Abstract
The bumblebee Bombus terrestris is used worldwide for crop pollination. Despite its positive impact on crop yield, it has become a widespread threat to biodiversity due to its interactions with local bumblebee populations. Commercial subspecies introduced to the Iberian Peninsula since the 1990s without any regulation have colonized the environment, with evidence of naturalization and introgression with the endemic subspecies Bombus terrestris lusitanicus. We have used mitochondrial and nuclear genetic data to describe the current genetic diversity of the Iberian population and to estimate the expansion of commercial bumblebees. Samples from the natural distribution range of the commercial subspecies, the natural intergradation area between the two subspecies and from a period prior to the use of commercial colonies (i.e., before the 1990s) have been used for comparison. Our results show that the mitochondrial haplotype of the commercial breeds has spread throughout the territory, which, together with subtle changes observed in the nuclear genetic diversity of the populations, indicates that hybridization and consequent introgression are occurring in most of the peninsula. It is, therefore, necessary to improve the existing legislation concerning the management and exportation of commercial bumblebees to conserve locally adapted populations.
Similar content being viewed by others
Introduction
In the context of the current human-induced biodiversity crisis1, scientific knowledge about the drivers of decline and how they affect organisms is necessary to propose adequate conservation measures2. In the last decades, given their economic and environmental importance, concern for the welfare of pollinators has risen3. Many studies have explored the main potential causes of their decline4. In addition to those identified as major causes (e.g., pesticides and loss of habitat5), anthropic translocations of domesticated taxa like honey bees or bumblebees within their natural ranges are receiving more attention6,7. Such human-mediated movement may harm other pollinators through competition for resources and the spread of parasites (e.g., microsporidia and trypanosomatids8,9,10). More research on these effects is needed to encourage regulatory authorities to implement scientifically supported conservation measures.
The West Palearctic bumblebee species Bombus terrestris (Linnaeus, 1758) began to be used commercially in greenhouses for crop pollination in the 1980s in Belgium, and the subspecies B. t. terrestris and B. t. dalmatinus Dalla Torre, 1882 are now widely used in artificial pollination11. Commercial breeds can escape from greenhouses into natural habitats when they seek floral resources12 or nesting places and can colonize suitable environments due to their high thermic resistance, generalist diet and wide climatic tolerance13,14. The exportation of Bombus terrestris outside its Palearctic distribution range has made this bumblebee an invasive species15,16, driving native species to local decline or extinction due to competition, pathogen spillover and reproductive interference2,9,17.
Many countries have already taken legislative measures to manage this situation. In Japan, B. terrestris was classified as an invasive species in 2006 and its importation is now severely restricted17. In Argentina, the importation of B. terrestris and B. impatiens Cresson, 1863 is also strongly regulated to protect the native South American species B. dahlbomii Guérin-Méneville, 183516, although commercial species can still spread from Chile15. Within the natural distribution range of the species, specific regulations to protect endemic subspecies of B. terrestris have also been approved, as commercial breeds could become a potential competition to the wild populations based on their characteristics (e.g., higher gyne production, greater foraging ability and larger colony size)8. Norway and the Canary Islands have strict importation policies on foreign bumblebees11, while in the United Kingdom the trade of non-native subspecies is restricted, favouring the use of the endemic subspecies B. t. audax (Harris, 1776)9. However, no regulation has been enacted to protect the endemic subspecies B. t. lusitanicus (Krüger, 1956) in the Iberian Peninsula, where there is a high density of greenhouse agriculture that use bumblebees as managed pollinators (Supplementary Fig. S1, online).
B. terrestris presents eight subspecies based on colour patterns, distribution and pheromonal analysis18,19. B. t. lusitanicus, B. t. terrestris and B. t. dalmatinus are described with a yellow and black band pattern and a white tail at the end of the abdomen. However, B. t. lusitanicus also presents a variable colouration of ferruginous brown in the ventral area of the gaster, pleurae and corbiculae that allows its differentiation18,20.
The natural distribution range of B. t. lusitanicus includes the Iberian Peninsula, a current hotspot of biodiversity after its role as a glacial refugium in the Quaternary period21, and the Pyrenees and south of France, where it overlaps with the central European subspecies B. t. terrestris. The similar composition of the cephalic labial gland secretions of the two subspecies13 facilitates gyne preference towards interbreeding and, therefore, a natural intergradation area occurs in the distribution overlap18,20. The colouring of hybrid specimens varies, but they typically show a variable mixture of light and dark pubescence on the ventral area of the gaster, pleurae and, especially, the legs and corbiculae.
Since the establishment of rearing companies in the 1990s in the region of Andalucia in southern Spain, there are records of the naturalization of managed commercial bumblebees in the environment22. Nowadays, a new anthropic intergradation area has developed in the south of the peninsula, where hybridization events and introgression with wild populations have been confirmed with genetic evidence23,24,25.
To preserve endemic taxa, the genetic diversity and structure of local populations must first be investigated26. Conservation genetics provides a theoretical framework of the appropriate methodologies to reveal differences between taxa27, populations28 and historical data29. The use of mitochondrial and nuclear markers in population genetics is advisable due to the different inheritance mechanisms of these genomes30, especially in haplodiploid species like bees that can show a strong mitochondrial bias in introgression rates31.
Mitochondrial DNA presents certain implications because of its maternal inheritance. In bumblebees, mitochondrial DNA analysis has confirmed the presence of exotic queens in the environment23 and of drifter workers that can lay unfertilized eggs in foreign colonies, resulting in male adults10. Moreover, based on empirical data, the maternal genotype is considered decisive for the inheritance of the colony traits32. Finally, the mitochondrial haplotype of the studied subspecies have already been characterized23. Alternatively, nuclear markers such as microsatellites are a validated population genetics tool to study the genetic diversity and structure of populations in the genus7,29,33.
