Abstract
Optimisation of protein binders relies on laborious screening processes. Investigation of sequence–function relationships of protein binders is particularly slow, since mutants are purified and evaluated individually. Here we developed peptide barcoding, a high-throughput approach for accurate investigation of sequence–function relationships of hundreds of protein binders at once. Our approach is based on combining the generation of a mutagenised nanobody library fused with unique peptide barcodes, the formation of nanobody–antigen complexes at different ratios, their fine fractionation by size-exclusion chromatography and quantification of peptide barcodes by targeted proteomics. Applying peptide barcoding to an anti-GFP nanobody as a model, we successfully identified residues important for the binding affinity of anti-GFP nanobody at once. Peptide barcoding discriminated subtle changes in KD at the order of nM to sub-nM. Therefore, peptide barcoding is a powerful tool for engineering protein binders, enabling reliable one-pot evaluation of sequence–function relationships.
Similar content being viewed by others
Introduction
Protein binders are indispensable molecules for therapeutics1, diagnostics2 and basic science3. However, current screening processes of protein binders are labour-intensive. Especially, investigating sequence–function relationships of a protein binder, which is essential for optimising protein binders, is slow because individual mutated binders are separately evaluated using low-throughput technologies4,5,6. Typically, diverse amino-acid-substituted mutants are constructed and purified separately, and their binding kinetics are individually evaluated by enzyme-linked immunosorbent assay7, surface plasmon resonance (SPR)8,9 or biolayer interferometry10,11.
Display technologies, such as phage12, yeast13, ribosome14 and messenger RNA15 display, are used as alternatives to low-throughput technologies. These technologies use protein binder display on a surface, which ensures physical genotype–phenotype linkage to retrieve genetic information after selection. Although display technologies enable high-throughput evaluation of highly diverse mutants of a protein binder, fusion of a protein binder on a surface inevitably causes negative effects on screening processes. Display on a surface increases stability of protein binders, and screened protein binders may lose their activities upon conversion into soluble forms16,17. Oligomeric antigens can cause avidity, leading to difficulties in accurate estimation of binding kinetics18,19. Fusion partners can cause nonspecific binding. For example, phage can nonspecifically bind to antigens20 and the huge size of it could lead to co-enrichment of aggregated molecules21. Consequently, display technologies are not appropriate for accurate evaluation of sequence–function relationships of protein binders.
Mass spectrometry (MS)-based approaches have been developed to identify protein binders without display on a surface22,23,24,25,26,27,28. These approaches require proteolytic digestion of protein binder pools and MS analysis of the peptides to estimate binding kinetics. They enable direct detection of free protein binders; however, their potential is limited because (1) unambiguous identification of unique antibody clones is difficult due to the high sequence homology within protein binder pools27 and (2) peptides derived from protein binders often exhibit low ionisation efficiencies, resulting in poor detectability29.
Recently, Egloff et al. and we developed a peptide barcoding approach for high-throughput, reliable identification of protein binders without being displayed on a surface8,30. In this approach, each protein binder is fused with a unique, genetically encoded peptide barcode that contains genotype information. These peptide barcodes are designed to have high detectability for MS, leading to low identification bias and a high identification rate. We used predetermined highly specific, sensitive and quantitative peptide barcodes and detected them using selected reaction monitoring (SRM)-based quantitative targeted proteomics. Egloff et al. used randomised peptide barcodes expected to have high ionisation and fragmentation efficiencies and detected them using an Orbitrap mass spectrometer; they succeeded in deep-mining a nanobody (Nb)-immune repertoire to identify functional protein binders and also in ranking mutant Nbs by their off-rates. These results show the feasibility of peptide barcoding; however, it is unclear whether this approach can accurately evaluate subtle differences in binding kinetics, which is necessary for investigating sequence–function relationships of protein binders.
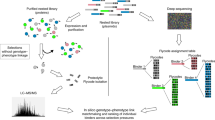
Here, we developed peptide barcoding 2.0 for accurate investigation of the binding kinetics of hundreds of free (non-immobilised) Nbs at once (Fig. 1). Our approach is based on combining the generation of a mutagenised Nb library fused with unique peptide barcodes, the formation of Nb–antigen complexes at different ratios, their fine fractionation by size-exclusion chromatography (SEC) and highly specific quantification of peptide barcodes by SRM-based proteomics. In the experiments described below, we demonstrated that peptide barcoding 2.0 is a powerful tool for engineering protein binders, enabling reliable one-pot evaluation of sequence–function relationships.
Peptide barcoding for one-pot evaluation of sequence–function relationships of free nanobodies (Nbs). (a) DNA library encoding combinatorial mutant Nbs. Each mutant Nb is fused with a unique peptide barcode. (b) One-pot production of mutant Nbs by Pichia pastoris. (c) One-pot separation of functional and nonfunctional Nbs by size-exclusion chromatography (SEC). Each fraction is collected for further analysis. (d) Evaluation of sequence–function relationships of free Nbs. Peptide barcodes are cleaved out from Nbs in each fraction and quantified by liquid chromatography–tandem mass spectrometry (LC–MS/MS). The relative amount of each peptide barcode in each fraction correlates to the binding kinetics of each mutant Nb. This figure was created using Illustrator CS2 (https://www.adobe.com/).
Results
Selection of peptide barcodes
We tried to select candidate peptide barcodes from the yeast SRMAtlas31, a compendium for highly specific, sensitive and quantitative targeted proteomics. We reasoned that we can easily screen peptides with appropriate profiles as peptide barcodes using the yeast SRMAtlas for SRM-based proteomic workflows. We selected 838 candidate peptides that were composed of 8–10 amino acids without methionine and cysteine32 and that showed optimal detectability via liquid chromatography–tandem mass spectrometry (LC–MS/MS). We confirmed that these 838 peptides showed high specificity and detectability in SRM analysis (Fig. 2a and Supplementary Table 1). We selected 107 peptides that are enough to cover all anti-green fluorescent protein (GFP) mutant Nbs with single amino acid substitution. The 107 peptide barcodes had diverse physiochemical properties in terms of the isoelectric point (pI)33, hydropathicity (the GRAVY score)34 and retention times (Fig. 2b and Supplementary Fig. 1) and showed no sequence similarity (Supplementary Fig. 2). Diverse hydropathicity is especially important to fully exploit the separation power of liquid chromatography (LC).
