Abstract
There is a lack of information highlighting the possible association between strain carrying genes of enteroaggregative Escherichia coli (EAEC) and environmental enteric dysfunction (EED) and on linear growth during childhood. Strain carrying genes of EAEC from stool samples collected from 1705 children enrolled in the MAL-ED birth cohort were detected by TaqMan Array Cards. We measured site-specific incidence rate by using Poisson regression models, identified the risk factors and estimated the associations of strain carrying genes of EAEC with the composite EED score and linear growth at 24 months of age. Overall highest incidence rate (43.3%) was found among children having infection with the aggR gene, which was the greatest in Tanzania (56.7%). Low maternal education, lack of improved floor, and ownership of domestic cattle were found to be risk factors for EAEC infection. In the multivariate models, after adjusting the potential covariates, strain carrying genes of EAEC showed strong positive associations with the EED scores and with poor linear growth at 24 months of age. Our analyses may lay the cornerstone for a prospective epidemiologic investigation for a potential vaccine development aimed at reducing the burden of EAEC infections and combat childhood malnutrition.
Similar content being viewed by others
Introduction
Childhood malnutrition is strongly associated with the risk of death from diarrhea, pneumonia, and other infectious diseases and is associated with growth failure, cognitive delay, and loss of productivity1. The main bacterial enteric pathogen in less-developed countries, particularly among children aged 2 to 24 months, is Escherichia coli2, 3. Among the major categories of diarrhoeagenic E. coli, enteroaggregative E. coli (EAEC) constitute a significant health risk and remains an important cause of infant mortality in developing countries4.
Evidence suggests that gastrointestinal infections with several enteropathogens, including EAEC, are linked with childhood malnutrition5,6,7. EAEC has been increasingly recognized as important enteropathogens since their initial discovery by patterns of adherence to HEp-2 (Human epithelial type 2) cells in E. coli isolates from Chilean children with diarrhea8. The genetic determinants and biological mechanisms for the virulence of EAEC have been reported to be mediated by a complex array of interacting traits that reside on both the chromosome and the plasmid9. As presently defined, EAEC is heterogeneous with regards to their genetic content10.
The aagR plasmid, harbored by most EAEC strains encodes a transcriptional activator of the AtaC/XylS class known as AggR11, which in turn, regulates the expression of virulence proteins involved in the adherence to mucosal secretions and thereby promoting intestinal colonization12, 13. AggR also activates the expression of the genes encoding dispersin (aap), the dispersin translocator Aat, the Aai chromosomal type IV secretion system (aaiA-Y), as well as the plasmid-borne aatA gene, which encodes an ABC (ATP-binding cassette) transporter14. Consequently, the aar gene of EAEC encodes a small protein named Aar (AggR activated regulator), whose expression is activated by AggR, and serves as a negative regulator of AggR15. Henceforth, there is a complex interplay between AggR and Aar, in relation to the virulence of EAEC and its consequent pathogenesis16.
Environmental Enteric Dysfunction (EED) is a subclinical intestinal disorder that is highly prevalent in low- and middle-income countries (LMICs)17 and is attributable to the infection by many environmental enteropathogens of bacterial, viral, and parasitic nature18, 19. Mechanisms contributing to growth failure in EED include intestinal leakiness and elevated gut permeability, gut inflammation, bacterial translocation, nutrient malabsorption, and systemic inflammation17. However, the definite contribution of different strains of this EAEC with linear growth faltering and EED remains poorly understood20.
The main goal of our study was to estimate the site-specific incidence rates of potential strain carrying genes of EAEC and their possible associations with the composite EED score and the consequent growth failure among children at 24 months of age.
Results
General characteristics of the study population and incidence rate of strain carrying genes of EAEC
A total of 34,622 monthly stool samples were collected from 1715 participants who completed the follow-up to 24 months. All the stool samples collected over this time from all the participants at the different study sites were assessed for the presence of strain carrying genes associated with EAEC using TaqMan Array Cards (TAC). The general characteristics of the study children are presented in Table 1.
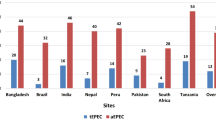
The incidence rates of the strain carrying genes of EAEC in the stool samples collected across all the 8 study sites over the 24 months’ study period have been shown in Table 2. The overall incidence rate of aggR was highest (43.3%). The incidence of strain carrying gene Aar associated with EAEC was highest in Tanzania (58.1%). It was also observed that the overall incidence of both aaiC and aatA strain carrying gene was lowest among all the sites.