Both mitochondrial and nuclear markers were analysed to estimate genetic diversity parameters and clustering patterns across Bombus terrestris populations in the Iberian Peninsula. To study the effect of hybridization, some analyses were repeated after filtering hybrid individuals (based on morphological determination and molecular analysis) to identify differential changes in the genetic diversity of populations. Male diploidy rates (2n♂) were also determined to estimate the inbreeding in bumblebee populations34,35. In haplodiploid species such as B. terrestris, male diploidy is caused by homozygosis at the sex determination locus (sl-CSD) in diploid individuals, which would develop into females if they were heterozygous36. The presence of diploid males implies a reduction in the number of workers, which can negatively affect the fitness of the colony28. Although male diploidy occurs naturally37, high rates have been linked with declines in effective population size and low genetic diversity38 and therefore would be expected to appear at a lower rate in hybrid populations.
This research aims to study the genetic diversity and structure of the Iberian B. terrestris populations from a broader perspective on both spatial (i.e., Iberian Peninsula) and temporal (before and after the 1990s) scales, taking into account the dispersion of commercial breeds (i.e., central European subspecies) into the wild and potential introgression events between wild and commercial populations. We hypothesize that, due to the dispersion of commercial subspecies from greenhouses, hybridization with the wild populations of the endemic subspecies is not limited to the south and east of the Iberian Peninsula23,24,25,39 and introgression is occurring across the territory. These events would have a more pronounced impact on the genetic diversity of the populations in the south of the peninsula, where many bumblebee breeding companies provide pollination services to the high quantity of greenhouse crops that depend on bumblebees40.
Results
Subspecies and hybrid distribution based on morphology
Of the 594 studied individuals, 39 were discarded as eight corresponded to a different species after BLAST analyses (seven B. lucorum [Linnaeus, 1761] and one B. soroeensis [Fabricius, 1777]), and 31 from pinned collections had low-quality DNA extractions. Of the remaining 555 individuals (365 females and 190 males), 487 were morphologically identified as B. t. lusitanicus (87.75%), 58 as B. t. terrestris (10.45%) and 10 as morphological hybrids (1.8%). Within the Iberian sampling, B. t. terrestris individuals and most hybrids were found in the south of the peninsula and in the Pyrenees (Table 1), (i.e. in the two main detected areas of intergradation). However, two hybrids were found in Sierra de Guadarrama (SP_SG3, central peninsula), where two subspecies are not believed to be in contact.
Subspecies and hybrid distribution based on mitochondrial haplotypes
Mitochondrial haplotype information was obtained for the 555 morphologically described individuals. The mitochondrial haplotype was determined by the nad2-RFLP approach in 433 individuals (78.01%), by sequencing the Bter_rrnL fragment in 33 individuals (5.95%) and by the 16S fragment in 89 individuals (16.04%) from previous studies23,39.
Two haplotypes were detected: a haplotype more frequent in the Spanish and Portuguese populations (the Iberian haplotype, SP + PT: 81.70%) and one more frequent in the Belgian and French populations (the central-European haplotype, REF_BL: 100%; REF_FR: 86.96%) (Fig. 1). A different haplotype from that expected based on individuals’ morphology (discrepant hybrid) was detected in 20.13% of the analysed individuals in the peninsula. B. t. lusitanicus individuals with the central-European haplotype (discrepant hybrids) were found in 12 of the 17 Iberian locations sampled (0% to 44%) and the Pyrenees (18.74%). 61.1% of the naturalized B. t. terrestris found in the south of the peninsula bore the Iberian haplotype. Higher percentages of discrepant hybrids were found in the two intergradation areas: the Pyrenees (PN_JA: 100%; PN_EY: 47.37%; PN_AM: 40.00%) and the southern peninsula (SP_MU2: 44.00%; SP_SN3: 35.94%), while three individuals from France presented the Iberian haplotype (REF_FR: 13.04%) and no hybrids were found in the Belgian reference group. In the historical population, the central-European haplotype was found only in one individual close to the intergradation area of the Pyrenees (Iberian haplotype in REF_SP80: 97.73%).
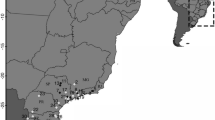
Bombus terrestris subspecies and haplotype distribution in the Iberian Peninsula based on their morphologic traits and the genetic haplotypes within the mitochondrial genes nad2 and rrnL. Subspecies information is shown by colour: yellow for B. t. lusitanicus and blue for B. t. terrestris; haplotype information is shown by pattern: plain for haplotype 1 (Iberian) and striped for haplotype 2 (central European). Morphological hybrids are represented by green and a spotted pattern. The data of the reference populations are shown on the top right. The radius of the circles is proportional to the number of individuals analysed in each location. The data from nearby locations have been merged for readability. This map has been adapted with Inkscape 0.92 from https://commons.wikimedia.org/wiki/File:Iberian_Peninsula_location_map.svg, licensed under the Attribution-ShareAlike 3.0 Unported license (https://creativecommons.org/licenses/by-sa/3.0/).
Microsatellite data validation
Of the ten microsatellite loci amplified, locus B10 was excluded due to a low amplification rate and difficulty in scoring. In addition, 27 individuals with > 30% missing data were removed from further analyses and 18 females were also removed after sibship inference analysis, leaving a total of 320 female individuals for population analysis. Null alleles due to homozygote excess were only found in the Iberian populations (SP + PT) and the historical reference (REF_SP80) at low frequencies (Oosterhout > 0.25%). Most loci presented a statistically significant deviation from HWE expectations due to heterozygote deficit. None of the microsatellite loci analysed showed significant linkage disequilibrium.
Subspecies and hybrid distribution based on microsatellite markers
Microsatellite information was only considered from locations with more than ten genotyped individuals (N = 254; PT_VC, SP_SG2, SP_SG3, SP_MU1, SP_MU2, SP_SN3, PN_EY, REF_SP80, REF_BL and REF_FR).