Selection of peptide barcodes. (a) Peak intensities of 838 candidate peptide barcodes analysed by liquid chromatography–tandem mass spectrometry (LC–MS/MS). Candidate peptide barcodes were selected using the yeast SRMAtlas. The vertical axis shows the sum of the intensities of each peptide obtained from three independent analyses. The intensity of each peptide was calculated as the sum of the peak areas of four transitions. The horizontal axis shows the number of corresponding proteins per cell calculated previously35. Blue dots indicate analysed peptides, and red dots indicate selected peptide barcodes among them. (b) Physiochemical properties of the selected 107 peptide barcodes. The hydropathicity (GRAVY) and isoelectric point (pI) of each peptide were mapped in a 2D map. (c) Physicochemical properties of nine representative peptide barcodes. The selected 107 peptide barcodes were classified into nine groups based on GRAVY and pI values, and 1 representative peptide was selected from each group. The left and right figures in parentheses indicate GRAVY and pI values, respectively. This figure was created using Illustrator CS2 (https://www.adobe.com/).
To determine whether fusion with peptide barcodes affect the function of Nbs, the 107 peptide barcodes were classified into nine categories in terms of hydropathicity and pI, 1 representative peptide was selected from each category (Fig. 2c) and anti-GFP Nb were fused with these representative nine peptide barcodes. We chose anti-GFP Nb as a model because it is a popular Nb used in various studies36,37,38,39,40 and the crystal structure of the anti-GFP Nb–GFP complex has been already solved41,42, which was an important resource to validate our peptide barcoding analysis. The binding kinetics of these peptide-barcoded Nbs were similar to that of wild-type (WT) Nb (Table 1 and Supplementary Fig. 3). This result indicated that most, if not all, short peptide barcodes with diverse physiochemical properties do not affect the function of anti-GFP Nb.
Production of anti-GFP alanine scanning mutant Nbs
We planned to carry out alanine scanning of anti-GFP Nb to prove the feasibility of one-pot evaluation of sequence–function relationships of free Nbs. Alanine scanning is an appropriate method to identify important residues of proteins and provide important information for investigating sequence–function relationships. Each non-alanine residue of anti-GFP Nb was substituted with alanine. We constructed a library of 107 plasmids encoding anti-GFP WT and mutant Nbs fused with unique peptide barcodes at their C-termini (Supplementary Table 2). The library was introduced into Pichia pastoris by electroporation. We obtained 2500 unique clones, which were sufficient to cover all 107 anti-GFP WT and mutant Nbs. The colonies were collected in one-pot and subjected to production and purification processes. Successful production and purification of anti-GFP WT and mutant Nbs were confirmed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie Brilliant Blue (CBB) staining (Supplementary Fig. 4). The purified anti-GFP mutant Nbs were subjected to SRM analysis, and we identified most of anti-GFP mutant Nbs (102/107) (Supplementary Table 3).
One-pot evaluation of sequence–function relationships of free anti-GFP mutant Nbs using peptide barcoding
We adopted SEC for one-pot separation of functional and nonfunctional anti-GFP mutant Nbs. First, we investigated the separation power of a Superdex 75 increase 10/300 GL column. We injected GFP (27 kDa) with or without equimolar anti-GFP WT Nb (16 kDa) into the column. The GFP peak clearly shifted with the addition of anti-GFP WT Nb to higher-molecular-weight fractions (Fig. 3a and Supplementary Fig. 5a), indicating successful separation of GFP and the anti-GFP WT Nb–GFP complex using SEC. Then, we carried out one-pot separation of functional and nonfunctional anti-GFP mutant Nbs by SEC. GFP alone, the anti-GFP mutant Nb library alone and an equimolar mixture of them were separately injected into the column (Fig. 3b and Supplementary Fig. 5b). Injection of anti-GFP mutant Nbs alone showed a single peak, suggesting that they were purified without aggregation. As shown in Fig. 3a, the GFP peak clearly shifted with the addition of the anti-GFP mutant Nb library to higher-molecular-weight fractions (Fig. 3b). The peak of the complex formed by Nb mutants was slightly shifted from the peak formed by WT Nb probably because of experimental errors derived from AKTA. The fractions (F1–F14) in these SEC experiments were collected and analysed by SDS-PAGE and silver staining (Fig. 3c–f). SDS-PAGE analysis showed that anti-GFP mutant Nbs were separated in two regions, indicating that fractions F4–F7 contained Nbs bound to GFP and fractions F11–F13 contained nonbound Nbs.
One-pot evaluation of affinities of the anti-green fluorescent protein (GFP) mutant nanobody (Nb) library. (a, b) Size-exclusion chromatography (SEC) for separation of functional and nonfunctional Nbs. To confirm separation of GFP and the GFP–Nb complex, GFP alone and a mixture of equimolar amounts of GFP and anti-GFP wild-type (WT) Nb were subjected to SEC in (a). For one-pot evaluation of affinities of the anti-GFP mutant Nb library, GFP alone, the anti-GFP mutant Nb library alone and a mixture of equimolar amounts of GFP and the anti-GFP mutant Nb library were subjected to SEC in (b). The purified sample from Pichia pastoris transformed with a backbone vector (pPIC9K) was used as a control. Fourteen fractions were collected in each experiment. (c–f) Sodium dodecyl sulphate–polyacrylamide gel electrophoresis and silver staining of collected fractions. Fractions from the SEC analysis of GFP (27 kDa) are shown in (c), anti-GFP WT Nb (16 kDa) in (d), equimolar amounts of GFP and anti-GFP WT Nb in (e) and equimolar amounts of GFP and the anti-GFP mutant Nb library in (f). Fraction numbers correspond to those of SEC analysis. These gels are cropped and full-length gels are presented in Supplementary Figs. 9–12. (g) Quantification of the relative amount of each peptide barcode in each fraction. The total amount of each peptide barcode in fractions F3–F7 and F11–F12 was defined as 1. Each dotted line indicates each peptide barcode. (h) Identification of nonfunctional anti-GFP mutant Nbs. The graph shows the relative amount of each peptide barcode in fractions F11 and F12 in which nonfunctional mutant Nbs were enriched. The total amount of each peptide barcode in fractions F3–F7 and F11–F12 was defined as 1. Five nonfunctional anti-GFP mutant Nbs whose peptide barcodes were mostly detected in fractions F11 and F12 (> 50%) are coloured in dark blue. Anti-GFP mutant Nbs whose peptide barcodes were not identified by mass spectrometry are not shown. The data shown are the first of two independent experiments, and the second showed equivalent results to the first (Supplementary Fig. 5). This figure was created using Illustrator CS2 (https://www.adobe.com/).
