Abstract
Due to their important phylogenetic position among extant vertebrates, sharks are an invaluable group in evolutionary developmental biology studies. A thorough understanding of shark anatomy is essential to facilitate these studies and documentation of this iconic taxon. With the increasing availability of cross-sectional imaging techniques, the complicated anatomy of both cartilaginous and soft tissues can be analyzed non-invasively, quickly, and accurately. The aim of this study is to provide a detailed anatomical description of the normal banded houndshark (Triakis scyllium) using computed tomography (CT) and magnetic resonance imaging (MRI) along with cryosection images. Three banded houndsharks were scanned using a 64-detector row spiral CT scanner and a 3 T MRI scanner. All images were digitally stored and assessed using open-source Digital Imaging and Communications in Medicine viewer software in the transverse, sagittal, and dorsal dimensions. The banded houndshark cadavers were then cryosectioned at approximately 1-cm intervals. Corresponding transverse cryosection images were chosen to identify the best anatomical correlations for transverse CT and MRI images. The resulting images provided excellent detail of the major anatomical structures of the banded houndshark. The illustrations in the present study could be considered as a useful reference for interpretation of normal and pathological imaging studies of sharks.
Similar content being viewed by others
Introduction
The past half-century has seen a sharp decline in the population of sharks due to indiscriminate fishing, and they are now facing a severe extinction crisis1,2,3,4. Sharks are apex predators and keystone species, playing an important role in maintaining the marine ecosystem. Thus, extinction of sharks in certain areas is expected to lead to marine food chain collapses, which could result in a sharp reduction in the marine food resources available for human beings5,6,7. As a result, the importance of protecting endangered shark species has been recognized worldwide, and efforts to study these species to understand and protect them are being made by various groups, including aquariums, fishery industries, and biologists7,8,9,10,11,12.
With the increasing availability of cross-sectional imaging techniques such as computed tomography (CT) and magnetic resonance imaging (MRI), the complicated and sophisticated anatomy of both skeletal and soft tissues can be analyzed non-invasively, quickly, and accurately13,14,15,16. CT and MRI have often been used not only for humans but also for various animal species13,17,18,19. Some of these studies were conducted on sharks and involved diverse fields such as physiology, anatomy, developmental biology, archaeology, and evolutionary biology9,20,21,22,23,24,25,26,27,28.
Although a wide variety of studies have been performed on sharks29, official publications describing the basic anatomy of sharks particularly in CT and/or MRI evaluations are currently unavailable. Accurate evaluation of CT and MRI images requires precise knowledge of the anatomy and physiology of the animal. In this regard, the development of accurate atlases for certain species by comparing CT and MRI images with actual cryosections is one of the primary steps that should be established by researchers and veterinarians30. Normal atlases of CT and MRI findings in various animals have already been set up in order to facilitate accurate image analysis30,31,32,33,34,35,36,37,38. In the same context, precise atlases of CT and MRI findings in shark species should also be established.
The banded houndshark (Triakis scyllium) is a relatively small-sized shark that is easily found in Korean water. They inhabit the Northwest Pacific Ocean and are classified as the “least concern” group by the International Union for Conservation of Nature (IUCN) red list4,39. The purpose of this study is to set up a detailed atlas of the banded houndshark by comparing CT and MRI images with actual cryosections. To the best of our knowledge, this is the first study reporting delicate comparison of CT, MRI, and cryosection findings for shark anatomy.
Results
In the present study, CT and MRI allowed identification of a broad selection of anatomical structures and correlated well with the corresponding cryosections of the banded houndshark. Clinically relevant anatomical structures are labeled in Figs. 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23. Figures 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14 (transverse images), Figs. 15, 16, 17 (sagittal images), and Figs. 18, 19, 20, 21, 22, 23 (dorsal images) present the findings in cranial to caudal, right to left, and dorsal to ventral progression, respectively. Three-dimensional (3D) reconstructed images of CT and MRI data sets showing major skeletons and organs of the banded houndshark were presented in Fig. 24. Transverse images of MRI and CT are shown continuously in Supplementary Video S1 and Supplementary Video S2, respectively, to make up for the shortcomings of discontinuous cross-sectional images.
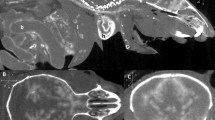
Transverse (a) CT, (b) MR, and (c) cryosection images of a banded houndshark (Triakis scyllium) at level 1 of Fig. 25. Al Ampullae of Lorenzini, Ic Internasal cartilage, Mc Meckel’s cartilage, Mt Mandibular teeth, Nc Nasal capsule cartilage, Oa Olfactory lamellae, Ob Olfactory bulb, Os Olfactory sac, Ov Oral cavity, Sc Supraorbital canal.
Transverse (a) CT, (b) MR, and (c) cryosection images of a banded houndshark (Triakis scyllium) at level 2 of Fig. 25. Al Ampullae of Lorenzini, Cb Cerebrum, Ic Internasal cartilage, Mc Meckel’s cartilage, Te Teeth, Nc Nasal capsule cartilage, Oa Olfactory lamellae, Ot Olfactory tract, Sc Supraorbital canal.