A total of 19 individuals from the historical population (REF_SP80) were classified as reference B. t. lusitanicus and 17 from Belgium and France (REF_BL and REF_FR) as reference B. t. terrestris by the assignment test using GenAlEx. The presence of hybridization was corroborated with microsatellite analysis. The GeneClass assignment test using Nei’s standard distance41 and the Bayesian method42 allowed us to observe introgression in all the Iberian localities analysed (Fig. 2). Comparing both methods, the Bayesian method assigned a greated percentage of individuals to the lusitanicus subspecies. However, despite the discrepancies in assignment between methods, likely reflecting their statistical differences and the difficulty of reliably detecting hybrids43, both methods approximately coincide in the percentages of hybrids obtained at PT_VC, SP_SG2, SP_MU1 and SP_MU2.
Assignment analysis showed that the percentage of hybrids identified at the nuclear level is higher than that detected with morphological determination and mitochondrial haplotype methods, especially in areas with percentages of hybrids higher than 20.00%. A positive correlation was observed (r2 = 0.593; p = 0.033) between the percentage of hybrids found by mitochondrial haplotype and by nuclear genetic parameters (Supplementary Fig. S2, online).
Genetic diversity and effect of hybridization
Allelic richness (Ar) ranged from 4.27 to 4.63 across the Iberian populations (Table 2). The lowest allelic richness was found in the Pyrenees (PN_EY: 4.16) and the highest value was found in Normandy (Ref_FR: 4.70). More private alleles were found in the overall Iberian population. Genetic diversity estimated from expected heterozygosity (HE) values varied from 0.630 to 0.716 (observed in the Pyrenean PN_EY and French Ref_FR populations, respectively). The fixation index (Fis) showed signs of inbreeding in both the Iberian and the historical reference populations.
To study introgression at the nuclear level in the Iberian populations, population parameters (dataset with hybrids, hIN) were compared with those obtained after the removal of individuals labelled as hybrids (whether by morphological description or because of their mitochondrial haplotype) in each location (dataset without hybrids, hOUT) (Table 2). The Wilcoxon signed-rank test showed a strong variation in the expected heterozygosity values after the removal of the hybrids, although it was not significant after Bonferroni correction (p = 0.014, V = 52). The results after random removal (hIN/rOUT) (p = 0.082, V = 45) suggest that the decrease of HE in hOUT might not be due to the mere removal of individuals (Table 3), but to the hybrid identity of the removed individuals.
Linear regressions showed that genetic diversity values did not depend directly on the percentage of hybrid individuals in each location (r2 = 0.012; p = 0.741) and were likely affected by other variables. However, the variation in HE due to the presence of hybrid individuals (ΔHE = HEhIN − HEhOUT) does depend significantly on the percentage of hybrids present in each location (r2 = 0.593; p = 0.005) (Fig. 3).
Linear regressions between (a) genetic diversity (HE) or (b) variation in genetic diversity (ΔHE) and the percentage of hybrids observed in each population. Genetic diversity does not decrease significantly as the percentage of hybrids rises (r2 = 0.012, p = 0.741), but when the percentage of hybrids increases, the variation in genetic diversity increases as well (r2 = 0.593, p = 0.005).
Genetic structure
Structure Harvester showed K = 4 as the most likely number of clusters (Supplementary Fig. S3, online), although this result is probably a consequence of the limitations of the program, which cannot detect values of K = 144. However, K = 2 was also studied to investigate possible structuration due to the subspecies (lusitanicus and terrestris) present (Supplementary Fig. S4, online). Visualization of Structure results indicates approximately equal contributions to each population (K), implying that the true value of K is indeed 1, with no clear structuring due to subspecies. The find.clusters function from the package adegenet 2.1.1 showed that BIC was minimised at K = 4, with K = 2 presenting the largest increase in BIC (Supplementary Fig. S5, online). Clusters observed at K = 2 to K = 4 were not related either to the subspecies identification or population (Supplementary Fig. S6, online). A final analysis in which populations were selected as clusters (Fig. 4) showed an overlapping distribution in the axis, indicating a low degree of genetic differentiation and corroborating the Structure results. However, the Iberian population from the historical reference group REF_SP80 presented a higher degree of clustering than the other populations.
Scatter plot from discriminant analysis of principal components. 80 principal components (PC) were selected after α-score estimations to avoid overfitting. Chosen discriminant analysis eigenvalues are depicted in the top right of the plot. Ellipses indicate the 95% confidence interval of assignment, yellow squares identify bumblebee individuals collected before the 1990s, orange rhombuses represent Portuguese samples, green represents individuals from Sierra de Guadarrama, pink represents individuals from the Sierra Nevada and blue represents samples from outside the Iberian Peninsula, as detailed in the figure legend at the bottom right.
Diploid male detection
A total of 13 diploid males were found out of the 181 male individuals sampled in the Iberian Peninsula (Table 1): one in the north (SP_VI: 1, ϕ = 0.3), seven in the central area (SP_SG2: 5, ϕ = 0.07; SP_MA: 2, ϕ = 0.66) and five in the south (SP_MU1: 2, ϕ = 0.29; SP_MU2: 2, ϕ = 0.29; SP_SN3: 1, ϕ = 0.04), with an overall ϕ for the Iberian Peninsula of 0.07. A male from the locality SP_SG2 was identified as a triploid (3n) after amplifying each microsatellite locus individually.