Functional and nonfunctional anti-GFP mutant Nbs were annotated by quantification of peptide barcodes derived from Nbs in the collected fractions. The proteins in each fraction were digested and the resultant peptides subjected to SRM analysis. The relative amount of each peptide barcode in each fraction was calculated against the total amount of each peptide barcode in all analysed fractions. The plot of the relative amount of each peptide barcode in each fraction showed that the majority of peptide barcodes were enriched in fraction F5 (Fig. 3g and Supplementary Fig. 5c). This result was consistent with SEC and SDS-PAGE analyses (Fig. 3b,f). No peaks of peptide barcodes were detected in the fractions of the control sample (GFP + pPIC9K in Fig. 3b), suggesting that SRM analysis enables highly specific quantification of peptide barcodes derived from anti-GFP mutant Nbs. To identify nonfunctional mutants, we explored the relative amount of each peptide barcode in the fractions containing nonbound Nbs (F11–F12). Peptide barcodes derived from five anti-GFP mutant Nbs (R35A, Y37A, W47A, G50A and E103A) were highly enriched in the nonbound fractions (Fig. 3h and Supplementary Fig. 5d). More than 90% of the peptide barcodes derived from R35A, Y37A, G50A and E103A and nearly 80% from W47A were found in the nonbound fractions (Fig. 3h). These results suggested that the five anti-GFP mutant Nbs have lower affinities against GFP than that of anti-GFP WT Nb and the difference in the relative amount of each peptide barcode in the nonbound fractions reflected differences in their affinities.
Stringent enrichment of nonfunctional anti-GFP mutant Nbs
We reasoned that we could separate low-affinity and nonfunctional Nbs by increasing the amount of GFP. We changed the stoichiometric balance of GFP and the anti-GFP mutant Nb library from 1:1 to 2:1 and subjected the protein mixture to SEC. Most of the anti-GFP mutant Nbs were detected in the first half of the fractions (Fig. 4a and Supplementary Fig. 6a), suggesting that most anti-GFP mutant Nbs are bound to GFP. The SEC analysis fractions were collected, and the peptide barcodes in each fraction were quantified by SRM analysis. The plot of the relative amount of each peptide barcode in each fraction showed that the majority of peptide barcodes had the strongest intensities in fraction F5 (Fig. 4b and Supplementary Fig. 6b). Almost no peptide barcode was detected in fractions F11 and F12, suggesting that most Nbs bound to GFP and were eluted at higher-molecular-weight fractions. To identify low-affinity or nonfunctional mutants, we determined the relative amount of each peptide barcode in fraction F7. Peptide barcodes derived from two anti-GFP mutant Nbs (R35A and E103A) were highly enriched in fraction F7 (Fig. 4c and Supplementary Fig. 6c), and 78% of peptide barcodes derived from R35A and 68.5% from E103A were detected in fraction F7 (Fig. 4c). Other peptide barcodes were not enriched in fraction F7. These results suggested that R35A and E103A have very weak or no affinity against GFP.
Separation of low-affinity and nonfunctional nanobodies (Nbs) by varying the green fluorescent protein (GFP)/Nb molar ratio. (a) Size-exclusion chromatography (SEC) for separation of low-affinity and nonfunctional Nbs. For one-pot evaluation of affinities of the anti-GFP mutant Nb library, GFP alone and a mixture of GFP and the anti-GFP mutant Nb library (2:1 molar amount) were subjected to SEC. The purified sample from Pichia pastoris transformed with a backbone vector (pPIC9K) was used as a control. Fourteen fractions were collected in each experiment. (b) Quantification of the relative amount of each peptide barcode in each fraction. The total amount of each peptide barcode in fractions F3–F7 was defined as 1. Each dotted line indicates each peptide barcode. (c) Identification of nonfunctional anti-GFP mutant Nbs. The graph shows the relative amount of each peptide barcode in fraction F7 in which nonfunctional mutant Nbs were enriched. The total amount of each peptide barcode in fractions F3–F7 was defined as 1. Two nonfunctional anti-GFP mutant Nbs whose peptide barcodes were mostly detected in fraction F7 (> 50%) are coloured in dark blue. Anti-GFP mutant Nbs whose peptide barcodes were not identified by mass spectrometry (including G50A) are not shown. The data shown are the first of two independent experiments, and the second showed equivalent results to the first (Supplementary Fig. 6). This figure was created using Illustrator CS2 (https://www.adobe.com/).
Validation of peptide barcoding analyses by SPR analysis
We tried to validate the peptide barcoding analyses by SPR analysis. Five anti-GFP mutant Nbs (R35A, Y37A, W47A, G50A and E103A) without peptide barcodes were produced by P. pastoris and purified with Ni–nitrilotriacetic acid (Ni–NTA) agarose. SPR analysis showed that R35A, G50A and E103A did not bind to GFP and Y37A and W47A had lower affinities compared with anti-GFP WT Nb (Table 2 and Supplementary Fig. 7). This result was highly consistent with our peptide barcoding analyses. The anti-GFP mutant Nbs annotated as nonfunctional protein binders in the stringent SEC analysis showed no binding to GFP in SPR. In addition, anti-GFP mutant Nbs annotated as weak protein binders in the first SEC analysis showed weak affinities against GFP in SPR. These results showed that peptide barcoding 2.0 is useful for evaluating the affinities of free Nbs in a multiplex manner.