Factors associated with strain carrying genes of EAEC
Factors associated with strain carrying genes associated with EAEC across all study sites were identified using Poisson regression (Table 3). The incidence rate for infection of EAEC in female children was comparable with male children. Additionally, household ownership of cattle among children with infections by the genomic strains of aaiC [IRR: 0.90 (95% CI: 0.83, 0.97); p = 0.010], Aar [IRR: 0.94 (95% CI: 0.88, 1.0); p = 0.042], and aggR [IRR: 0.94 (95% CI: 0.88, 1.0); p = 0.048]; improved floor among aaiC strain infected children [IRR: 0.93 (95% CI: 0.87, 0.99; p = 0.029]; monthly income less than 150 USD among children infected with the genomic strains of Aar and aggR; maternal education in years among children infected with aaiC [IRR: 0.99 (95% CI: 0.99, 1.0); p = 0.030] and Aar [IRR: 0.99 (95% CI: 0.99, 1.0); p = 0.006] genomic strains were associated and found to be statistically significant.
The incidence rate of aaiC and aatA positive strains was higher for the sites of India, Nepal, Peru, and Tanzania, while that of the genomic strain of Aar was the greatest in Tanzania. Consequently, the incidence rate of aatA genomic strain was the lowest in Brazil and that of aggR was the lowest in Nepal, South Africa, Brazil, and Pakistan. For concomitant infection by the positive strains of aaiC and aatA, the incidence rate was greater in India, Nepal, Peru, Tanzania compared to the Bangladesh site (Table 3).
Association between the strain carrying genes of EAEC and child growth
Infections with the strain carrying genes of EAEC, namely: Aar, aatA, aggR, and the concomitant presence of aaiC and aatA strains were associated with poor linear growth (difference in 24 months LAZ: length-for-age z score), with a stronger association being observed for all the study sites (Table 4). In Bangladesh, aaiC [− 1.32 difference in 24 months LAZ (95% CI: − 2.48, − 0.16); p < 0.027] and both aaiC and aatA carrying genes [− 1.36 difference in 24 months LAZ (95% CI: − 1.73, − 0.99; p < 0.001] had a negative association with LAZ. In Peru, Aar strain [− 0.96 difference in 24 months LAZ (95% CI: − 1.84, − 0.09); p = 0.030] was negatively associated with LAZ. In South Africa aatA [− 1.02 difference in 24 months LAZ (95% CI: − 2.00, − 0.05); p = 0.040]; in Tanzania, aatA [− 2.04 difference in 24 months LAZ (95% CI: − 3.55, − 0.53); p = 0.009] was negatively associated with LAZ. For the other four countries (South Africa, Brazil, Nepal and India), strain carrying genes associated with EAEC were negatively associated with LAZ but were not statistically significant.
Association between strain carrying genes associated with EAEC and enteric inflammation
After adjusting for the potential covariates like age, sex, WAMI index (water/sanitation, assets, maternal education, and income); enrollment length-for-age z score; maternal BMI; the number of children in the household, presence of poultry/cattle in the household, seasonality, serum zinc level, AGP (alpha-1-acid glycoprotein), presence of co-pathogens (Campylobacter, LT-ETEC, ST-ETEC, Shigella/EIEC, and Giardia), site for the overall estimate and age as the time variable in GEE model for strain carrying genes of EAEC infection were also clearly and consistently associated with increased EED score (Table 5) across all sites except Pakistan and Brazil. In case of the overall effect, the presence of all the strain carrying genes was associated with the EED score except aaiC. The same findings were observed in India, Nepal and Peru individually except aaiC strain carrying gene. In Pakistan and Brazil, there was no statistically significant relationship found among all the strain carrying genes. In Tanzania, only aaiC gene was non-significantly associated with the EED score. Aar, and aggR, strain carrying genes had a positive relationship with EED score among the children from Bangladesh. In South Africa except for Aar and aaiC, all other genes were significantly associated with EED score (Table 5). In Nepal and Peru both aaiC and aatA strain carrying genes had a statistically significant positive relationship with EED score.
Discussion
To our knowledge, this is the first study investigating the association between different strain carrying genes of EAEC with linear growth and enteric inflammation in children from birth until 2 years of age. Several EAEC strain carrying genes have been used in case–control and epidemiological studies in recent years, with aatA, aap, aagR, astA, and aafA being among the most common for EAEC diagnosis21,22,23.