Discussion
This research aimed to study the genetic diversity of the Iberian B. terrestris populations under threat of introgression from managed non-native populations at both the spatial and temporal scales. Our results, based on extensive sampling throughout the Iberian Peninsula (Spain and Portugal), indicate that the mitochondrial haplotype associated with central European populations of B. terrestris has expanded beyond both the natural intergradation area (i.e., the Pyrenees) and the anthropic one (i.e., the southeast region), reaching most of the north–south gradient with some exceptions in the northwest. These results contrast with the haplotype distribution in historical populations (i.e., REF_SP80, specimens collected in Spain 40 years ago) in which all individuals presented the Iberian haplotype, except one individual detected close to the natural intergradation area in the Pyrenees, where natural hybridization occurs. These results support the idea that the endemic Iberian haplotype was predominant in the territory and that, due to hybridization and introgression events with naturalized commercial breeds, the central European haplotype has recently spread into the environment. This change in the haplotype distribution of the peninsula is a first signal of how this expansion is modifying the genetic pool of the populations, which may soon develop into losses of local adaptation6,45. Another hypothesis to explain the present genetic structure could be migration, although unlikely. In the context of climate change, the only plausible hypothesis is a migration from the Southern area to the north1. However, based on our knowledge, the European subspecies was never recorded in the Southern region of Spain and we did not detect any individuals with the central European haplotype in the historical data set but one, close to the natural intergradation area in Pyrenees.
Our results expand the current knowledge about the genetic integrity of B. t. lusitanicus in the Iberian Peninsula. Previous records of B. t. terrestris and hybrid individuals have occurred only in the south23,25,39 and west24 (putative genetic hybrids) of the Iberian Peninsula, whereas we have found evidence of introgression in most of the territory (see Table 1 and Fig. 1). The presence of the central European haplotype in the central area of the peninsula, where the subspecies B. t. terrestris has not been detected, could be due to the dispersion of individuals from areas where commercial breeds are being used (greenhouses in the region or an expansion from the south), as B. terrestris dispersion can be assisted by strong winds to cross long distances15,33. On the other hand, in the most northwestern localities sampled (SP_PO, SP_BR and SP_PA), the central European haplotype was not found, which suggests that these populations of B. t. lusitanicus are less affected by introgression, probably due to a lower density of greenhouses in the area, as well as a larger distance from the two main areas of intergradation40 (Supplementary Fig. S1 Online).
This study confirms that the inclusion of samples from old collections is crucial to assess the evolution of the genetic diversity of local populations, as has been done before in other species of the genus Bombus (Latreille, 1802)29. Furthermore, the use of old samples as historical references in population genetics will be especially important in subsequent years, as we monitor the expected changes in the distribution of not only B. terrestris subspecies but also other organisms that are affected by climatic change and anthropisation. The distribution range of B. t. lusitanicus, instead of shifting or reducing, is expanding, both in altitude and latitude13,18,46, while the populations of B. t. audax in the UK have become bivoltine7. Given the subspecies’ similar resistance to heat stress14, further changes due to climatic change are expected to occur. Even under these conditions, although we must assume that hybridization may play a key role in evolution47,48, there is a consensus that human-induced introgression of non‐indigenous organisms harms native gene pools45,49,50.
As previous studies on the genetic diversity of the species have reported, the heterozygosity values obtained in this analysis, the assignment test (GeneClass) and the clusters inferred (Structure and DAPC) suggest an intense gene flow among B. terrestris populations, which leads to a reduced population structure in a continuum that is not only peninsular but also continental33,51,52. When comparing the Iberian populations with their historical reference, only slight variations can be observed in the allelic frequencies, although the data show an overall decrease in allelic richness and expected heterozygosity values. Conversely, the Wilcoxon test suggests that hybridization and introgression events are affecting the native populations by increasing the genetic diversity of those populations with hybrid individuals, which is expected from the admixture of different gene pools45. Given the low structure of bumblebee populations, the effect of this change is difficult to measure, but if this dynamic continues, the loss of endemicity and increased homogenization of European populations will be recurring concerns in the future.
The analysis of genetic diversity in the Iberian Peninsula showed the lowest values in the central peninsular populations (Sierra de Guadarrama: SP_SG2, SP_SG3), where the highest number of diploid males and one triploid male were found. These results could represent a sign of a potential threat of inbreeding depression in the populations35, although it has been previously discussed that B. terrestris is able to withstand inbreeding in its populations56. In this sense, B. terrestris does not show mating preferences between related and unrelated individuals35, so there are no mechanisms preventing diploidy or even triploidy in males53. However, these results should be taken into account in future studies on the species, as the ploidy values obtained in this study were higher than those of populations of endangered species such as B. florilegus Panfilov, 1956 (2.7%)54 or B. muscorum Linnaeus, 1758 (5%)55.
Given that B. terrestris can escape from greenhouses23,24 and the great expansion capacity of the managed populations in the environment15, we emphasize the importance of the propagation of the commercial B. terrestris subspecies from greenhouses across the Iberian Peninsula as a driver of population change. Moreover, the inadequate management of colonies by breeding companies and farmers after colony use due to a lack of information about the consequences of their dispersal in the environment is another little-addressed factor contributing to the emergence and dispersion of commercial breeds (implied by the presence of the central European haplotype) in the environment. Some of the mispractices that occur in the territory include the outdoor use of colonies, incomplete removal or abandonment of colonies24 and attempts to manage the nesting of commercial queens in the environment (personal observation). To avoid escapes, bumblebee nests should be placed only inside greenhouses and destroyed at the end of the crop pollination campaign before new sexual individuals emerge. The arrival of commercial breeds of B. terrestris into new environments and their subsequent colonization is guaranteed unless stricter regulations on their transport and management are adopted16, both outside and within the species’ natural distribution range8,13. To our concern, there is still no legislation regarding the management of bumblebee colonies for agricultural use in the Iberian Peninsula beyond the advice and instructions of the provider companies. Because the Iberian Peninsula is a large producer and exporter of fruits and vegetables, and therefore an intensive user of commercial pollination services, we endorse stricter legislation on the importation of foreign subspecies and support for companies breeding local subspecies11, as the breeding of B. t. lusitanicus does not involve additional costs compared with other subspecies of B. terrestris (de Jonghe; Rasmont; personal communication). Such regulations are already in effect in other regions within the natural distribution range of the species, such as the United Kingdom, Norway and the Canary Islands. Authorization of the importation of exotic taxa should only be legalised after studying the survival rate, expansion capacity and environmental impact of the managed populations. It is important to coordinate an update of the existing legislation with more active dissemination to first-hand users on the correct management of commercial breeds and the benefits of using local subspecies.