Mechanistic estimation of sequence–function relationships of anti-GFP Nb
We investigated the crystal structure of the anti-GFP Nb–GFP complex (Protein Data Bank accession codes 3OGO [PDBID 3OGO])42 to validate our results and to infer why the identified mutations (R35A, Y37A, W47A, G50A and E103A) led to decreased affinities. The crystal structure showed that four residues (R35, Y37, W47 and E103) among the five identified ones are located at the binding surface to GFP42 (Fig. 5a,b). We also calculated the binding free energy of the anti-GFP Nb–GFP complex (Fig. 5c). The three residues (Y37, W47 and E103) had high negative ΔG and stabilised interaction with GFP. R35 had positive ΔG, but it forms a salt bridge with E142 of GFP and contributes to specific binding to GFP42. The crystal structure and binding free energy analyses supported the results of our peptide barcoding analyses.
Mechanistic estimation of effects of important residues on binding affinities. (a, b) Crystal structure of the green fluorescent protein (GFP)–anti-GFP wild-type (WT) Nb complex (PDBID 3OGO)42. GFP is coloured in green and Nb in light blue and yellow. The GFP–anti-GFP WT Nb interface was enlarged. (c) Calculation of the binding free energy of anti-GFP WT Nb. The binding free energy of each residue against GFP was calculated and shown as the mean ± standard error of the mean (SEM) (n = 5) from 50 ns of the prediction run. The important residues annotated by peptide barcoding are shown in dark blue. (d) Simulated structure of anti-GFP G50A mutant Nb binding to GFP. The GFP–anti-GFP G50A mutant Nb interface was enlarged. GFP is coloured in green and Nb in light blue and yellow. (e) Calculation of the binding free energy of the simulated anti-GFP G50A mutant Nb (light blue). The binding free energy of each residue against GFP was calculated and shown as the mean ± SEM (n = 5) from 50 ns of the prediction run. The data of anti-GFP WT Nb are shown in grey for comparison. This figure was created using Illustrator CS2 (https://www.adobe.com/).
Interestingly, G50 was not located at the binding surface to GFP (Fig. 5d), and the binding free energy of G50 was estimated to be almost zero (Fig. 5c). To determine why anti-GFP G50A mutant Nb showed low-affinity, we calculated the binding free energy of a simulated structure of anti-GFP G50A mutant Nb. The binding free energy of R35, an important residue for the specific binding, fluctuated (Fig. 5e). The crystal structure showed that the methyl group of the G50A side chain could change the configuration of R35, leading to decreased affinity (Fig. 5d).
Binding free energy analysis also suggested that R57 and F102 might be important for the function of anti-GFP Nb (Fig. 5c), while peptide barcoding analyses showed no remarkable changes in affinity (Figs. 3h and 4c). We evaluated the affinities of purified anti-GFP R57A and F102A mutant Nbs using SPR and found that they have similar affinities as anti-GFP WT Nb (Table 3 and Supplementary Fig. 8), indicating that peptide barcoding 2.0 can provide highly reliable and multiplex evaluation of the affinities of free Nbs.
Discussion
In this study, we successfully developed peptide barcoding 2.0, a simple, fast, quantitative, and reliable approach for investigating sequence–function relationships of hundreds of free Nbs at once. Using anti-GFP Nb as a model, we identified five important residues for target binding (R35, Y37, Y47, G50 and E103). The result was validated by SPR analysis and the crystal structure of the anti-GFP Nb–GFP complex (PDBID 3OGO)42. The identified residues R35, Y37, Y47 and E103 are located at the anti-GFP Nb–GFP interface. Binding free energy analysis showed that the residues Y37, W47 and E103 have high negative ΔG for complex formation. Binding free energy analysis also suggested that R57 and F102 might be important for target binding, while peptide barcoding and SPR analyses of these two mutants showed no remarkable changes in affinity. This result indicated that peptide barcoding 2.0 can provide highly reliable and multiplex evaluation of the affinities of free Nbs.
Peptide barcoding 2.0 successfully discriminated subtle change in KD at the order of nM to sub-nM. NestLink30 enables ranking of mutant Nbs by their off-rates using an antigen trap column. However, the difference in off-rates is evaluated in a narrow range of 10−2–10−3 s−1. Nbs used for research tools or diagnostics have KD of the order of nM to sub-nM, and their off-rates are often less than 10−3 s−12,43,44. This motivated us to develop an improved peptide barcoding methodology to discriminate subtle change in KD of the order of nM to sub-nM. Peptide barcoding 2.0 identified five mutants, and follow-up SPR analysis showed that our methodology successfully distinguished anti-GFP WT Nb (KD ≈ 10−11 M), weakly attenuated mutants (Y37A, KD ≈ 10−8 M; W47A, KD ≈ 10–10 M) and nonfunctional mutants (R35A, G50A and E103A). This result indicated that peptide barcoding 2.0 can be useful for Nb engineering.
Peptide barcoding 2.0 has advantages and disadvantages compared to conventional methods. In comparison with display technologies, peptide barcoding 2.0 enables direct evaluation of free protein binders but has limited throughput. Compared to ELISA and SPR, peptide barcoding 2.0 enables a highly multiplex evaluation of mutants in a single analysis. Taking these points into account, peptide barcoding 2.0 is a useful method for accurate evaluation of free binders at a moderate throughput.
We focused on proving peptide barcoding can accurately evaluate subtle differences in binding kinetics, however, it will be essential to improve the scalability of peptide barcoding to enable high-throughput evaluation of a large number of clones for binder selection. Peptide barcoding 2.0 is a potentially scalable method that can evaluate more than 105 clones. More than 100,000 peptides for highly specific, sensitive and quantitative targeted proteomics are ready to use in the SRMAtlas31,45. Regarding library construction, DropSynth46,47 is used to synthesise thousands of genes at once. Alternatively, when the library size is limited compared with the number of available peptide barcodes, library nesting, in which each Nb gene is linked to numerous unique peptide barcodes in a controlled manner, is used30. In library nesting, the linked peptide barcodes are unique because the experimental peptide barcode diversity largely exceeds the number of linked Nbs, enabling unambiguous identification of Nbs. As the library size increases, we need higher separation power of LC. In this case, highly reproducible micro-pillar array columns (µPAC™) may be a better choice48.