We documented an overall high incidence rate of the concomitant presence of aggR gene carrying strains of EAEC across all eight study sites. The different clinical and nutritional outcomes associated with EAEC infection included: poor child growth and development during the study period and changes in the status of intestinal inflammation and provide stern challenges for understanding pathobiology and proposing potential therapeutic approaches for EAEC8. In this scenario, EAEC strain heterogeneity is a key contributor to these effects, but very little is known about the individual genomic strains responsible for this8.
In our study, the overall incidence rate for the genomic strain of aaiC was the lowest across all the study sites. Previously, an incidence rate of 38% for the aaiC gene among children under 5 years of age was reported in the Global Enteric Multicenter Study (GEMS)10. The incidence of aaiC, aggR, aatA, aaR, and the concomitant presence of aaiC and aatA strain carrying genes were higher among the study participants in Tanzania. Similar findings were observed in the case of traveler’s diarrhea in Guatemala and Mexico24. Many plasmid-encoded (AAFs, AggR itself, Aap, and Aat) and chromosomal-encoded (pheU pathogenicity island) virulence determinants are regulated by aggR15. Reduced viability of infected G. mellonella larvae with an aggR mutant strain and elevated virulence of atypical EAEC strains were discovered in a study, suggesting that EAEC virulence is linked to the AggR regulon25.
The low incidence of the aaiC gene in our study compared to the other strain carrying genes of EAEC may be additional support for the plasmid-mediated gene transfer of the pAA. In several studies, the authors suggested that aggR was not a feasible virulence marker for the diagnosis of EAEC infections since this gene alone was not a considerably sensitive target in comparison to the aatA gene26, 27. Our study findings regarding aaiC being an incongruent marker for EAEC identification were found to be consistent with a study conducted in southern Mozambique23.
The association between strain carrying genes associated with EAEC and malnutrition is not entirely clear, although plausible models for pathogenesis can be proposed. The Aar gene, which has been hypothesized to act directly or indirectly as a virulence suppressor, was present among the children in Tanzania28. Similar findings have also been reported in Mali and Brazil and these findings may strongly support a significant role for the Aar gene in the epidemiology of EAEC infection23. We also observed that there was poor linear growth among the children from Peru who were infected with the genomic strain of Aar; which may be pointed towards increased pathogenicity of Aar gene.
Consequently, the aggR gene mediates the expression of a large number of other EAEC genes responsible for virulence. The first genes found to be regulated by aggR were those encoding the aggregative adherence fimbriae (AAF)11. Our study findings demonstrate that an overall high incidence of the genomic strain of aggR was found across all the sites and the linear growth of children was negatively associated with the presence of aggR gene. However, though the role of aggR of EAEC in child growth remains unknown.
Our study findings also illustrate that the concomitant presence of aaiC and aatA strain carrying genes of EAEC was negatively associated with childhood linear growth except Nepal and Tazania. Other studies suggested that the presence of different virulence genes from within and outside the plasmid AA is necessary for complete EAEC virulence. Such aforementioned finding is in line with the findings of another concurrent study where the diagnosis of EAEC without overt diarrhea is associated with length/height-for-age z score decrements and chronic inflammation in children from Brazil29, 30.
Our data shows presence of improved floors in the house has a protective effect against infection of EAEC by aaiC genomic strain. We found higher maternal education was protective for aaiC and Aar genomic strain of EAEC. Findings from several other studies conducted in the MAL-ED settings also found an association of these factors with EAEC and Campylobacter infection31, 32.
The presence of cattle in the household was associated with aaiC, Aar, and aggR strain carrying genes. Still, there is no evidence regarding the association of the presence of an animal in the household with EAEC infection. The transfer of Giardia lamblia genes, any E. coli virulence gene, and unique E. coli virulence genes from animal feces to the hands of the mothers upon animal handling were reported in a study conducted in rural Bangladesh. As a result, domestic animals played a significant role in the spread of enteric pathogens in households33.
Previous findings have reported that EAEC detection was associated with higher levels of MPO (a marker for intestinal inflammation), NEO (intestinal inflammation), and AAT (permeability) among all 8 sites of MAL-ED32. In our study, the overall concomitant presence of aaiC and aatA strain carrying genes were more strongly associated with increased EED score, implying a higher intestinal inflammation. The relevance of elevated intestinal inflammation associated with virulence-related genes associated with EAEC is not yet clear and there is no evidence that virulence-related genes associated with EAEC were associated with elevated intestinal inflammatory biomarkers and subsequently an increased EED score. Henceforth, our study is the first attempt undertaken towards the generation of evidence-based understanding of the contribution of the different virulent genetic strains of EAEC with regards to enteric inflammation and poor child growth in LMIC settings.