Material and methods
Spatial and temporal framework
Individuals were collected along the Iberian Peninsula, the Pyrenees, north of France, and Belgium (Fig. 1). The sampling was focused on the National Parks of Sierra de Guadarrama and Sierra Nevada (central and southern regions of the Iberian Peninsula, respectively), which are immersed in very different landscape contexts. In this sense, agriculture in the surroundings of Sierra de Guadarrama is more dependent on irrigated and rain-fed crops (greenhouse crops in the surrounding provinces cover less than 200 ha), while in the vicinity of Sierra Nevada, greenhouse crops have expanded over the last 50 years to more than 30,000 ha (Supplementary Fig. S1 Online), some of them less than 20 km from the boundaries of the National Park38. To determine the impact of the introduction of commercial bumblebees, a set of individuals collected in Spain in the 1980s before the use of bumblebee colonies started in the peninsula in the 1990s, was included in the analyses. In the current sampling we considered as a population the set of individuals sampled within a radius of 10 km based on the maximum flight distance recorded for B. terrestris (according to Kraus et al.57) taking into account geographic barriers (e.g. if mountains were present).
Sample collection
A total of 594 individuals were initially selected for this work. From them, 437 individuals were sampled (256 females and 181 males) in a north–south gradient in 17 locations of the Iberian Peninsula (Supplementary Table S1 online). Two sets of reference populations were additionally considered for spatial comparisons: 78 individuals from the Pyrenees as it is the natural intergradation area between the two subspecies, 23 individuals from France (Normandy) and 12 from Belgium as they are within the natural distribution range of B. t. terrestris. As a historical reference, 44 individuals collected in Spain before the introduction of commercial B. terrestris39 were included (Table 1, Supplementary Fig. S7 online).
Individuals from the Iberian Peninsula, Belgium and France were net-sampled and maintained individually in absolute ethanol at -20 ºC until processed. Individuals from the Pyrenees and the historical reference sampling were obtained from the pinned collections of the University of Mons and the Complutense University of Madrid (UCME), respectively. A first subspecies identification of individuals was performed through their morphological characters18,20 (i.e. ferruginous brown coloration only present in B. t. lusitanicus).
DNA extraction
Total DNA was extracted from one leg of each individual following Ivanova et al.58 protocol. Concentration and purity of DNA extractions from pinned individuals were quantified, as DNA obtained from antique individuals presents a lesser quality29,59.
Mitochondrial haplotype screening
Haplotype information of B. terrestris individuals was obtained with the RFLP approach on the NADH dehydrogenase subunit 2 gene (nad2) following Cejas et al.23. To improve the amplification success, Bovine Serum Albumin (BSA, 1.2 mg/mL) was added to the PCR reactions.
DNA is often highly fragmented in samples from old collections, making it difficult to amplify larger sequences59. Therefore, for the Pyrenean individuals, a new primer set (F: 5’-TCAACATCGAGGTCGCAATCA-3’ and R: 5’-TGGCTGCGGTATAATTGACTGT-3’) was designed with Primer360 to obtain a shorter fragment (388 bp) within the 16S ribosomal RNA (rrnL) gene. The same PCR conditions were used as in Hines et al.61. Amplicons were sequenced in Secugen (Madrid, Spain). We added the haplotype information from the samples processed in Cejas et al.23,39 by sequencing the 16S fragment as described in Hines et al.61.
Microsatellite amplification and sample genotyping
Ten microsatellite loci designed for B. terrestris51,62 were amplified in all individuals with two multiplex sets following Cejas et al.63: RB1 (B10, B11, B100, B96, B124 and B126), and RB2 (B118, B119, B121 and B132). PCR products were sent to the Servei Central de Suport a la Investigació Experimental (University of Valencia, Spain) for capillary electrophoresis on an ABI Prism 3700 (Applied Biosystems). Allele scoring was performed using GENEMAPPER 4.8 (Applied Biosystems) by comparing alleles with an internal size standard (GeneScan-500 Liz, Applied Biosystems). A second allele scoring quality control was made manually to avoid the wrong assignation as alleles of fluorescence peaks during the automated allele scoring process. Individuals with missing data > 30% were discarded. Siblings were inferred with Colony 2.0.6.464 with a Full-Likelihood analysis under default parameters for haplodiploid species. We retained only one individual from every kinship with a greater probability of 70%.
Genetic diversity and effect of hybridization
The individual assignment test was performed with the program GeneClass 2.065 to find the proportion of hybrid individuals among subspecies in the Iberian populations, using Nei’s standard distance41 and a Bayesian method42 with MCMC method66 as the criteria for computation. For these analyses, the purest individuals from reference populations (REF_SP80 for B. t. lusitanicus, REF_FR and REF_BL for B. t. terrestris) detected in the assignment test (assignation by logarithmic likelihood) in GenAlEx 6.567 were selected as reference groups. The assignment of the Iberian samples was evaluated.
To evaluate the potential effect of the hybridization and introgression between populations, genetic diversity parameters were calculated (1) in Iberian locations with an appropiate number of samples (≥ 14 individuals, 7 sampling locations), (2) in the Iberian Peninsula as one population (SP + PT), and (3) in the four reference populations (dataset recorded as hIN). The same genetic diversity parameters were estimated again after removing all detected hybrids (morphologically identified hybridsand thoseindividuals with discrepant mitochondrial haplotype) and naturalized B. t. terrestris individuals (dataset recorded as hOUT). Same remarks were taken into consideration for the French population, (Supplementary Table S2 online). Finally, to avoid type-I error the analyses were conducted again after removing a random set of the same size (dataset recorded as rOUT).