There are potential biases in peptide barcoding 2.0. For example, differences in production levels of nanobody mutants could lead to identification biases. In this study, we detected 95.3% of peptide barcodes (102/107) in the SEC fractions, indicating some mutants were not produced enough by P. pastoris. Concomitant use of other production hosts such as mammalian cells or E. coli may mitigate the production bias. In addition, there are potential biases in trypsin digestion and ionization efficiencies. These biases can be mitigated by using tandem Lys-C/trypsin proteolysis49 and predetermined peptide barcodes with high specificity and sensitivity45.
Peptide barcoding 2.0 and NestLink have some resemblances. Both methods enrich free target protein binders using SEC and detect peptide barcodes by MS spectrometer. These strategies enable a functional evaluation of free binders. However, these methods have some differences. Peptide barcoding 2.0 collects fractions more finely, leading to the detection of subtle differences in affinities among binder mutants. Peptide barcoding uses predetermined peptide barcodes and NestLink randomized peptide barcodes. Predetermined peptide barcodes enable highly selective, sensitive and quantitative analysis of peptide barcodes. Randomized peptide barcodes do not necessarily guarantee selective, sensitive and quantitative analysis but enable more scalable detection of peptide barcodes.
In conclusion, we successfully developed peptide barcoding 2.0, a simple, fast, quantitative, and reliable approach to investigating sequence–function relationships of hundreds of free Nbs at once, which is difficult by conventional low-throughput technologies, where individual protein binders are separately evaluated. Our methodology is also applicable to affinity maturation of Nbs because it can detect subtle differences in affinities of the order of nM to sub-nM, which is often required for diagnostic reagents or drugs. In addition, our methodology is applicable to not only protein binders but also other types of proteins; for example, it will be possible to screen proteins with different profiles, which leads to differences in mobility in SEC analysis.
Methods
Selection of candidate peptide barcodes
We selected 12,038 candidate peptides for peptide barcodes from the yeast SRMAtlas31. Of these, 838 peptides composed of 8–10 amino acid residues with strong intensities and optimal detectability in the previous LC–MS/MS analysis were selected31,32. Peptides containing methionine or cysteine were excluded because of possible oxidation and cross-linking32.
Protein extraction from yeast
We analysed the Saccharomyces cerevisiae BY4741 proteome to validate the yeast SRMAtlas data. Briefly, S. cerevisiae BY4741 was cultured in yeast extract peptone dextrose (YPD) medium until the optical density at 600 nm (OD600) was 1. Three methods were used to lyse yeast cells. First, cells were suspended in lysis buffer A (100 mM NaOH, 50 mM ethylenediaminetetraacetic acid [EDTA], 2% SDS) and incubated at 90 °C for 10 min. Second, cells were suspended in lysis buffer B (100 mM NaCl, 1 mM EDTA, 2% SDS, 10 mM Tris–HCl, pH 8) and disrupted with sonication using a Bioruptor 2 ultrasonic crusher (Sonic Bio Co., Ltd., Kanagawa, Japan). Third, cells were suspended in lysis buffer B and disrupted with grass beads using a shaker. These disrupted cell solutions were centrifuged at 16,000×g for 20 min, and proteins were extracted by methanol/chloroform precipitation.
Protein reduction, alkylation and digestion
Protein reduction, alkylation and digestion were conducted using a phase transfer surfactant50. Briefly, the extracted proteins were diluted with solubilise buffer (12 mM sodium deoxycholate, 12 mM N-lauroylsarcosinate, 50 mM tetraethylammonium bromide [TEAB]) to obtain 100 μL of 1 mg/mL protein solution. The solution was reduced using dithiothreitol (DTT) (final concentration 50 mM) at 37 °C for 30 min and then alkylated by adding iodoacetamide (final concentration 50 mM). The solution was further diluted five times using 50 mM TEAB and digested with lysyl endopeptidase (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) and trypsin (Promega Corporation, Madison, WI, USA) at 37 °C overnight for proteins extracted from yeast and the trypsin/Lys-C Mix, Mass Spec Grade (Promega Corporation) at 37 °C overnight for fractionated Nbs. The detergents were removed with ethyl acetate containing 0.5% trifluoroacetic acid and the resultant solutions were freeze-dried, desalted using MonoSpin C18 (GL Sciences Inc., Tokyo, Japan) and lyophilised. The dried pellets were re-solved with 50 μL of 0.1% formic acid for proteins extracted from yeast and 50 μL of 0.1% formic acid and 0.005% polyethylene glycol 20,00051 for fractionated Nbs and then filtered by Ultrafree-MC-HV Centrifugal Filters Durapore PVDF 0.45 μm (Merck Millipore, Burlington, MA, USA). Finally, 1 μL of the aliquot was subjected to LC–MS/MS analysis for proteins extracted from yeast and 5 μL for fractionated Nbs.
LC–MS/MS analysis
Peptides were analysed by an UltiMate 3000 LC (Thermo Fisher Scientific, Waltham, MA, USA) and LC–MS-8060 triple quadrupole mass spectrometer (Shimadzu Corporation, Kyoto, Japan) system equipped with a C18 monolithic silica capillary column (50 cm, 100 μm internal diameter). Peptides were separated by reverse-phase chromatography using the C18 column, which was kept at 40 °C, with a flow rate of 500 nL/min and injected into the MS system through a nano-electrospray ion source. A gradient was generated by changing the mixing ratio of the two eluents: A (0.1% [v/v] formic acid) and B (acetonitrile containing 0.1% [v/v] formic acid). The gradient programme was as follows: 5% B for 5 min, 5–45% B for 25 min, 45–95% B for 10 min, 95% B for 10 min, and 5% B for 10 min. The heat block and desolvation line temperatures were set at 200 °C and 250 °C, respectively. SRM methods for the peptides were constructed using Skyline (Supplementary Tables S1 and S2)52. During peptide barcode selection, four transitions were analysed with a 0.055 m/z permissible error. The loop time and dwell time were 8.78–11.34 s and 4–28 ms, respectively. During peptide barcode analyses, four transitions per peptide were analysed, and these transitions were scheduled in a 2 min window before and after the obtained retention time from the analysis of S. cerevisiae BY4741 with a 0.055 m/z permissible error. The loop time and dwell time were 1.47 s and 5 ms, respectively.