Transverse (a) CT, (b) MR, and (c) cryosection images of a banded houndshark (Triakis scyllium) at level 3 of Fig. 25. Al Ampullae of Lorenzini, Am Adductor mandibularis, Ba Basihyal cartilage, Hc Hyomandibular canal, Ic Infraorbital canal, Io Inferior obliquus, Le Lens, Mc Meckel’s cartilage, Mt Mandibular teeth, Ne Neurocranium, Ni Nictitating fold, Ol Optic lobe, On Optic nerve, Pa Palatoquadrate, Pr Posterior rectus, Sc Supraorbital canal, So Superior obliquus, Th Thalamus, Ty Thyroid, Vi Vitreous humor.
Transverse (a) CT, (b) MR, and (c) cryosection images of a banded houndshark (Triakis scyllium) at level 4 of Fig. 25. Al Ampullae of Lorenzini, Am Adductor mandibularis, Cc Ceratohyal cartilage, Ch Coracohyoideus, Co Coracomandibularis, Hc Horizontal semicircular canal, Hs Hyoidean sinus, Me Medulla Oblongata, Mc Meckel’s cartilage, Ne Neurocranium, Ot Otolith, Pa Palatoquadrate, Sp Spiracle, Va Ventral aorta.
Transverse (a) CT, (b) MR, and (c) cryosection images of a banded houndshark (Triakis scyllium) at level 5 of Fig. 25. Aa Afferent branchial artery, Ab Adductor branchial, As Anterior cardinal sinus, Ba Basibranchial, Br Branchial rays, Ca Conus arteriosus, Cb Coracobranchialis, Ch Coracohyoideus, Co Coracomandibularis, Ds Dorsal skeletogenous septum, Ea Epaxial, Ec Epibranchial and ceratobranchial cartilage, Gf Gill filament, Hp Hypobranchial cartilage, Nu Neural arch, Vc Vertebral centrum.
Transverse (a) CT, (b) MR, and (c) cryosection images of a banded houndshark (Triakis scyllium) at level 6 of Fig. 25. As Anterior cardinal sinus, At Atrium, Cr Coracoid bar, Da Dorsal aorta, Ds Dorsal median septum, Ea Epaxial, Ec Epibranchial and ceratobranchial cartilage, Ha Hypaxial, Sc Spinal cord, Tr Trapezius, Ve Ventricle, Vc Vertebral centrum.
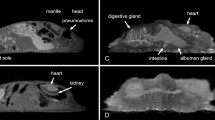
Transverse (a) CT, (b) MR, and (c) cryosection images of a banded houndshark (Triakis scyllium) at level 7 of Fig. 25. Da Dorsal aorta, Dp Depressor pectoralis, Ds Dorsal median septum, Ea Epaxial, Ga Gall bladder, Ha Hypaxial, Lp Levator pectoralis internus, Nu Neural arch, Lr Liver, Rc Radial cartilage, Sc Spinal cord, Sm Stomach, Vc Vertebral centrum, Vv Ventral abdominal vein.
Transverse (a) CT, (b) MR, and (c) cryosection images of a banded houndshark (Triakis scyllium) at level 8 of Fig. 25. Ds Dorsal median septum, Du Duodenum, Ea Epaxial, Ha Hypaxial, Id Inclinator dorsalis, Kd Kidney, Lc Lateral line canal, Lr Liver, Pc Pancreas, Pf Pectoral fin, Rc Radial cartilage, Sc Spinal cord, Sm Stomach, Ve Vertebra, Vi Valvular intestine, Vs Visceral cavity, Vv Ventral abdominal vein.
Transverse (a) CT, (b) MR, and (c) cryosection images of a banded houndshark (Triakis scyllium) at level 9 of Fig. 25. Ds Dorsal median septum, Du Duodenum, Ea Epaxial, Ha Hypaxial, Id Inclinator dorsalis, Kd Kidney, Lc Lateral line canal, Lr Liver, Pc Pancreas, Rc Radial cartilage, Sc Spinal cord, Sm Stomach, Ve Vertebra, Vi Valvular intestine, Vs Visceral cavity, Vv Ventral abdominal vein.
Transverse (a) CT, (b) MR, and (c) cryosection images of a banded houndshark (Triakis scyllium) at level 10 of Fig. 25. Ds Dorsal median septum, Du Duodenum, Ea Epaxial, Ha Hypaxial, Id Inclinator dorsalis, Kd Kidney, Lc Lateral line canal, Lr Liver, Rc Radial cartilage, Sc Spinal cord, Sm Stomach, Sn Spleen, Ve Vertebra, Vi Valvular intestine, Vv Ventral abdominal vein.