To avoid bias due to the haplodiploidy of the species, only genotypes of female individuals were considered. Micro-Checker 2.2.368 was used to examine genotyping errors such as stutter bands and the frequency of null alleles. Departure of Hardy–Weinberg equilibrium (HWE) and estimates of linkage disequilibrium (LD) were calculated for the Iberian and reference populations using Genepop69. The level of significance for the analyses was adjusted with Bonferroni corrections (p-value = 0.005 and 0.0002, respectively). Observed and expected heterozygosity (HO and HE) were estimated with GenAlEx 6.5. Allele richness (Ar) and private allelic richness (pAr) were calculated with Hp-Rare 1.170, and rarefacted to a sample size of 10 gene copies. The inbreeding coefficient (Fis) was obtained with FSTAT 2.9.471.
Obtained parameters from datasets hIN and hOUT were compared with the non-parametric Wilcoxon signed-rank test for related samples using package stats 3.6.0 (function wilcox.test, paired = TRUE) in R72. For this test, data was paired according to their label and ranked by their absolute differences. The level of significance for the analysis was adjusted under Bonferroni corrections (p-value = 0.0083). Wilcoxon signed-rank test was repeated against the random removal of individuals (datasets hIN and rOUT), in order to investigate whether the significance of the results was due to the removal of individuals itself or to the identity of those excluded. The Iberian Peninsula as a population (SP + PT) was not included in this analysis to avoid oversampling.The percentage of hybrid individuals found in each location was compared with the values of expected heterozygosity (HE) and with the change in heterozygosity (ΔHE), after removing the hybrids (hOUT─hIN), to estimate the effect of introgression on the genetic diversity of the populations through linear regressions in R with package stats v3.6.272.
Genetic structure
A clustering analysis was carried out on Structure73 with a burn-in period of 100,000 steps and 1,000,000 MCMC repetitions. Potential clustering values (K) 1 to 10 were tested under 10 iterations each. Structure Harvester42 was used to select the best fitting K by calculating ΔK74. Software Clumpak75 was used to reach a consensus between the different iterations of each selected K, following the Markov clustering algorithm. Furthermore, a graphical representation was printed with Distruct 1.176.
Discriminant analyses of principal components (DAPC) was performed using R (R Core Team, 2013) package adegenet 2.1.177. Function find.clusters was used to identify the number of genetic clusters according to the Bayesian Inference Criterion (BIC). Nevertheless, from a biogeographic perspective, other K numbers were also analysed. Finally, populations were selected as clusters to investigate their identity and relationship. The number of principal components for the analysis was selected after α-score estimations to explain the data while avoiding overfitting. Results were graphically represented in a scatter plot.
Diploid male monitoring
The proportion (ϕ) of diploid males (2n ♂) was assessed for each sampling location. Males with two or more diploid loci were considered as diploids. Diploidy proportion was calculated following Zayed et al.34 by dividing the number of diploid males between the total number of sampled male individuals in the location.
Data availability
Microsatellite genotypes of the populations for analysis hIN and hOUT have been detailed in Supplementary Table S3.
References
Sage, R. F. Global change biology: A primer. Glob. Change Biol. 26, 3–30 (2020).
Sutherland, W. J. et al. A horizon scan of emerging issues for global conservation in 2019. Trends Ecol. Evol. 34, 83–94 (2018).
Porto, R. G. et al. Pollination ecosystem services: A comprehensive review of economic values, research funding and policy actions. Food Secur. 12, 1425–1442 (2020).
Potts, S. G. et al. Safeguarding pollinators and their values to human well-being. Nature 540, 1–10 (2016).
Goulson, D., Nicholls, E., Botías, C. & Rotheray, E. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 347, 1255957 (2015).
Ellis, J. S. et al. Introgression in native populations of Apis mellifera mellifera L: implications for conservation. J. Insect Conserv. 22, 377–390 (2018).
Hart, A. F., Maebe, K., Brown, G., Smagghe, G. & Ings, T. Winter activity unrelated to introgression in British bumblebee Bombus terrestris audax. Apidologie 52, 315–327 (2021).
Ings, T. C., Ward, N. L. & Chittka, L. Can commercially imported bumble bees out-compete their native conspecifics?. J. Appl. Ecol. 43, 940–948 (2006).
Graystock, P., Blane, E. J., McFrederick, Q. S., Goulson, D. & Hughes, W. O. Do managed bees drive parasite spread and emergence in wild bees?. IJP-PAW 5, 64–75 (2016).
Chandler, D., Cooper, E. & Prince, G. Are there risks to wild European bumble bees from using commercial stocks of domesticated Bombus terrestris for crop pollination?. J. Apic. Res. 58, 1–17 (2019).
Velthuis, H. H. W. & Doorn, A. A century of advances in bumblebee domestication and the economic and environmental aspects of its commercialization for pollination. Apidologie 37, 421–451 (2006).
Trillo, A. et al. Contrasting occurrence patterns of managed and native bumblebees in natural habitats across a greenhouse landscape gradient. Agric. Ecosyst. Environ. 272, 230–236 (2019).
Lecocq, T., Rasmont, P., Harpke, A. & Schweiger, O. Improving international trade regulation by considering intraspecific variation for invasion risk assessment of commercially traded species: The Bombus terrestris case. Conserv. Lett. 9, 281–289 (2015).
Martinet, B. et al. Global effects of extreme temperatures on wild bumblebees. Conserv. Biol. 35(5), 1507–1518 (2021).
Schmid-Hempel, R. et al. The invasion of southern South America by imported bumblebees and associated parasites. J. Anim. Ecol. 83, 823–837 (2014).
Aizen, M. A. et al. Coordinated species importation policies are needed to reduce serious invasions globally: The case of alien bumblebees in South America. J. Appl. Ecol. 56, 100–106 (2018).