Despite some potentially promising nature of our study findings, there are several possible limitations associated with our analysis. As an observational cohort study, the causality of the associations between infection with various genomic strains of EAEC and both intestinal inflammation and linear growth cannot be proven but can be hypothesized based on several factors, including the appropriate adjustment of the models for possible confounders, the strength and consistency of the associations, and the biological plausibility. We were unable to establish a temporal relationship between infections and the outcomes, which would require structured longitudinal models.
In conclusion poor maternal education, lack of improved floor, and ownership of cattle in the household are possible risk factors for EAEC infections by different genomic strains and thereby leading to a compromised linear growth in childhood. The burden of strain carrying genes associated with EAEC was associated with increased enteric inflammation among children in the first 2 years of life. EAEC virulence related genomic strains of aaiC, Aar, aatA, and the concomitant presence of aaiC and aatA genomic strains had a stronger association with growth failure among children of Bangladesh, whereas the association with inflammation was strongest for aggR, strain carrying genes.
Method
Study design and participants
MAL-ED (Etiology, Risk Factors, and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health) was a birth cohort study performed across eight sites in South America, sub-Saharan Africa, and Asia. The MAL-ED study design and methodology have been described elsewhere34. Briefly, 1715 children were enrolled from November 2009 to February 2012 from the community within 17 days of birth at all eight sites, namely: Bangladesh, India, Nepal, Pakistan, South Africa, Tanzania, Brazil, and Peru. In our current analysis, data from all 1715 participants were available from enrolment soon after birth up to 24 months of age.
Data collection
Anthropometric measurements were done at monthly intervals up to the age of 24 months using standard scales (Seca gmbh & co. kg., Hamburg, Germany). Length-for-age z score (LAZ), weight-for-age z score (WAZ), and weight-for-length z score (WLZ) were calculated through the use of the 2006 WHO standards for children35. Anthropometric measurements were performed monthly. Details of illness and child feeding practices were collected during twice-weekly household visits36. Additionally, household demographics, presence of siblings, maternal characteristics and other data on the child’s birth and anthropometry were obtained at enrollment34. Beginning at 6 months of age, socioeconomic data were collected every 6 months. The WAMI score (Water, sanitation, hygiene, Asset, Maternal education, and Income index, ranging from 0 to 1) is a socioeconomic status index that includes access to improved water and sanitation, eight selected assets, maternal education, and household income as a representative of the socioeconomic status of the households37. A better socioeconomic status is indicated by a higher WAMI score38. Improved water and sanitation were defined following World Health Organization guidelines39. Treatment of drinking water was defined as filtering, boiling, or adding bleach40.
Collection of stool and blood samples
Non-diarrheal stool samples were collected monthly (at least 3 days before or after a diarrhea episode) from birth to age 2 years and venous/peripheral blood was collected at 7, 15, and 24 months of age41. Raw stool aliquots and blood samples were processed at all sites using harmonized protocols and kept at − 80 °C freezers before subsequent laboratory analyzes42.
In this study, plasma zinc was assessed as the measure of zinc status at the age of 7, 15, and 24 months. Plasma zinc concentration is a proxy marker and recommended for the assessment of population zinc status, especially for children in low-income countries43. Plasma alpha-1-acid glycoprotein (AGP) level was considered as a biomarker for systemic inflammation and was also assessed at 7, 15, and 24 months44.
Assessment of enteropathogens by TaqMan array cards (TAC)
Total nucleic acid (both DNA and RNA) was extracted from the fecal samples using the QIAamp Fast DNA Stool Mini kit (Qiagen), following the manufacturer’s guidelines. Two external controls, namely: MS2 bacteriophage and Phocine herpesvirus (PhHV) were added to the samples for the confirmation of nucleic acid extraction and amplification efficiency45.
