kiers, J. L. Tempe fermentation, innovation and functionality: Update into the third millenium. J. Appl. Microbiol. 98(4), 789–805 (2005).
van Veen, A. G. & Steinkraus, K. H. Nutritive value and wholesomeness of fermented foods. J. Agric. Food Chem. 18(4), 576–578 (1970).
Shurtleff, W. & Aoyagi, A. History of Tempeh. A Special Report on the History of Traditional Fermented Soyfoods. A Chapter from the Unpublished Manuscript, History of Soybeans and Soyfoods: 1100 B.C. to the 1980s (Soyinfo Center, 2007).
Shurtleff, W. & Aoyagi, A. History of Tempeh and Tempeh Products (1815–2011): Extensively Annotated Bibliography and Sourcebook (Soyinfo Center, 2011).
van Veen, A. G. & Schaefer, G. The influence of the tempeh fungus on the soya bean. Doc. Neerl. Indones. Morbis Trop. 2, 270–281 (1950).
Steinkraus, K. H., Hwa, Y. B., van Buren, J. P., Provvidenti, M. I. & Hand, D. B. Studies on tempeh—An Indonesian fermented soybean food. N. Y. State Agric. Exp. Stn. Cornell Univ. 1176, 777–788 (1959).
Inui, T., Takeda, Y. & Iizuka, H. Taxonomical studies on genus Rhizopus. J. Gen. Appl. Microbiol. 11, 1–121 (1965).
Schipper, M. A. A. A revision of the genus Rhizopus. I. The Rhizopus stolonifer-group and Rhizopus oryzae. Stud. Mycol. 25, 1–19 (1984).
Schipper, M. A. A. & Stalpers, J. A. A revision of the genus Rhizopus. II. The Rhizopus microsporus-group. Stud. Mycol. 25, 20–34 (1984).
Abe, A., Oda, Y., Asano, K. & Sone, T. Rhizopus delemar is the proper name for Rhizopus oryzae fumaric-malic acid producers. Mycologia 99(5), 714–722 (2007).
Liu, X. Y., Huang, H. & Zheng, R. Y. Molecular phylogenetic relationships within Rhizopus based on combined analyses of ITS rDNA and pyrG gene sequences. Sydowia 59, 235–253 (2007).
Zheng, R. Y., Chen, G. Q., Huang, H. & Liu, X. Y. A monograph of Rhizopus. Sydowia 59(2), 273–372 (2007).
Dolatabadi, S., de Hoog, G. S., Meis, J. F. & Walther, G. Species boundaries and nomenclature of Rhizopus arrhizus (syn. R. oryzae). Mycoses 57(Suppl. 3), 108–127 (2014).
Gryganskyi, A. P. et al. Phylogenetic and phylogenomic definition of Rhizopus species. Genes Genomes Genet. 8(6), 2007–2018 (2018).
Ellis, J. J. Species and varieties in the Rhizopus arrhizus–Rhizopus oryzae group as indicated by their DNA complementarity. Mycologia 77, 243–247 (1985).
Abe, A., Oda, Y., Asano, K. & Sone, T. The molecular phylogeny of the genus Rhizopus based on rDNA sequences. Biosci. Biotechnol. Biochem. 70, 2387–2393 (2006).
Abe, A., Asano, K. & Sone, T. A molecular phylogeny-based taxonomy of the genus Rhizopus. Biosci. Biotechnol. Biochem. 74(7), 1325–1331 (2010).
Dolatabadi, S., Walther, G., Gerrits van den Ende, A. H. G. & de Hoog, G. S. Diversity and delimitation of Rhizopus microsporus. Fungal Divers. 64, 145–163 (2013).
Gryganskyi, A. P. et al. Structure, function, and phylogeny of the mating locus in the Rhizopus oryzae complex. PLoS ONE 5(12), e15273 (2010).
Gandjar, I. & Santoso, I. The role of Rhizopus spp. in biotechnology. In Proceedings International Tempe Symposium, July 13–15. Bali, Indonesia (eds Sudarmadji, S. & Raharjo, S.) 55–63 (Indonesian Tempe Foundation, 1997).
Ilyas, N., Peng, A. C. & Gould, W. A. Tempeh, an Indonesian fermented soybean food. Hortic. Ser. 394, 40 (1973).
Dwidjoseputro, D. & Wolf, F. T. Microbiological studies of Indonesian fermented foodstuffs. Mycopathol. Mycol. Appl. 41, 211–222 (1970).
Hesseltine, C. W. A millennium of fungi, food, and fermentation. Mycologia 57, 149–197 (1965).
Wang, H. L. & Hesseltine, C. W. Wheat tempeh. Cereal Chem. 43, 563–570 (1966).
Febriani, R., Sjamsuridzal, W., Oetari, A., Santoso, I. & Roosheroe, I. G. ITS regions of rDNA sequence and morphological analyses clarify five Rhizopus strains from tempeh as Rhizopus oryzae. AIP Conf. Proc. 2023, 020159. https://doi.org/10.1063/1.5064156 (2018).