Purification of anti-GFP Nbs for SPR analysis
DNA fragments encoding anti-GFP WT Nb or anti-GFP mutant Nbs fused with or without peptide barcodes were inserted into the pPIC9K_6 × His vector (Supplementary Table 2 and “Supplementary Information”) digested by EcoRI and SpeI using the In-Fusion® HD Cloning Kit (Takara Bio Inc., Shiga, Japan). These plasmids were digested by SacI, column-purified and introduced into the P. pastoris GS115 strain (Thermo Fisher Scientific) using the Frozen EZ Yeast Transformation II Kit (Zymo Research Corporation, Irvine, CA, USA). Transformants were selected on MD solid medium and cultured in 20 mL of buffered glycerol-complex medium (BMGY) at 30 °C for 48 h, and the cells were inoculated in 10 mL of buffered methanol-complex medium (BMMY) and cultured at 30 °C for 24 h. The culture media were filtered using a 0.45 μm filter. The Nbs obtained were purified using His SpinTrap™ (Cytiva, Marlborough, MA, USA). The solvent of the purified solutions was replaced with HBS-EP + buffer (Cytiva) using Amicon Ultra-0.5 Centrifugal Filters Ultracel-3K (Merck Millipore). The purity of Nbs was confirmed using SDS-PAGE and CBB staining, and their concentrations were estimated with A280 extinction coefficients calculated using Benchling software (Benchling, San Francisco, CA, USA).
GFP production in Escherichia coli
GFP was produced by E. coli BL21 (DE3) harbouring the GFP gene. The cells were cultured in 80 mL of LBK medium at 37 °C until OD600 was 0.6. Isopropyl-β-d-thiogalactopyranoside was added at a final concentration of 0.1 M, and the cells were further incubated for 4 h. Next, the cells were disrupted by sonication using a Bioruptor 2 (Sonic Bio Co., Ltd.), and the supernatant was incubated with 100 μL of Ni–nitrilotriacetic acid (NTA) agarose (FUJIFILM Wako Pure Chemical Corporation). GFP was eluted with 200 μL of elution buffer containing 250 mM imidazole. The purity of GFP was confirmed using SDS-PAGE and CBB staining, and its concentration was estimated with A280 extinction coefficients calculated using Benchling software.
Surface plasmon resonance
Kinetic parameters of Nbs were measured using Biacore T-200 (Cytiva) with the multicycle method. According to the manufacturer’s protocol, 265–274 response units of recombinant GFP were immobilised on Series S Sensor Chip CM5 (Cytiva). Solutions containing anti-GFP Nbs were diluted with HBS-EP + buffer to prepare five diluted series. The flow rate and temperature were 30 μL/min and 25 °C, respectively, and the running buffer was HBS-EP + buffer. Contact time and dissociation time varied depending on the measurements. The GFP-coated chip was regenerated using 50 mM NaOH (flow rate 30 μL/min and contact time 20 s).
Preparation of the anti-GFP mutant Nb library for SEC analysis
We constructed plasmids encoding peptide-barcoded anti-GFP Nb mutants with single alanine substitutions by primer-based mutagenesis. In brief, two complement primers were designed to introduce alanine (GCT codon) substitution at the target residue. In addition, one primer encoding unique peptide barcode was designed to anneal the 3′ end of anti-GFP Nb and another primer to anneal the 5′ end of anti-GFP Nb. PCR was performed using these four primers to prepare two DNA fragments, and the fragments were inserted into the pPIC9K_6 × His vector digested by EcoRI and SpeI using the In-Fusion® HD Cloning Kit (Takara Bio Inc.). A plasmid library containing all mutant plasmids was transformed into the P. pastoris GS115 strain, as previously described53. Briefly, the plasmid library was digested by SacI and purified to a concentration of 5 ng/μL. P. pastoris was grown in YPD medium until OD600 was 1.5, and 8 × 108 cells were collected, suspended in a buffer (100 mM lithium acetate, 10 mM DTT, 600 mM sorbitol and 10 mM Tris–HCl, pH 7.5) and incubated at room temperature for 30 min. Next, the cells were washed thrice with ice-cold 1 M sorbitol and resuspended (~ 1010 cells/mL) in 80 μL of 1 M sorbitol containing 20 ng of purified DNA fragments. The cells were then transferred to a 0.2 cm gap vial, and a pulse was applied at 1.5 kV, 25 µF and 200 Ω using the Gene Pulser® II Electroporation System (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The cells were immediately diluted with 1 mL of ice-cold 1 M sorbitol, inoculated in 10 mL of SC-His medium and cultured at 30 °C overnight. The transformants were selected on RDB solid medium. The transformation efficiency was calculated from RDB solid medium on which 20 μL aliquots were plated and incubated at 30 °C for 3 days. The colonies were collected with 20 mL of BMGY and cultured at 30 °C for 24 h. Next, cells in 1 mL of the culture medium were stored at − 80 °C with an equal amount of 30% glycerol. The glycerol stock was thawed, and the cells were suspended in 100 mL of BMGY for Nb production. After cultivation at 30 °C for 24 h, the cells were transferred to 50 mL of BMMY and cultured at 30 °C for 24 h. The culture supernatant was collected and filtered using a 0.45 μm filter. Nbs in the supernatant were adsorbed on 250 μL of Ni–NTA agarose (FUJIFILM Wako Pure Chemical Corporation) and eluted with 500 μL of elution buffer containing 250 mM imidazole. Finally, the purity of Nbs was confirmed using SDS-PAGE and CBB staining, and their concentration was estimated with A280 extinction coefficients calculated using Benchling software.