For the detection of enteropathogens, a quantitative polymerase chain reaction (qPCR) with the use of a customized TaqMan Array Card (TAC) involving compartmentalized probe-based real-time PCR assays was used for the detection of a possible 29 pathogens from each of the samples41. Ct (quantification cycle) value of 35 was set as a threshold for analysis, whereby a Ct > 35 was considered as negative, as mentioned elsewhere45. In MAL-ED study, they investigated the occurrence of putative virulence-related genes (VRG) of EAEC, namely: aatA, aggR, Aar, and aaiC. To diagnose EAEC, the genes aatA (dispersin transporter protein), and aaiC (secreted protein) were targeted by PCR. Primers specific for EAEC identification were aaiC and aatA. Samples were considered positive for EAEC if they could detect either one of the two diagnostic genes or both. Only EAEC positive samples were further analyzed by multiplex PCRs to identify 20 EAEC VRG46. Moreover, cases positive for concomitant presence of both aaiC and aatA genotypes were further analyzed for co-infection and the list of pathogens in this study, as described elsewhere42. In MAL-ED Brazil site they compared the combinations of EAEC VRGs from positive samples presenting both diagnostic genes (aaiC and aatA). Their choice was based on the fact that only the samples presenting both genes were statistically associated with malnourished children 46.
Assessment of biomarkers of intestinal inflammation
Intestinal inflammation was evaluated by measuring the levels of the biomarkers: alpha-1-anti-trypsin (Biovendor, Chandler, NC), neopterin (GenWay Biotech, San Diego, CA), and myeloperoxidase (Alpco, Salem, NH) in the stool samples collected from the study participants at the 3, 6, 9, 15, and 24 months of age time points by quantitative ELISA, using manufacturer’s guidelines34.
Statistical analysis
All statistical analyses were performed in STATA 15 (Stata Corporation, College Station, TX). Descriptive statistics such as proportion, mean and standard deviation (SD) for symmetric data, and median with inter‐quartile range (IQR) for asymmetric quantitative variables were used to summarize the data. Incidence rates were calculated using Poisson regression where outcome variables were the number of infections of EAEC (different strain carrying genes) and offset variables were a log of number of follow up. The factors associated with strain carrying genes of EAEC in the monthly stool samples were calculated using Poisson regression models. In the final multiple Poisson regression model, the following variables were considered for inclusion using stepwise forward selection: child sex, birth weight, duration of exclusive breastfeeding in months, enrollment weight for age z-score, length for age z score, maternal age in years, maternal education, mother having less than 3 living children, maternal BMI, routine treatment of drinking water, improved sanitation, household ownership of cattle/poultry, and less than 2 people live in per room. We excluded children from the Pakistan site for growth analysis, owing to bias noted in a subset of this cohort within the study period. Myeloperoxidase (MPO), neopterin (NEO), and alpha-1-antitrypsin (AAT) values were log‐transformed before the analysis. At each time point, the composite EED score ranging from 0 to 10 was calculated from the three fecal markers, as described in the previous literature by MAL-ED researchers20, 47. Categories were assigned values as 0 (low), 1 (medium), or 2 (high). The formula for the composite EED score is as follows48:
Associations between strain carrying genes of EAEC and composite EED score was estimated using generalized estimating equations (GEE) to fit regression models after adjusting for sex, age, water/sanitation, assets, maternal BMI, and (WAMI) index; enrollment length-for-age and weight-for-age z score, maternal height; poultry/ cattle in house, serum zinc level, inflammatory biomarker AGP (alpha-1-acid glycoprotein), presence of co-pathogens (Campylobacter, LT-ETEC, ST-ETEC, Shigella/EIEC, and Giardia), seasonality, and site for overall estimate and age in the month as time variable49. To assess and compare the associations of strain carrying genes of EAEC infection burden on growth at 24 months of age, we used multivariable linear regression after adjusting for the site and the necessary covariates. To detect multicollinearity, the variance inflation factor (VIF) was calculated, and no variable producing a VIF value > 5 was found in the final model. We calculated the strength of association by estimating the coefficient and its 95% CI (confidence interval). A p-value of < 0.05 was considered statistically significant during the multivariable analysis.
Ethical consideration
The study was approved by the ethical committees at each of the participating institutes across each of the eight study sites34. The study was approved by the Research Review Committee and the Ethical Review Committee, icddr,b (BGD); Committee for Ethics in Research, Universidade Federal do Ceara; National Ethical Research Committee, Health Ministry, Council of National Health (BRF); Institutional Review Board, Christian Medical College, Vellore; Health Ministry Screening Committee, Indian Council of Medical Research (INV); Institutional Review Board, Institute of Medicine, Tribhuvan University; Ethical Review Board, Nepal Health Research Council; Institutional Review Board, Walter Reed Army Institute of Research (NEB); Institutional Review Board, Johns Hopkins University; PRISMA Ethics Committee; Health Ministry, Loreto (PEL); Ethical Review Committee, Aga Khan University (PKN); Health, Safety and Research Ethics Committee, University of Venda; Department of Health and Social Development, Limpopo Provincial Government (SAV); Medical Research Coordinating Committee, National Institute for Medical Research; Chief Medical Officer, Ministry of Health and Social Welfare (TZH)32.Written informed consent was obtained from the parents or legal guardians of every child.