Khasanah, M., Sjamsuridzal, W., Oetari, A., Santoso, I. & Roosheroe, I. G. Phylogenetic analyses based on ITS regions of rDNA identified five Rhizopus strains from tempeh as R. delemar and R. oryzae. AIP Conf. Proc. 2023, 020141. https://doi.org/10.1063/1.5064138 (2018).
Vebliza, Y., Sjamsuridzal, W., Oetari, A., Santoso, I. & Roosheroe, I. G. Re-identification of five strains of Rhizopus arrhizus from tempeh based on ITS regions of rDNA sequence data. AIP Conf. Proc. 2023, 020167. https://doi.org/10.1063/1.5064164 (2018).
White, T. J., Bruns, T., Lee, S. & Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications (eds Innis, M. A. et al.) 315–322 (Academic Press, 1990).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215(3), 403–410 (1990).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality. Analysis tools. Nucleic Acids Res. 25(24), 4876–4882 (1997).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Saitou, N. & Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic tree. Mol. Biol. Evol. 4(4), 406–425 (1987).
Rzhetsky, A. & Nei, M. A simple method for estimating and testing minimum-evolution trees. Mol. Biol. Evol. 9, 945–967 (1992).
Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 17, 368–376 (1981).
Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39(4), 783–791 (1985).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequence. J. Mol. Evol. 16(2), 111–120 (1980).
Walther, G. et al. DNA barcoding in Mucorales: An inventory of biodiversity. Persoonia 30, 11–47 (2013).
Arbianto, P. Pengembangan industri fermentasi tradisional umumnya, tempe khususnya: suatu gagasan. Prosiding Pengembangan Tempe dalam Industri Pangan Modern, 182–188. (Yayasan Tempe Indonesia, 1995).
Samson, R. A., van Kooij, J. A. & De Boer, E. Microbiological quality of commercial tempeh in the Netherlands. J. Food Prot. 50(2), 92–94 (1987).
Nout, M. J., Martoyuwono, T. D., Bonné, P. C. J. & Odamtten, G. T. Hibiscus leaves for the manufacture of usar, a traditional inoculum for tempe. J. Sci. Food Agric. 58, 339–346 (1992).
Hartanti, A. T., Rahayu, G. & Hidayat, I. Rhizopus species from fresh tempeh collected from several regions in Indonesia. Hayati J. Biosci. 22, 136–142 (2015).
Anggriawan, R. Microbiological and Food Safety Aspects of Tempeh Production in Indonesia. Dissertation, Faculty of Agricultural Sciences, Georg-August-University Göttingen, Göttingen, Germany (2017).
Prihatna, C. & Suwanto, A. Phenotypic, metabolic, and genetic diversity of the Indonesian isolates of Rhizopus oligosporus. Microbiol. Indones. 1(1), 27–32 (2007).
Barus, T., Halim, R., Hartanti, A. T. & Saputra, P. K. Genetic diversity of Rhizopus microsporus from traditional inoculum of tempeh in Indonesia based on ITS sequences and RAPD marker. Biodiversitas 20(3), 847–852 (2019).
Sukardi, W. & Purwaningsih, I. Uji coba penggunaan inoculum tempe dari kapang Rhizopus oryzae dengan substrat tepung beras dan ubikayu pada unit produksi tempe Sanan Kodya Malang. J. Teknol. Pertan. 9(3), 207–215 (2008).
Jennessen, J., Schnürer, J., Olsson, J., Samson, R. A. & Dijksterhuis, J. Morphological characteristics of sporangiospores of the tempe fungus Rhizopus oligosporus differentiate it from other taxa of the R. microsporus group. Mycol. Res. 112(5), 547–563 (2008).
Vebliza, Y. Re-identifikasi Lima Strain Rhizopus arrhizus Fischer Koleksi UICC Berdasarkan Data Sequence Daerah Internal Transcribed Spacer Ribosomal DNA. B.Sc. Final Project; (Universitas Indonesia, Depok, Jawa Barat, Indonesia, 2016).
Acknowledgements
The authors gratefully acknowledge Dr. Peter Green from the Society for Applied Microbiology of the United Kingdom (SfAM UK) Endangered Collection Grant for providing financial support to UICC in 2007. We thank Fitrianingsih (Universitas Indonesia) for editing the manuscript and Dhian Chitra Ayu Fitria Sari (Universitas Indonesia) for assistance with microscopic observation.
Funding
This work was supported by Hibah Publikasi Artikel di Jurnal Internasional Kuartil Q1 dan Q2 (Q1Q2), Universitas Indonesia, Tahun Anggaran 2019 [Grant Number NKB-0280/UN2.R3.1/HKP.05.00/2019].
Author information
Authors and Affiliations
Contributions
W.S. conceived the study, analyzed the data and wrote the manuscript. M.K., R.F., Y.V. performed data collection. I.S. and A.O. supervised data collection. I.G. edited the manuscript. All authors approved the final version of manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declared no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sjamsuridzal, W., Khasanah, M., Febriani, R. et al. The effect of the use of commercial tempeh starter on the diversity of Rhizopus tempeh in Indonesia. Sci Rep 11, 23932 (2021). https://doi.org/10.1038/s41598-021-03308-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-03308-6
This article is cited by
-
The contribution of fungi to the global economy
Fungal Diversity (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.