SEC analysis
SEC analysis was conducted using ÄKTAexplorer 10S (Cytiva) equipped with a Superdex 75 increase 10/300 GL column (Cytiva). A mixture of GFP and anti-GFP Nbs (2 nmol each of GFP and anti-GFP Nbs in Fig. 3 and 2 nmol of GFP and 1 nmol of anti-GFP Nbs in Fig. 4) in phosphate-buffered saline (PBS, pH 7.4) was prepared to a total volume of 400 μL and injected into the column. Next, six-fifth of the column volume of PBS (pH 7.4) was loaded at a flow rate of 0.8 mL/min. The eluate was collected in 0.5 mL portions. Next, 10 μL of the collected fractions were subjected to SDS-PAGE and silver staining using Sil-Best Stain One (Nacalai Tesque, Inc., Kyoto, Japan). Finally, the fractionated solutions were lyophilised and stored at − 80 °C.
Binding free energy analysis
The initial coordinates of the anti-GFP Nb–GFP complex were obtained from PDBID 3OGO42. The G50A structure was generated by introducing a point mutation into the Nb structure. All molecular dynamics simulations were performed using the AMBER 16 program54 on TSUBAME (Global Scientific Information and Computing Center, Tokyo Institute of Technology, Japan). For binding free energy calculations using the anti-GFP Nb–GFP complex, the systems were fully solvated with explicit solvent and six Na+ counterions were added to obtain electrostatic neutrality. We used the AMBER ff14SB force field for proteins and the TIP3P model for water molecules. Next, the systems were optimised by energy minimisation and equilibrated with backbone restraints. Production runs were performed for 100 ns. The binding free energy of anti-GFP Nbs and GFP during the final 50 ns of the production run was calculated using the molecular mechanics/generalised Born surface area (MM/GBSA) module.
Data availability
MS data generated and/or analysed in this study are available in the jPOST repository55 (JPST001198). Sequences of plasmids used in the study are shown in Supplementary Information. The other datasets generated and analysed during the current study are available from the corresponding author on reasonable request.
References
Duggan, S. Caplacizumab: First global approval. Drugs 78, 1639–1642 (2018).
Jailkhani, N. et al. Noninvasive imaging of tumor progression, metastasis, and fibrosis using a nanobody targeting the extracellular matrix. Proc. Natl. Acad. Sci. USA 116, 14181–14190 (2019).
Rasmussen, S. G. F. et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469, 175–180 (2011).
Qiu, Y. L., He, Q. H., Xu, Y., Wang, W. & Liu, Y. Y. Modification of a deoxynivalenol-antigen-mimicking nanobody to improve immunoassay sensitivity by site-saturation mutagenesis. Anal. Bioanal. Chem. 408, 895–903 (2016).
Tiller, K. E. et al. Facile affinity maturation of antibody variable domains using natural diversity mutagenesis. Front. Immunol. 8, 986. https://doi.org/10.3389/fimmu.2017.00986 (2017).
Lee, K. B., Dunn, Z. & Ge, X. Reducing proteolytic liability of a MMP-14 inhibitory antibody by site-saturation mutagenesis. Protein Sci. 28, 643–653 (2019).
Valenzuela Nieto, G. et al. Potent neutralization of clinical isolates of SARS-CoV-2 D614 and G614 variants by a monomeric, sub-nanomolar affinity nanobody. Sci. Rep. 11, 3318. https://doi.org/10.1038/s41598-021-82833-w (2021).
Miyamoto, K. et al. Peptide barcoding for establishment of new types of genotype–phenotype linkages. PLoS ONE 14, e0215993. https://doi.org/10.1371/journal.pone.0215993 (2019).
Schoof, M. et al. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science 370, 1473–1479 (2020).
Cao, L. et al. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science 370, 426–431 (2020).
Esparza, T. J., Martin, N. P., Anderson, G. P., Goldman, E. R. & Brody, D. L. High affinity nanobodies block SARS-CoV-2 spike receptor binding domain interaction with human angiotensin converting enzyme. Sci. Rep. 10, 22370; https://doi.org/10.1038/s41598-020-79036-0 (2020).
Smith, G. P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 228, 1315–1317 (1985).
Boder, E. T. & Wittrup, K. D. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15, 553–557 (1997).
Hanes, J. & Plückthun, A. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. USA 94, 4937–4942 (1997).
Roberts, R. W. & Szostak, J. W. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. USA 94, 12297–12302 (1997).
Weaver-Feldhaus, J. M., Miller, K. D., Feldhaus, M. J. & Siegel, R. W. Directed evolution for the development of conformation-specific affinity reagents using yeast display. Protein Eng. Des. Sel. 18, 527–536 (2005).
Wang, Z., Mathias, A., Stavrou, S. & Neville, D. M. Jr. A new yeast display vector permitting free scFv amino termini can augment ligand binding affinities. Protein Eng. Des. Sel. 18, 337–343 (2005).
Terskikh, A. V. et al. “Peptabody”: A new type of high avidity binding protein. Proc. Natl. Acad. Sci. USA 94, 1663–1668 (1997).
Sheehan, J. & Marasco, W. A. Phage and yeast display. Microbiol. Spectr. 3,3.1.11. https://doi.org/10.1128/microbiolspec.AID-0028-2014 (2015).
Lamboy, J. A. et al. Phage wrapping with cationic polymers eliminates nonspecific binding between M13 phage and high pI target proteins. J. Am. Chem. Soc. 131, 16454–16460 (2009).
Burkovitz, A. & Ofran, Y. Understanding differences between synthetic and natural antibodies can help improve antibody engineering. MAbs 8, 278–287 (2016).
Cheung, W. C. et al. A proteomics approach for the identification and cloning of monoclonal antibodies from serum. Nat. Biotechnol. 30, 447–452 (2012).
Sato, S. et al. Proteomics-directed cloning of circulating antiviral human monoclonal antibodies. Nat. Biotechnol. 30, 1039–1043 (2012).