Transverse (a) CT, (b) MR, and (c) cryosection images of a banded houndshark (Triakis scyllium) at level 11 of Fig. 25. Ds Dorsal median septum, Ea Epaxial, Ha Hypaxial, Id Inclinator dorsalis, Kd Kidney, Lc Lateral line canal, Lr Liver, Rc Radial cartilage, Sc Spinal cord, Sm Stomach, Sn Spleen, Ve Vertebra, Vi Valvular intestine, Vv Ventral abdominal vein.
Transverse (a) CT, (b) MR, and (c) cryosection images of a banded houndshark (Triakis scyllium) at level 12 of Fig. 25. Cd Caudal artery, Co cloaca, Cv Caudal vein, Ds Dorsal skeletogenous septum, Dv Depressor ventralis, Ea Epaxial, Ha Hypaxial, Kd Kidney, Le Levator ventralis externus, Li Levator ventralis internus, Ll Lateral line canal, Rc Radial cartilage, Sc Spinal cord.
Transverse (a) CT, (b) MR, and (c) cryosection images of a banded houndshark (Triakis scyllium) at level 13 of Fig. 25. Ds Dorsal skeletogenous septum, Ea Epaxial, Ha Hypaxial, Id Inclinator dorsalis, Rc Radial cartilage, Sc Spinal cord, Ve Vertebra, Vs Ventral skeletogenous septum.
Transverse (a) CT, (b) MR, and (c) cryosection images of a banded houndshark (Triakis scyllium) at level 14 of Fig. 25. Cv Caudal vessel, Ea Epaxial, Ha Hypaxial, Hh Haemal arch, Rc Radial cartilage, Sc Spinal cord, Ve Vertebra.
Sagittal (a) CT and (b) MR images of a banded houndshark (Triakis scyllium) at level 15 of Fig. 25. Al Ampullae of Lorenzini, Cc Ceratohyal cartilage, Cr Coracoid bar, Ec Epibranchial and ceratobranchial cartilage, Es Esophagus, Ga Gall bladder, Mc Meckel’s cartilage, Oa Olfactory lamellae, Oc Otic capsule, Pa Palatoquadrate, Sm Stomach, Nv Nasal cavity.
Mid-sagittal (a) CT and (b) MR images of a banded houndshark (Triakis scyllium) at level 16 of Fig. 25. Al Ampullae of Lorenzini, At Atrium, Ba Basihyal cartilage, Cb Cerebrum, Ce Cerebellum, Cl cloaca, Co Coracomandibularis, Cr Coracoid bar, Ch Coracohyoideus, Ct Ceratotrichia, Da Dorsal aorta, Ea Epaxial, Es Esophagus, Ga Gall bladder, Ha Hypaxial, Ic Internasal cartilage, Kd Kidney, Lr Liver, Me Medulla oblongata, Ne Neurocranium, Nu Neural arch, Oa Olfactory lamellae, Ol Optic lobe, Rc Radial cartilage, Sc Spinal cord, Te Teeth, Ts Transverse septum, Vi Valvular intestine, Ve Ventricle, Vt Ventral constrictors, Vc Vertebral centrum, Sm Stomach, Sv Spiral valve.
Sagittal (a) CT and (b) MR images of a banded houndshark (Triakis scyllium) at level 17 of Fig. 25. Al Ampullae of Lorenzini, Am Adductor mandibularis, Ba Basihyal cartilage, Cc Ceratohyal cartilage, Es Esophagus, Hc Hyomandibular canal, Ic Infraorbital canal, Io Inferior obliquus, Lc Levator cranium maxillae, Le Lens, Mc Meckel’s cartilage, Mp Metapterygium, Mt Mandibular teeth, Ne Neurocranium, Ni Nictitating fold, Ol Optic lobe, Op Optic nerve, Ov Oral cavity, Pa Palatoquadrate, Po Preorbitalis, Pr Posterior rectus, Sc Supraorbital canal, Su Scapular cartilage, So Superior obliquus, Th Thalamus, Ty Thyroid, Vi Vitreous humor.
Dorsal (a) CT and (b) MR images of a banded houndshark (Triakis scyllium) at level 18 of Fig. 25. Al Ampullae of Lorenzini, Ce Cerebellum, Ea Epaxial, Sg Scleral cartilage, Ln Lens, Me Medulla oblongata, Mp Metapterygium, Nc Neurocranium, Oc Otic capsule, Ol Optic lobe, Re Retina, Rp Rectus posterior.
Dorsal (a) CT and (b) MR images of a banded houndshark (Triakis scyllium) at level 19 of Fig. 25. Al Ampullae of Lorenzini, Cb Cerebrum, Ij Intervertebral junction, Mp Metapterygium, Nc Neurocranium, Oa Olfactory lamellae, Oc Otic capsule, Ol Optic lobe, Ot Olfactory tract, Sp Spiracle, Tp Transverse process, Vc Vertebral centrum.