Data availability
A publicly available MAL-ED dataset was analyzed in this study. This data can be obtained from here: ClinEpiDB [https://clinepidb.org/ce/app/record/dataset/DS_841a9f5259].
References
Syed, S., Ali, A. & Duggan, C. Environmental enteric dysfunction in children. J. Pediatr. Gastroenterol. Nutr. 63(1), 6–14 (2016).
Mondal, D., Haque, R., Sack, R. B., Kirkpatrick, B. D. & Petri, W. A. Jr. Attribution of malnutrition to cause-specific diarrheal illness: Evidence from a prospective study of preschool children in Mirpur, Dhaka, Bangladesh. Am. J. Trop. Med. Hyg. 80(5), 824–826 (2009).
Neto, U. F. & Scaletsky, I. C. A. Escherichia coli infections and malnutrition. Lancet 356, S27 (2000).
Deborah Chen, H. & Frankel, G. Enteropathogenic Escherichia coli: Unravelling pathogenesis. FEMS Microbiol. Rev. 29(1), 83–98 (2005).
Petri, W. A. et al. Enteric infections, diarrhea, and their impact on function and development. J. Clin. Investig. 118(4), 1277–1290 (2008).
Fagundes-Neto, U. & Scaletsky, I. C. A. The gut at war: the consequences of enteropathogenic Escherichia coli infection as a factor of diarrhea and malnutrition. Sao Paulo Med. J. 118(1), 21–29 (2000).
Mondal, D. et al. Contribution of enteric infection, altered intestinal barrier function, and maternal malnutrition to infant malnutrition in Bangladesh. Clin. Infect. Dis. 54(2), 185–192 (2012).
Jensen, B. H., Olsen, K. E. P., Struve, C., Krogfelt, K. A. & Petersen, A. M. Epidemiology and clinical manifestations of enteroaggregative Escherichia coli. Clin. Microbiol. Rev. 27(3), 614–630 (2014).
Dudley, E. G. & Rasko, D. A. Genomic and virulence heterogeneity of enteroaggregative Escherichia coli. Popul. Genet. Bacteria A Tribute to Thomas S Whittam 2011, 181–198 (2011).
Boisen, N. et al. Redefining enteroaggregative Escherichia coli (EAEC): Genomic characterization of epidemiological EAEC strains. PLoS Negl. Trop. Dis. 14(9), e0008613 (2020).
Nataro, J. P., Yikang, D., Yingkang, D. & Walker, K. AggR, a transcriptional activator of aggregative adherence fimbria I expression in enteroaggregative Escherichia coli. J. Bacteriol. 176(15), 4691–4699 (1994).
Bernier, C., Gounon, P. & Le Bouguénec, C. Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect. Immun. 70(8), 4302–4311 (2002).
Poole, N. M., Rajan, A. & Maresso, A. W. Human intestinal enteroids for the study of bacterial adherence, invasion, and translocation. Curr. Protocols Microbiol. 50(1), e55 (2018).
Dudley, E. G., Thomson, N. R., Parkhill, J., Morin, N. P. & Nataro, J. P. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol. Microbiol. 61(5), 1267–1282 (2006).
Morin, N., Santiago, A. E., Ernst, R. K., Guillot, S. J. & Nataro, J. P. Characterization of the AggR regulon in enteroaggregative Escherichia coli. Infect. Immun. 81(1), 122–132 (2013).
Mickey, A. S. & Nataro, J. P. Dual function of Aar, a member of the new AraC negative regulator family, in Escherichia coli gene expression. Infect. Immun. 88(6), e00100-00120 (2020).
Owino, V. et al. Environmental enteric dysfunction and growth failure/stunting in global child health. Pediatrics 138(6), e20160641 (2016).
Kosek, M. et al. Assessment of environmental enteropathy in the MAL-ED cohort study: Theoretical and analytic framework. Clin. Infect. Dis. 59, S239–S247 (2014).
Salazar-Lindo, E. et al. Intestinal infections and environmental enteropathy: Working Group report of the second world congress of pediatric gastroenterology, hepatology, and nutrition. J. Pediatr. Gastroenterol. Nutr. 39, S662–S669 (2004).