Tsuchida, K., Yamaguchi, A., Kanbe, Y. & Goka, K. Reproductive interference in an introduced bumblebee: Polyandry may mitigate negative reproductive impact. Insects 10, 59 (2019).
Rasmont, P., Coppée, A., Michez, D. & De Meulemeester, T. An overview of the Bombus terrestris (L. 1758) subspecies (Hymenoptera: Apidae). Ann. Soc. Entomol. Fr. (N.S.) 44, 243–250 (2008).
Lecocq, T. et al. The alien’s identity: Consequences of taxonomic status for the international bumblebee trade regulations. Biol. Conserv. 195, 169–176 (2016).
Ornosa, C. & Ortiz-Sánchez, F. Hymenoptera: Apoidea I. In Fauna Ibérica Vol. 23 (eds Ramos, M. A. et al.) (Museo Nacional de Ciencias Naturales, CSIC, 2004).
Hewitt, G. M. Mediterranean Peninsulas: The Evolution of Hotspots. In Biodiversity Hotspots (eds Zachos, F. & Habel, J.) 123–147 (Springer-Verlag, 2011).
Ortiz-Sánchez, F. Introducción de Bombus terrestris terrestris (Linnaeus, 1758) en el Sur de España para la polinización de cultivos en invernadero (Hymenoptera, Apidae). Boln. Asoc. Esp. Ent. 16, 247–248 (1992).
Cejas, D., López-López, A., Muñoz, I., Ornosa, C. & De la Rúa, P. Unveiling introgression in bumblebee (Bombus terrestris) populations through mitogenome-based markers. Anim. Genet. 51, 70–77 (2020).
Seabra, S. G. et al. Genomic signatures of introgression between commercial and native bumblebees, Bombus terrestris, in western Iberian Peninsula—Implications for conservation and trade regulation. Evol. Appl. 12, 1–13 (2019).
Bartomeus, I., Molina, F. P., Hidalgo-Galiana, A. & Ortego, J. Safeguarding the genetic integrity of native pollinators requires stronger regulations on commercial lines. Ecol. Solut. Evid. 1(1), e12012 (2020).
Coates, D. J., Byrne, M. & Moritz, C. Genetic diversity and conservation units: Dealing with the species-population continuum in the age of genomics. Front. Ecol. Evol. 6, 165 (2018).
Williams, P. H. et al. Genes suggest ancestral colour polymorphisms are shared across morphologically cryptic species in arctic bumblebees. PLoS ONE 10, e0144544 (2015).
Gosterit, A. Adverse effects of inbreeding on colony foundation success in bumblebees, Bombus terrestris (Hymenoptera: Apidae). Appl. Entomol. Zool. 51, 521–526 (2016).
Maebe, K., Karise, R., Meeus, I., Mänd, M. & Smagghe, G. Pattern of population structuring between Belgian and Estonian bumblebees. Sci. Rep. 9, 1–8 (2019).
Allio, R., Donega, S., Galtier, N. & Nabholz, B. Large variation in the ratio of mitochondrial to nuclear mutation rate across animals: Implications for genetic diversity and the use of mitochondrial DNA as a molecular marker. Mol. Biol. Evol. 34, 2762–2772 (2017).
Patten, M. M., Carioscia, S. A. & Linnen, C. R. Biased introgression of mitochondrial and nuclear genes: A comparison of diploid and haplodiploid systems. Mol. Ecol. 24, 5200–5210 (2015).
Gosterit, A. & Baskar, V. C. Impacts of commercialization on the developmental characteristics of native Bombus terrestris (L.) colonies. Insectes Soc. 63, 609–614 (2016).
Moreira, A. S., Horgan, F. G., Murray, T. E. & Kakouli-Duarte, T. Population genetic structure of Bombus terrestris in Europe: Isolation and genetic differentiation of Irish and British populations. Mol. Ecol. 24, 3257–3268 (2015).
Zayed, A. Bee genetics and conservation. Apidologie 40, 237–262 (2009).
Schenau, E. & Jha, S. High levels of male diploidy but low levels of genetic structure characterize Bombus vosnesenskii populations across the Western US. Conserv. Genet. 18, 597–605 (2017).
Van Wilgenburg, E., Driessen, G. & Beukeboom, L. W. Single locus complementary sex determination in Hymenoptera: An “unintelligent” design?. Front. Zool. 3, 1–15 (2006).
Bogo, G. et al. No evidence for an inbreeding avoidance system in the bumble bee Bombus terrestris. Apidologie 49, 473–483 (2018).
Kent, C. F. et al. Conservation genomics of the declining North American bumblebee Bombus terricola reveals inbreeding and selection on immune genes. Front. Genet. 9, 316 (2018).
Cejas, D., Ornosa, C., Muñoz, I. & De la Rúa, P. Searching for molecular markers to differentiate Bombus terrestris (Linnaeus) subspecies in the Iberian Peninsula. Sociobiology 65, 558–565 (2018).
Ministerio de Agricultura, Pesca y Alimentación de España. Encuesta sobre Superficies y Rendimientos de Cultivos (ESYRCE). https://cpage.mpr.gob.es N.I.P.O.: 001-19-051-9 (2021).
Nei, M. Genetic distance between populations. Am. Nat. 106, 283–292 (1972).
Rannala, B. & Mountain, J. L. Detecting immigration by using multilocus genotypes. PNAS 94, 9197–9201 (1997).
Anderson, E. C. Bayesian inference of species hybrids using multilocus dominant genetic markers. Philos. Trans. R. Soc. B 363(1505), 2841–2850 (2008).
Earl, D. A. & von Holdt, B. M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361 (2012).
Facon, B. et al. Can things get worse when an invasive species hybridizes? The harlequin ladybird Harmonia axyridis in France as a case study. Evol. Appl. 4, 71–88 (2011).
Ornosa, C., Torres, F. & De la Rúa, P. Updated list of bumblebees (Hymenoptera: Apidae) from the Spanish Pyrenees with notes on their decline and conservation status. Zootaxa 4237, 41–77 (2017).