Dorsal (a) CT and (b) MR images of a banded houndshark (Triakis scyllium) at level 20 of Fig. 25. Al Ampullae of Lorenzini, Cb Cerebrum, Ea Epaxial, Eb Epibranchial, Hm Hyomandibular, Ij Intervertebral joint, Lc Levator cranium maxillae, Mp Metapterygium, Nc Nasal capsule, Oa Olfactory lamellae, Ob Olfactory bulb, Oc Optic capsule, On Optic nerve, Ov Obliquus ventralis, Pb Pharyngobranchial, Po Preorbitalis, Ri Rectus inferior, Sp Spiracle, Th Thalamus, Vc Vertebral centrum.
Dorsal (a) CT and (b) MR images of a banded houndshark (Triakis scyllium) at level 21 of Fig. 25. Al Ampullae of Lorenzini, Cb Cerebrum; Da Dorsal aorta, Eb Epibranchial, Hm Hyomandibular, Ij Intervertebral junction, Lc Levator cranium maxillae, Mp Metapterygium, Nc Nasal capsule, Nv Nasal cavity, Oa Olfactory lamellae, Ob Olfactory bulb, Oc Optic capsule, On Optic nerve, Ov Obliquus ventralis, Pb Pharyngobranchial, Tp Transverse process, Ri Rectus inferior, Sp Spiracle, Vc Vertebral centrum.
Dorsal (a) CT and (b) MR images of a banded houndshark (Triakis scyllium) at level 22 of Fig. 25. Al Ampullae of Lorenzini, Am Adductor mandibularis, Ba Basihyal cartilage, Bc Branchial cartilage, Cc Ceratohyal cartilage, Ch Coracohyoideus, Ha Hypaxial, Hb Hypobranchial, Gb Gall bladder, Kd Kidney, Lr Liver, Mc Meckel’s cartilage, Ms Mesopterygium, Mp Metapterygium, Nv Nasal cavity, Oa Olfactory lamellae, Pa Palatoquadrate, Pc Pericardial cavity, Vt Ventral constrictors, Rc Rostral cartilage, Te Teeth, Sm Stomach, Sy Symphysis.
Dorsal (a) CT and (b) MR images of a banded houndshark (Triakis scyllium) at level 23 of Fig. 25. Am Adductor mandibularis, Co Coracomandibularis, Cr Coracoid bar, Ch Coracohyoideus, Ga Gall bladder, Gf Gill filament, Ha Hypaxial, Lr Liver, Mc Meckel’s cartilage, Ms Mesopterygium, Rc Radial cartilage, Sm Stomach, Te Teeth, Ts Transverse septum, Ve Ventricle, Vi Valvular intestine.
Three dimensional reconstructed images of (a) CT and (b) MRI showing major skeletal structures and organs respectively of a banded houndshark (Triakis scyllium). Cr Coracoid bar, Ec Epibranchial and ceratobranchial cartilage, Ic Internasal cartilage, Mc Meckel’s cartilage, Ne Neurocranium, Ot Otolith, Pa Palatoquadrate, Rc Radial cartilage, Vc Vertebral centrum.
Transverse images
Figures 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14.
Sagittal images
Dorsal images
Discussion
The MRI signal intensity is proportional to the hydrogen density of the structures, ensuring excellent definition of soft tissues, organs, and cavitary structures in banded houndsharks. These structures included major muscles; cardiovascular system structures, including the atrium, ventricle, and major arteries and veins; digestive system structures, including the stomach, gall bladder, and valvular intestine; excretory system structures, including the kidney; nervous system structures, including the cerebrum, optic lobe, and thalamus. Due to the absence of hydrogen atoms, most of the cartilaginous skeletons in banded houndsharks did not provide sufficient magnetic resonance signal and showed a hypointense (black/dark grey) pattern. Nonetheless, some cartilaginous structures, such as the jaws and coracoid bar, could still be observed due to the contrast between the cartilage and the adjacent soft tissues.
In contrast to MRI, excellent discrimination of cartilaginous structures was evident in the CT images. The higher electron density of cartilages caused increased attenuation compared to soft tissue, making them appear more whitish in the CT images. The cartilage margins were better defined by means of CT than cryosections or MRI, particularly in assessing joint structures. Soft tissues were vaguely distinguished using CT examinations due to their similar attenuation properties. Nevertheless, attenuation differences between lipid-rich tissues (e.g., liver) and muscles (e.g., stomach and valvular intestine) were shown in varying shades of grey allowing definition of some organs in the visceral cavity.
CT and MRI offer considerable advantages over traditional radiographic approaches on identification of the anatomical structures of sharks. In traditional radiography, images are presented as superimposed two-dimensional projections of three-dimensional structures, which result in limitation to exhibit high conspicuity. The tomographic nature of CT and MRI allows organs to be examined in thin sections, eliminating superimposition of overlying structures that may hinder the specific interpretation of anatomical structures.
Another major advantage of CT and MRI is the ability to reformat the datasets in any imaging plane or as 3D projections, allowing better understanding of the spatial relationships of anatomical structures in sharks. These techniques have added detail of the anatomical structures and can be used as an anatomic reference for imaging studies of the banded houndshark and other shark species. It is recommended to reduce CT/MRI slice thickness as much as possible in order to achieve excellent quality and high detail of 3D reconstructed images, since increasing slice thicknesses may require more interpolation between slices when rendering 3D images, which results in a loss of resolution40.