Kosek, M. et al. Fecal markers of intestinal inflammation and permeability associated with the subsequent acquisition of linear growth deficits in infants. Am. J. Trop. Med. Hyg. 88(2), 390–396 (2013).
Ikumapayi, U. N. et al. Identification of subsets of enteroaggregative Escherichia coli associated with diarrheal disease among under 5 years of age children from Rural Gambia. Am. J. Trop. Med. Hyg. 97(4), 997–1004 (2017).
Petro, C. D. et al. Genetic and virulence profiles of enteroaggregative Escherichia coli (EAEC) isolated from deployed military personnel (DMP) with Travelers’ Diarrhea. Front. Cell. Infect. Microbiol. 10, 200 (2020).
Mandomando, I. et al. Escherichia coli ST131 clones harbouring AggR and AAF/V fimbriae causing bacteremia in Mozambican children: Emergence of new variant of fimH27 subclone. PLoS Negl. Trop. Dis. 14(5), e0008274 (2020).
Bamidele, O., Jiang, Z.-D. & Dupont, H. Occurrence of putative virulence-related genes, aatA, aggR and aaiC, of Enteroaggregative Escherichia coli (EAEC) among adults with travelers’ diarrhea acquired in Guatemala and Mexico. Microb. Pathog. 128, 97–99 (2019).
Guerrieri, C. G. et al. Typical and atypical enteroaggregative Escherichia coli are both virulent in the Galleria mellonella model. Front. Microbiol. 10, 1791 (2019).
Okeke, I. N. & Nataro, J. P. Enteroaggregative Eescherichia coli. Lancet. Infect. Dis 1(5), 304–313 (2001).
Lima, I. F. N. et al. Prevalence of enteroaggregative Escherichia coli and its virulence-related genes in a case–control study among children from north-eastern Brazil. J. Med. Microbiol. 62(Pt 5), 683 (2013).
Moyo, S. J., Maselle, S. Y., Matee, M. I., Langeland, N. & Mylvaganam, H. Identification of diarrheagenic Escherichia coli isolated from infants and children in Dar es Salaam, Tanzania. BMC Infect. Dis. 7, 92 (2007).
Chattaway, M. A. et al. Investigating the link between the presence of enteroaggregative Escherichia coli and infectious intestinal disease in the United Kingdom, 1993 to 1996 and 2008 to 2009. Eurosurveillance 18(37), 20582 (2013).
Steiner, T. S., Lima, A. A. M., Nataro, J. P. & Guerrant, R. L. Enteroaggregative Escherichia coli produce intestinal inflammation and growth impairment and cause interleukin-8 release from intestinal epithelial cells. J. Infect. Dis. 177(1), 88–96 (1998).
Haque, M. A. et al. Determinants of Campylobacter infection and association with growth and enteric inflammation in children under 2 years of age in low-resource settings. Sci. Rep. 9(1), 1–8 (2019).
Rogawski, E. T. et al. Epidemiology of enteroaggregative Escherichia coli infections and associated outcomes in the MAL-ED birth cohort. PLoS Negl. Trop. Dis. 11(7), e0005798-e5801 (2017).
Fuhrmeister, E. R. et al. Predictors of enteric pathogens in the domestic environment from human and animal sources in rural Bangladesh. Environ. Sci. Technol. 53(17), 10023–10033 (2019).
The MAL-ED Network Investigators. The MAL-ED study: A multinational and multidisciplinary approach to understand the relationship between enteric pathogens, malnutrition, gut physiology, physical growth, cognitive development, and immune responses in infants and children up to 2 years of age in resource-poor environments. Clin. Infect. Dis. 59, S193–S206 (2014).
Onis, M. D. et al. Development of a WHO growth reference for school-aged children and adolescents. Bull. World Health Org. 85, 660–667 (2007).
Richard, S. A., Barrett, L. J., Guerrant, R. L., Checkley, W. & Miller, M. A. Disease surveillance methods used in the 8-site MAL-ED cohort study. Clin. Infect. Dis. 59, S220–S224 (2014).
Das, R. et al. Evaluating association of vaccine response to low serum zinc and vitamin D levels in children of a birth cohort study in Dhaka. Vaccine 39(1), 59–67 (2021).
Das, S., Alam, M. A., Mahfuz, M., El Arifeen, S. & Ahmed, T. Relative contributions of the correlates of stunting in explaining the mean length-for-age z-score difference between 24-month-old stunted and non-stunted children living in a slum of Dhaka, Bangladesh: Results from a decomposition analysis. BMJ Open 9(7), e025439 (2019).