Allendorf, F. W., Leary, R. F., Spruell, P. & Wenburg, J. K. The problems with hybrids: Setting conservation guidelines. Trends Ecol. Evol. 16, 613–622 (2001).
Arnold, M. L. & Kunte, K. Adaptive genetic exchange: A tangled history of admixture and evolutionary innovation. Trends Ecol. Evol. 32, 601–611 (2017).
Mallet, J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 20, 229–237 (2005).
De la Rúa, P. et al. Conserving genetic diversity in the honeybee: Comments on Harpur et al. (2012). Mol. Ecol. 22, 3208–3210 (2013).
Estoup, A., Solignac, M., Cornuet, J. M., Goudet, J. & Scholl, A. Genetic differentiation of continental and island populations of Bombus terrestris (Hymenoptera: Apidae) in Europe. Mol. Ecol. 5, 19–31 (1996).
Silva, S. E. et al. Population genomics of Bombus terrestris reveals high but unstructured genetic diversity in a potential glacial refugium. Biol. J. Linn. Soc. 129, 259–272 (2020).
Ayabe, T., Hoshiba, H. & Ono, M. Cytological evidence for triploid males and females in the bumblebee, Bombus terrestris. Chromosome Res. 12, 215–223 (2004).
Takahashi, J., Ayabe, T., Mitsuhata, M., Shimizu, I. & Ono, M. Diploid male production in a rare and locally distributed bumblebee, Bombus florilegus (Hymenoptera, Apidae). Insectes Soc. 55, 43–50 (2008).
Darvill, B., Ellis, J. S., Lye, G. C. & Goulson, D. Population structure and inbreeding in a rare and declining bumblebee, Bombus muscorum (Hymenoptera: Apidae). Mol. Ecol. 15, 601–611 (2006).
Gerloff, C. U. & Schmid-Hempel, P. Inbreeding depression and family variation in a social insect, Bombus terrestris (Hymenoptera: Apidae). Oikos 111, 67–80 (2005).
Kraus, F. B., Wolf, S. & Moritz, R. F. A. Male flight distance and population substructure in the bumblebee Bombus terrestris. J. Anim. Ecol. 78, 247–252 (2009).
Ivanova, N., Dewaard, J. & Herbert, D. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol. Ecol. Notes 6, 998–1002 (2006).
Wandeler, P., Hoeck, P. E. & Keller, L. F. Back to the future: museum specimens in population genetics. Trends Ecol. Evol. 22, 634–642 (2007).
Rozen, S. & Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Bioinform. Methods Protoc. 132, 365–386 (2000).
Hines, H., Cameron, S. & Williams, P. Molecular phylogeny of the bumble bee subgenus Pyrobombus (Hymenoptera: Apidae: Bombus) with insights into gene utility for lower-level analysis. Invertebr. Syst. 20, 289–303 (2006).
Estoup, A., Scholl, A., Pouvreau, A. & Solignac, M. Monoandry and polyandry in bumble bees (Hymenoptera; Bombinae) as evidenced by highly variable microsatellites. Mol. Ecol. 4, 89–94 (1995).
Cejas, D., Ornosa, C., Muñoz, I. & De la Rúa, P. Preliminary report on cross-species microsatellite amplification for bumblebee biodiversity and conservation studies. Arch. de Zootec. 68, 422–426 (2019).
Wang, J. Computationally efficent sibship and parentage assignment from multilocus marker data. Genetics 191, 183–194 (2012).
Piry, S. et al. Geneclass2: A software for genetic assignment and first generation migrant detection. Heredity 95(6), 536–539 (2004).
Cornuet, J. M., Piry, S., Luikart, G., Estoup, A. & Solignac, M. New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics 153, 1989–2000 (1999).
Peakall, R. & Smouse, P. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 28, 2537–2539 (2012).
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P. & Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4, 535–538 (2004).
Rousset, F. genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106 (2008).
Kalinowski, S. T. HP-RARE 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes 5, 187–189 (2005).
Goudet, J. FSTAT (version 1.2): A computer program to calculate F-statistics. J. Hered. 86, 485–486 (1995).
R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, http://www.R-project.org (2008).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 14, 2611–2620 (2005).
Kopelman, N. M., Mayzel, J., Jakobsson, M., Rosenberg, N. A. & Mayrose, I. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 15, 1179–1191 (2015).
Rosenberg, N. A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 4, 137–138 (2004).
Jombart, T. & Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 24, 1403–1405 (2011).
Acknowledgements
We would like to thank Pierre Rasmont and the University of Mons for the provision of samples and insight, and Ana Asensio and Nuria Blasco for their technical support. Sampling permissions were obtained from the corresponding authorities (National Parks of Sierra Nevada and Sierra de Guadarrama). This research was funded by Fundación Séneca (grant19908/GERM/2015). IM was supported by a MINECO Spanish postdoctoral grant “Juan de la Cierva-Incorporación” (JCI2018-036614-I). DM and DC were partly supported by the "Fonds de la Recherche Scientifique” (FNRS) and the “Fonds Wetenschappelijk Onderzoek” (FWO) under EOS project named CLIPS (n°3094785).
Author information
Authors and Affiliations
Contributions
Design of the work: D.C, I.M., C.O. & P.D.l.R.; Data acquisition: D.C, C.O., D.M. & P.D.l.R.; Laboratory Work: D.C.; Data analysis: D.C& I.M.; Draft: D.C.; Manuscript revision: D.C., I.M. & P.D.l.R. with input from C.O. and D.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cejas, D., De la Rúa, P., Ornosa, C. et al. Spatial and temporal patterns of genetic diversity in Bombus terrestris populations of the Iberian Peninsula and their conservation implications. Sci Rep 11, 22471 (2021). https://doi.org/10.1038/s41598-021-01778-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-01778-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.