Due to their important phylogenetic position among extant vertebrates, sharks are an invaluable group in evolutionary developmental biology studies. However, the presence of high levels of urea accelerates the decomposition of shark flesh41. Moreover, because of its cartilaginous nature, shark skeleton has a high tendency to warp, crack, and shrink during specimen preparation42. These factors represent major difficulties in preserving anatomical data from sharks. In the present study, CT and MRI were used to identify and document soft tissues and cartilaginous structures in banded houndsharks. The non-invasive nature of CT and MRI prevented the irretrievable destruction of specimens during traditional dissection. The spatial relationships among organs, soft tissues, and cartilage can be selectively observed in situ in their natural locations43,44. After examinations, all images are permanently recorded in the Digital Imaging and Communications in Medicine (DICOM) format and could be recalled at will. The images are digitally transferable, which can also facilitate discussion and opinion sharing among professionals worldwide even if the specimens cannot be physically provided.
Banded houndshark is a relatively small-sized shark species that can be adequately fit into the bore of medical CT and MRI used in the present study. However, to the best of the authors’ knowledge, only the head of large-sized shark species has been subjected to imaging examination possibly because of limited gantry size45,46,47. Sharks with body girth larger than the maximum diameter of field of view (FOV) of medical CT and MRI may also induce out-of-field artifacts, anatomical structures that lies beyond the FOV may be truncated. With the rapid advancement of medical imaging technologies, wide bore CT and MRI have become available for large animals48,49,50. Although the current availability of wide bore CT and MRI scanners are limited and the set up cost is high, the use of wide bore CT and MRI may offer a feasible alternative for imaging examination of large-sized shark species in future.
Conclusions
The illustrations in the present study are the first to provide a comprehensive atlas, using CT and MRI, to evaluate the anatomical structures of the banded houndshark. Both imaging modalities provide good contrast for the anatomical structures of the banded houndshark. With the rapid advancement and increasing availability of medical imaging technologies, CT and MRI are enhancing our knowledge of the anatomy of sharks together with dissected and necropsy materials. The findings of the present study could be considered as a useful reference of normal tomographic anatomy of sharks and used for the interpretation of normal and pathological imaging studies of sharks in future.
Material and methods
Animals
Three banded houndsharks were obtained commercially from Seoul, the Republic of Korea. The banded houndsharks were 1 male (total body length (TBL): 90 cm, age: estimated as 3.4 years) and 2 females (TBL: 91 cm, age: estimated as 3.3 years; TBL: 102 cm, age: estimated as 4.1 years) with body lengths ranging from 90 to 102 cm. Ages of the sharks were estimated based on their TBL and it was judged that the sharks were about to be sexually mature51. All animals were selected by a veterinarian (SWK) based on the lack of external evidence of trauma and disease. The banded houndsharks were euthanized via tricaine methanesulfonate overdose (500 ppm) right before imaging examinations in order to minimize post-mortem changes.
Magnetic resonance imaging (MRI)
MRI examinations were performed using a 3-T (T) MRI scanner (Achieva, Philips Healthcare, The Netherlands). A head coil was used to receive the signal using fast spin echo sequences in the T2-weighted (T2W) mode, in which signals from loosely bound protons (e.g., fluids) were enhanced:
-
1.
Transverse images were obtained with the following parameters: echo time (TE), 90 ms; repetition time (TR), 4920.8 ms; pixel bandwidth, 180 Hz; flip angle, 90°, and FOV, 123 × 123 mm with reconstructed spacing, 0.17 × 0.17 mm. Slice thickness was 2 mm.
-
2.
Sagittal images were obtained with the following parameters: TE, 90 ms; TR, 4870.9 ms; pixel bandwidth, 215 Hz; flip angle, 90°, and FOV, 76 × 76 mm with reconstructed spacing, 0.20 × 0.20 mm. Slice thickness was 3 mm.
-
3.
Dorsal images were obtained with the following parameters: TE, 70 ms; TR, 3865.9 ms; pixel bandwidth, 236 Hz; flip angle, 90°, and FOV, 81 × 81 mm with reconstructed spacing, 0.23 × 0.23 mm. Slice thickness was 1.5 mm.
Computed tomography (CT)
CT images were acquired with a 64-detector row spiral CT scanner (Aquilion, Toshiba Medical Systems, Nasu, Japan). The CT examination was performed at 120 kV and 80–100 mA with a 1-mm slice thickness. The scan field of view (sFOV) ranged from 20.3 to 30.2 cm. The transverse image datasets were acquired using the soft tissue and bone algorithm. CT images in the dorsal and sagittal dimensions were obtained using the multiplanar reconstruction (MPR) function. MRI and CT images were assessed using open-source DICOM viewer software (Horos Project, version 3.3.6; www.horosproject.org).