Unicef WHO. WHO Joint Monitoring programme for water supply and sanitation. Progress on drinking water and sanitation (2012).
Amour, C. et al. Epidemiology and impact of campylobacter infection in children in 8 low-resource settings: results from the MAL-ED study. Clin. Infect. Dis. 63(9), 1171–1179 (2016).
Rogawsk, E. T. et al. Use of quantitative molecular diagnostic methods to investigate the effect of enteropathogen infections on linear growth in children in low-resource settings: longitudinal analysis of results from the MAL-ED cohort study. Lancet Global Health 109(18), 30351 (2018).
Houpt, E. et al. Microbiologic methods utilized in the MAL-ED cohort study. Clin. Infect. Dis. 59, S225–S232 (2014).
De Benoist, B., Darnton-Hill, I., Davidsson, L., Fontaine, O. & Hotz, C. Conclusions of the joint WHO/UNICEF/IAEA/IZiNCG interagency meeting on zinc status indicators. Food Nutr. Bull. 28, S480–S484 (2007).
Gannon, B. M. et al. A point-of-care assay for alpha-1-acid glycoprotein as a diagnostic tool for rapid, mobile-based determination of inflammation. Curr. Res. Biotechnol. 1, 41–48 (2019).
Liu, J. et al. Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. Lancet. Infect. Dis 14(8), 716–724 (2014).
Havt, A. et al. Prevalence and virulence gene profiling of enteroaggregative Escherichia coli in malnourished and nourished Brazilian children. Diagn. Microbiol. Infect. Dis. 89(2), 98–105 (2017).
Arndt, M. B. et al. Fecal markers of environmental enteropathy and subsequent growth in Bangladeshi children. Am. J. Trop. Med. Hyg. 95(3), 694–701 (2016).
Fahim, S. M. et al. Association of Fecal markers of environmental enteric dysfunction with zinc and iron status among children at first two years of life in Bangladesh. Am. J. Trop. Med. Hyg. 99(2), 489–494 (2018).
Sanin, K. I. et al. Micronutrient adequacy is poor, but not associated with stunting between 12–24 months of age: A cohort study findings from a slum area of Bangladesh. PLoS ONE 13(3), e0195072 (2018).
Acknowledgements
We acknowledge with gratitude the commitment of The Etiology, Risk Factors, and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health and Development Project (MAL-ED) as a collaborative project supported by the Bill and Melinda Gates Foundation, the Foundation for the National Institutes of Health, and the National Institutes of Health, Fogarty International Center. The authors are grateful to MAL-ED staff, parents, and children for their contributions. We acknowledge the contribution of icddr,b’s core donors including the Government of the People’s Republic of Bangladesh, Global Affairs Canada (GAC), Canada; Swedish International Development Cooperation Agency and Foreign, Commonwealth and Development Office (FCDO), UK for their continuous support and commitment to icddr,b’s research efforts.
Funding
This work was supported, in whole or in part, by the Bill & Melinda Gates Foundation [Grant Number: OPP47075]. Under the grant conditions of the Foundation, a Creative Commons Attribution 4.0 Generic License has already been assigned to the Author Accepted Manuscript version that might arise from this submission.
Author information
Authors and Affiliations
Contributions
T.A. conceptualized and designed the study. R.D., P.P., M.A.H., and A.S.G.F conceived the hypothesis. M.A.H. managed the data set and provided technical support. R.D. and M.A.H. analyzed the data. R.D. developed the tables/graphs presented here and wrote the first draft of the manuscript. P.P. wrote the methodology part on laboratory investigations of stool samples. P.P., M.A.H., M.M., A.S.G.F., and T.A. critically reviewed the manuscript and provided intellectual inputs. T.A. gave the final approval for publication. All authors contributed to the final version of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Das, R., Palit, P., Haque, M.A. et al. Site specific incidence rate of virulence related genes of enteroaggregative Escherichia coli and association with enteric inflammation and growth in children. Sci Rep 11, 23178 (2021). https://doi.org/10.1038/s41598-021-02626-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-02626-z
This article is cited by
-
Correlation between gut microbiota composition, enteric infections and linear growth impairment: a case–control study in childhood stunting in Pidie, Aceh, Indonesia
Gut Pathogens (2023)
-
Gut biomolecules (I-FABP, TFF3 and lipocalin-2) are associated with linear growth and biomarkers of environmental enteric dysfunction (EED) in Bangladeshi children
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.