Wine, Y. et al. Molecular deconvolution of the monoclonal antibodies that comprise the polyclonal serum response. Proc. Natl. Acad. Sci. USA 110, 2993–2998 (2013).
Fridy, P. C. et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 11, 1253–1260 (2014).
Lavinder, J. J. et al. Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc. Natl. Acad. Sci. USA 111, 2259–2264 (2014).
Boutz, D. R. et al. Proteomic identification of monoclonal antibodies from serum. Anal. Chem. 86, 4758–4766 (2014).
Xiang, Y. et al. Integrative proteomics identifies thousands of distinct, multi-epitope, and high-affinity nanobodies. Cell Syst. 12, 220–234 (2021).
Liigand, P., Kaupmees, K. & Kruve, A. Influence of the amino acid composition on the ionization efficiencies of small peptides. J. Mass Spectrom. 54, 481–487 (2019).
Egloff, P. et al. Engineered peptide barcodes for in-depth analyses of binding protein libraries. Nat. Methods 16, 421–428 (2019).
Picotti, P. et al. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature 494, 266–270 (2013).
Uchida, Y. et al. A study protocol for quantitative targeted absolute proteomics (QTAP) by LC-MS/MS: Application for inter-strain differences in protein expression levels of transporters, receptors, claudin-5, and marker proteins at the blood-brain barrier in ddY, FVB, and C57BL/6J mice. Fluids Barriers CNS 10, 21. https://doi.org/10.1186/2045-8118-10-21 (2013).
Bjellqvist, B. et al. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 14, 1023–1031 (1993).
Kyte, J. & Doolittle, R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 (1982).
Ho, B., Baryshnikova, A. & Brown, G. W. Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst. 6, 192–205 (2018).
Rothbauer, U. et al. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteom. 7, 282–289 (2008).
Ries, J., Kaplan, C., Platonova, E., Eghlidi, H. & Ewers, H. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat. Methods 9, 582–584 (2012).
Shin, Y. J. et al. Nanobody-targeted E3-ubiquitin ligase complex degrades nuclear proteins. Sci. Rep. 5, 14269. https://doi.org/10.1038/srep14269 (2015).
Schellenberg, M. J., Petrovich, R. M., Malone, C. C. & Williams, R. S. Selectable high-yield recombinant protein production in human cells using a GFP/YFP nanobody affinity support. Protein Sci. 27, 1083–1092 (2018).
Stapornwongkul, K. S., de Gennes, M., Cocconi, L., Salbreux, G. & Vincent, J.-P. Patterning and growth control in vivo by an engineered GFP gradient. Science 370, 321–327 (2020).
Kirchhofer, A. et al. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17, 133–138 (2010).
Kubala, M. H., Kovtun, O., Alexandrov, K. & Collins, B. M. Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci. 19, 2389–2401 (2010).
Zavrtanik, U., Lukan, J., Loris, R., Lah, J. & Hadži, S. Structural basis of epitope recognition by heavy-chain camelid antibodies. J. Mol. Biol. 430, 4369–4386 (2018).
Koenig, P. et al. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science 371, eabe6230. https://doi.org/10.1126/science.abe6230 (2021).
Kusebauch, U. et al. Human SRMAtlas: A resource of targeted assays to quantify the complete human proteome. Cell 166, 766–778 (2016).
Plesa, C., Sidore, A. M., Lubock, N. B., Zhang, D. & Kosuri, S. Multiplexed gene synthesis in emulsions for exploring protein functional landscapes. Science 359, 343–347 (2018).
Sidore, A. M., Plesa, C., Samson, J. A., Lubock, N. B. & Kosuri, S. DropSynth 2.0: High-fidelity multiplexed gene synthesis in emulsions. Nucleic Acids Res. 48, e95. https://doi.org/10.1093/nar/gkaa600 (2020).
De Beeck, J. O. et al. A well-ordered nanoflow LC–MS/MS approach for proteome profiling using 200 cm long micro pillar array columns. https://doi.org/10.1101/472134v2 (2019).
Glatter, T. et al. Large-scale quantitative assessment of different in-solution protein digestion protocols reveals superior cleavage efficiency of tandem Lys-C/trypsin proteolysis over trypsin digestion. J. Proteome Res. 11, 5145–5156 (2012).
Masuda, T., Tomita, M. & Ishihama, Y. Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J. Proteome Res. 7, 731–740 (2008).
Stejskal, K., Potěšil, D. & Zdráhal, Z. Suppression of peptide sample losses in autosampler vials. J. Proteome Res. 12, 3057–3062 (2013).
MacLean, B. et al. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Wu, S. & Letchworth, G. J. High efficiency transformation by electroporation of Pichia pastoris pretreated with lithium acetate and dithiothreitol. Biotechniques 36, 152–154 (2004).
Case, D. A. et al. AMBER 2017. University of California, San Francisco (2017).
Okuda, S. et al. jPOSTrepo: An international standard data repository for proteomes. Nucleic Acids Res. 45, D1107–D1111 (2017).
Acknowledgements
We thank the Kyoto Integrated Science & Technology Bio-Analysis Center (KIST-BIC) for technical support for the SPR and SEC experiments. This research was supported by JST CREST (Grant number JPMJCR16G2), JST COI-NEXT (Grant number JPMJPF2008), and JST FOREST (Grant Number JPMJFR204K), Japan.
Author information
Authors and Affiliations
Contributions
Y.M., W.A., N.M., and M.U. conceived the project. Y.M., T.M., K.K., and T.K. performed experiments and data analysis. K.K. and N.K. contributed to data interpretation. S.A. and Y.O. contributed to development of the SRM methods. H.M. contributed to technical and material support. The manuscript was prepared by Y.M., W.A., T.K., and M.U. and edited by all co-authors.
Corresponding author
Ethics declarations
Competing interests
Kyoto Monotech provided a monolithic column and support in the form of salaries for H.M. The other authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Matsuzaki, Y., Aoki, W., Miyazaki, T. et al. Peptide barcoding for one-pot evaluation of sequence–function relationships of nanobodies. Sci Rep 11, 21516 (2021). https://doi.org/10.1038/s41598-021-01019-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-01019-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.