Cryosectioning
All specimens were frozen (− 22 °C) in the ventral recumbency position until cryosectioning could be carried out. The banded houndsharks were sectioned using an electrical band saw along the transverse plane at approximately 1-cm intervals. Slices were numbered, cleaned, and photographed on both sides. Fourteen transverse section levels were selected. For each section level, the corresponding transverse MRI and CT images were chosen to identify the best anatomical correlation. Lines corresponding to these levels were superimposed on three-dimensional reconstructed images of an banded houndshark (Fig. 25).
Three-dimensional surface reconstructions of a banded houndshark (Triakis scyllium) from computed tomography (CT) scans, indicating approximate levels of transverse, sagittal, and dorsal sections corresponding to Figs. 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, Figs. 15, 16, 17, and Figs. 18, 19, 20, 21, 22, 23, respectively. Slices 1–4 are approximately 2 cm intervals, slices 4–9 are approximately 5 cm intervals, slices 9–11 are approximately 6 cm intervals, slices 11–14 are approximately 12 cm intervals, slices 15–17 are approximately 3 cm intervals, and slices 18–23 are approximately 1 cm intervals.
Three-dimensional (3D) reconstruction
3D reconstructions of the skeletons were made on acquired CT data sets using intensity-based segmentation methods of the inbuilt software of 64-detector row spiral CT scanner (Aquilion, Toshiba Medical Systems, Nasu, Japan). The reconstructed skeletons were visualised after excluding remaining residual soft tissue structures. 3D reconstructions of major organs were made on MRI data sets using intensity-based and manual segmentation methods of an open-source DICOM viewer—InVesalius 3.0 (CTI—Center for Information and Technology Renato Archer, Campinas, São Paulo, Brazil).
Ethical approval
The study was approved by the Seoul National University Institutional Animal Care and Use Committee (approval number: SNU-190925-2). All experiments were performed in accordance with the IACUC guidelines and regulations. This study was also performed in accordance with the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines: to reflect the scarcity of banded houndshark over 90 cm TBL in the fish markets of the Republic of Korea, sample size was decided to three; to minimize the potential confounders, both male and female sharks were used; to show the developmental stages, TBL and estimated age of each shark were provided.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Lack, M. & Sant, G. The future of sharks: a review of action and inaction. TRAFFIC International and the Pew Environment Group. https://www.pewtrusts.org/~/media/legacy/uploadedfiles/peg/publications/report/the20future20of20sharkspdf.pdf (2011).
Barker, M. J. & Schluessel, V. Managing global shark fisheries: Suggestions for prioritising management strategies. Aquat. Conserv. 15, 325–347 (2005).
Baum, J. K. & Blanchard, W. Inferring shark population trends from generalized linear mixed models of pelagic longline catch and effort data. Fish Res. 102, 229–239 (2010).
International Union for Conservation of Nature (IUCN). The IUCN Red List of Threatened Species. Version 2018–1. http://www.iucnredlist.org (2018).
Heithaus, M. R., Wirsing, A. J. & Dill, L. M. The ecological importance of intact top-predator populations: A synthesis of 15 years of research in a seagrass ecosystem. Mar. Freshw. Res. 63(11), 1039–1050 (2012).
Ferretti, F., Worm, B., Britten, G. L., Heithaus, M. R. & Lotze, H. K. Patterns and ecosystem consequences of shark declines in the ocean. Ecol. Lett. 13, 1055–1071 (2010).
Myers, R. A., Baum, J. K., Shepherd, T. D., Powers, S. P. & Peterson, C. H. Cascading effects of the loss of apex predatory sharks from a coastal ocean. Science 315, 1846–1850 (2007).
Dulvy, N. K. et al. Challenges and priorities in shark and ray conservation. Curr. Biol. 27(11), R565–R572 (2017).
Summers, A. P., Ketcham, R. A. & Rowe, T. Structure and function of the horn shark (Heterodontus francisci) cranium through ontogeny: Development of a hard prey specialist. J. Morphol. 260(1), 1–12 (2004).
Stevens, J. D., Bonfil, R., Dulvy, N. K. & Walker, P. A. The effects of fishing on sharks, rays, and chimaeras (chondrichthyans), and the implications for marine ecosystems. Ices. J. Mar. Sci. 57(3), 476–494 (2000).
Dulvy, N. K. et al. Extinction risk and conservation of the world’s sharks and rays. eLife 3, e00590 (2014).
Davidson, L. N. K., Krawchuk, M. A. & Dulvy, N. K. Why have global shark and ray landings declined: Improved management or overfishing?. Fish Fish. 17(2), 438–458 (2016).
Lauridsen, H. et al. Inside out: Modern imaging techniques to reveal animal anatomy. PLoS ONE 6(3), e17879 (2011).
Dirnhofer, R., Jackowski, C., Vock, P., Potter, K. & Thali, M. J. VIRTOPSY: Minimally invasive, imaging-guided virtual autopsy. Radiographics. 26(5), 1305–1334 (2006).
Kot, B. C. W., Chan, D. K. P., Yuen, A. H. L. & Tsui, H. C. L. Diagnosis of atlanto-occipital dissociation: Standardised measurements of normal craniocervical relationship in finless porpoises (genus Neophocaena) using postmortem computed tomography. Sci Rep. 8, 8474 (2018).
Furukawa, A. et al. Cross-sectional imaging in crohn disease. Radiographics 24(3), 689–702 (2004).
Gumpenberger, M. & Henninger, W. The use of computed tomography in avian and reptile medicine. Semin. Avian Exot. Pet. Med. 10(4), 174–180 (2001).
Preziosi, R. et al. Spinal deformity in a sandtiger shark, Carcharias taurus Rafinesque: A clinical–pathological study. J. Fish Dis. 29(1), 49–60 (2005).
Van Bonn, W., Jensen, E. & Brook, F. Radiology, computed tomography, and magnetic resonance imaging. In CRC Handbook of Marine Mammal Medicine (eds Dierauf, L. A. & Gulland, F. M. D.) 557–591 (CRC Press Inc., Boca Raton, 2001).
Jambura, P. L. et al. Micro-computed tomography imaging reveals the development of a unique tooth mineralization pattern in mackerel sharks (Chondrichthyes; Lamniformes) in deep time. Sci. Rep. 9, 9652 (2019).
Abel, R. L. et al. Functional morphology of the nasal region of a hammerhead shark. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 155(4), 464–475 (2010).
Wroe, S. et al. Three-dimensional computer analysis of white shark jaw mechanics: How hard can a great white bite?. J. Zool. 276(4), 336–342 (2008).
Timm, L. L. & Fish, F. E. A comparative morphological study of head shape and olfactory cavities of sharks inhabiting benthic and coastal/pelagic environments. J. Exp. Mar. Biol. Ecol. 414–415, 75–84 (2012).
Kamminga, P., Bruin, P. W. D., Geleijns, J. & Brazeau, M. D. X-ray computed tomography library of shark anatomy and lower jaw surface models. Sci. Data. 4, 170047 (2017).
Mollen, F. H., Wintner, S. P., Iglésias, S. P., Sommeran, S. R. V. & Jagt, J. W. M. Comparative morphology of rostral cartilages in extant mackerel sharks (Chondrichthyes, Lamniformes, Lamnidae) using CT scanning. Zootaxa. 3340(1), 29–43 (2012).
Mara, K. R., Motta, P. J., Martin, A. P. & Hueter, R. E. Constructional morphology within the head of hammerhead sharks (sphyrnidae). J. Morphol. 276(5), 526–539 (2015).
Geraghty, P. T., Jones, A. L., Stewart, J. & Macbeth, W. G. Micro-computed tomography: An alternative method for shark ageing. J. Fish Biol. 80(5), 1292–1299 (2012).
Moyer, J. K., Hamilton, N. D., Seeley, R. H., Riccio, M. L. & Bemis, W. E. Identification of shark teeth (Elasmobranchii: Lamnidae) from a historic fishing station on Smuttynose island, maine, using computed tomography imaging. Northeast Nat. 22(3), 585–597 (2015).
Meng, Q. & Li, W. Monographs of Fishes of China. No. 3 Anatomy of Shark and Skate (Ocean Publishing, 1992). Meng, Q. & Li, W. Monographs of Fishes of China. No. 3 Anatomy of Shark and Skate (Ocean Publishing, 1992). Meng, Q. & Li, W. Monographs of Fishes of China. No. 3 Anatomy of Shark and Skate (Ocean Publishing, 1992).
Samii, V. F., Biller, D. S. & Koblik, P. D. Normal cross-sectional anatomy of the feline thorax and abdomen: Comparison of computed tomography and cadaver anatomy. Vet. Radiol. Ultrasound. 39(6), 504–511 (1998).
Alonso-Farre, J. M. et al. Cross-sectional anatomy, computed tomography and magnetic resonance imaging of the thoracic region of Common Dolphin (Delphinus delphis) and striped Dolphin (Stenella coeruleoalba). Anat. 43, 221–229 (2014).
Alonso-Farre, J. M. et al. Cross-sectional anatomy, computed tomography and magnetic resonance imaging of the head of common Dolphin (Delphinus delphis) and Striped Dolphin (Stenella coeruleoalba). Anat. 44, 13–21 (2015).
Ibrahim, A. A. H., Adam, Z. E. & Tawfiek, M. G. Cross-sectional anatomy, magnetic resonance imaging and computed tomography of fetlock joint in camel. J. Vet. Med. Res. 26(2), 258–270 (2019).
Wyman, A. C., Lawson, T. L. & Goodman, L. R. Transverse Anatomy of the Human Thorax, Abdomen and Pelvis in An Atlas of Anatomic Radiologic Computed Tomographic and Ultrasonic Correlation 1st edn. (Brown and Co., Little, 1978).
Feeney, D.A., Fletcher, T.F. & Hardy, R.M. Atlas of correlative imaging anatomy of the normal dog: Ultrasound and computed tomography. 1st ed. (WB Saunders, 1991).
Smallwood, J. E. & George, T. F. Anatomic atlas for computed tomography in the mesaticephalic dog: Thorax and cranial abdomen. Vet. Radiol. 34, 65–84 (1993).
Smallwood, J. E. & George, T. F. Anatomic atlas for computed tomography in the mesaticephalic dog: Caudal abdomen and pelvis. Vet. Radiol. 34, 143–167 (1993).
Valente, A. L. S. et al. Computed tomography of the vertebral column and coelomic structures in the normal loggerhead sea turtle (Caretta caretta). Vet. J. 174(2), 362–370 (2007).
Compagno, L. J. V. Sharks of the world: An annotated and illustrated catalogue of shark species known to date. FAO Species Catal. 4, 432 (1984).
Ford, J. M. & Decker, S. J. Computed tomography slice thickness and its effects on three-dimensional reconstruction of anatomical structure. J. Forens. Radiol. Imaging. 4, 43–46 (2016).
Kandoran, M. K., Govindan, T. K. & Suryanarayana Rao, S. V. Some aspects of curing of sharks and rays. Fish Technol. 2(2), 193–197 (1965).
Enault, S., Auclair, C., Adnet, S. & Debiais-Thibaud, M. A complete protocol for the preparation of chondrichthyan skeletal specimens. J. Appl. Ichthyol. 32(3), 409–415 (2016).
Montie, E. W., Manire, C. A. & Mann, D. A. Live CT imaging of sound reception anatomy and hearing measurements in the pygmy killer whale, Feresa attenuata. J. Exp. Biol. 214, 945–955 (2011).
Lautenschlager, S., Bright, J. A. & Rayfield, E. J. Digital dissection-using contrast-enhanced computed tomography scanning to elucidate hard- and soft-tissue anatomy in the Common Buzzard Buteo buteo. J. Anat. 224, 412–431 (2014).
Mollen, F. H., Wintner, S. P., Iglesias, S. P., Van Sommeran, S. R. & Jagt, J. W. Comparative morphology of rostral cartilages in extant mackerel sharks (Chondrichthyes, Lamniformes, Lamnidae) using CT scanning. Zootaxa. 3340, 29–43 (2012).
Tomita, T., Sato, K., Suda, K., Kawauchi, J. & Nakaya, K. Feeding of the megamouth shark (Pisces: Lamniformes: Megachasmidae) predicted by its hyoid arch: a biomechanical approach. J. Morphol. 272, 513–524 (2011).
Wroe, S. et al. Three-dimensional computer analysis of white shark jaw mechanics: How hard can a great white bite?. J. Zool. 276, 336–342 (2008).
Nakamae, Y. et al. Displacement of the large colon in a horse with enterolithiasis due to changed positions observed by computed tomography. J. Equine. Sci. 29, 9–13 (2018).
Porter, E. G., Winter, M. D., Sheppard, B. J., Berry, C. R. & Hernandez, J. A. Correlation of articular cartilage thickness measurements made with magnetic resonance imaging, magnetic resonance arthrography, and computed tomographic arthrography with gross articular cartilage thickness in the equine metacarpophalangeal joint. Vet. Radiol. Ultrasound. 57, 515–525 (2016).
Kraitchman, D., Kamel, I., Weiss, C. & Georgiades, C. Elucidation of percutaneously accessible lymph nodes in swine: A large animal model for interventional lymphatic research. J. Vasc. Interv. Radiol. 28, 451–456 (2017).
Fujinami, Y. & Tanaka, S. Age, growth and reproduction of the banded houndshark Triakis scyllium around the tip of the Izu Peninsul, Japan. Nippon Suisan Gakk. 79(6), 968–976 (2013).
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2018R1D1A1A02086128). This research was also supported by the ‘Korea Research Fellowship Program’ of the National Research Foundation of Korea (NRF), Ministry of Science and ICT (KRF: 2016H1D3A1909005). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
A.H.L.Y. and S.W.K. designed and conceptualized the study. A.H.L.Y., C.T.C.P., and S.W.K. performed the experiment, curated and analyzed the data. M.K.O., J.O.H., and C.J.L. did the MRI scanning. K.T.K. and M.C.C. did the CT scanning. H.J.K., J.K., S.B.L., and S.G.K. helped with shark sampling, transportation, and scanning. C.T.C.P. visualized the figures. A.H.L.Y. and S.W.K. wrote the original manuscript draft, and S.C.P., S.S.G., S.W.K., and A.H.L.Y. revised and edited the manuscript. S.C.P. acquired the funding and supervised the whole study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, S.W., Yuen, A.H.L., Poon, C.T.C. et al. Cross-sectional anatomy, computed tomography, and magnetic resonance imaging of the banded houndshark (Triakis scyllium). Sci Rep 11, 1165 (2021). https://doi.org/10.1038/s41598-020-80823-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-80823-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.