Abstract
Rice is one of the staple crops in the world. Grain size is an important determinant of rice grain yield, but the genetic basis of the grain size remains unclear. Here, we report a set of chromosome segment substitution lines (CSSL) developed in the genetic background of the genome-sequenced indica cultivar Zhenshan 97. Genotyping of the CSSLs by single nucleotide polymorphism array shows that most carry only one or two segments introduced from the genome-sequenced japonica cultivar Nipponbare. Using this population and the high-density markers, a total of 43 quantitative trait loci were identified for seven panicle- and grain-related traits. Among these loci, the novel locus qGL11 for grain length and thousand-grain weight was validated in a CSSL-derived segregating population and finely mapped to a 25-kb region that contains an IAA-amido synthetase gene OsGH3.13, This gene exhibited a significant expression difference in the young panicle between the near-isogenic lines that carry the contrasting Zhenshan 97 and Nipponbare alleles at qGL11. Expression and sequence analyses suggest that this gene is the most likely candidate for qGL11. Furthermore, several OsGH3.13 mutants induced by a CRISPR/Cas9 approach in either japonica or indica exhibit an increased grain length and thousand-grain weight, thus enhancing the final grain yield per plant. These findings provide insights into the genetic basis of grain size for the improvement of yield potential in rice breeding programs.
Similar content being viewed by others
Introduction
Rice (Oryza sativa L.) is the staple food for more than half of the world’s population. Rice yield is highly correlated with number of panicles per plant, number of seeds per panicle, and thousand-grain weight (TGW). Grain size (or grain weight) is mainly determined by its three dimensions: the grain length (GL), the grain width (GW), and the grain thickness. Due to its importance in yield improvement, panicle and grain traits have been extensively studied by genetic and molecular analyses. To date, a large number of quantitative trait loci (QTL) for these traits have been reported in rice (http://www.gramene.org/). Numerous genes associated with grain size have been identified, involving in multiple regulation pathways, such as the ubiquitin–proteasome pathway, G-protein signaling, mitogen-activated protein kinase signaling, the HAIKU (IKU) pathway, phytohormone perception and homeostasis1,2,3,4. As an essential phytohormone, Auxin (IAA) regulates a very broad range of plant growth and developmental processes. Many auxin signaling and transport genes have been reported to control grain size. For example, qTGW3, a major QTL for grain weight, encodes a SHAGGY-like kinase 41 (OsSK41)/OsGSK5, which interacts with auxin response factor 4 (OsARF4) to negatively regulate grain size and grain weight by affecting cell expansion5. OsSK41 and OsARF4 regulate the expression of some auxin-response genes. BIG GRAIN1 (BG1) encodes a positive regulator of auxin response and transport, which markedly increased the indole-3-acetic acid (IAA) level in the panicle. The activation of BG1 results in the production of large grains6. In addition, TGW6, a major QTL for thousand-grain weight, encodes an IAA-glucose hydrolase that produces free IAA7. Loss of function of TGW6 enhances grain weight and increases yield in rice. Although some genes in the auxin pathway have been reported as influencing grain size, the current understanding of the role of auxin (IAA) inactivation in the regulation of grain size is limited.
The identification of QTLs is a first step toward dissecting the genetic and molecular bases of grain size. Several studies have proven that chromosome segment substitution lines (CSSL) as a permanent genetic resource serve as a powerful platform for the identification, fine mapping and cloning of QTLs underlying complex traits8,9,10,11. Each CSSLs contains one or a few substitution segments of a particular donor genome within the common genetic background, providing a great advantage in identifying minor-effect QTLs, mainly due to minimization of the genetic interaction or background noise. CSSL could also facilitate the rapid development of near-isogenic lines (NIL) and secondary segregating populations for further fine mapping and cloning of the QTLs of interest12,13,14,15. This research process can be accelerated by high-throughput single nucleotide polymorphism (SNP) approaches. Therefore, a large number of CSSL populations have been developed with great effort in rice and other crops. However, most of the populations are composed of CSSLs each with multiple substituted segments16, although many populations were constructed with high-density mapping17,18.
In this study, we developed a set of genotype-defined CSSLs derived from a cross of two genome-sequenced varieties, Nipponbare (NIP), as a japonica cultivar19, and Zhenshan 97 (ZS97), as an indica cultivar20. Using the developed CSSLs with high-density SNP markers, we performed QTL analysis for seven panicle and grain-related traits and identified a number of QTLs with minor effects on the assayed traits. Furthermore, a novel QTL (qGL11) for grain length was validated and finely mapped to a small region containing OsGH3.13, an IAA-amido synthetase, specifically catalyzing the conversion of active IAA to its inactive form via the conjugation of IAA with amino acids21,22. The CRISPR-generated OsGH3.13 mutants exhibited increased grain length and grain weight, indicating that the regulation of grain size involves the IAA metabolic pathway.
Results
Development of the CSSL population with SNP-defined genotypes
Previously, a NIP/ZS97 CSSL population consisting of 143 lines (named original population) was constructed using an marker-assisted selection (MAS) backcross scheme23,24. However, most of the CSSLs contained two or more NIP segments when genotyped using an Infinium RICE6K array24. Therefore, we further used the MAS backcross scheme to improve the uniformity of the backgrounds of the CSSLs. Thirty-four lines, each with fewer than 4 substitution segments, were preferentially selected to backcross with ZS97 and self-cross to generate BC1F2. In each BC1F2 population, approximately 100 individuals were genotyped with simple sequence repeats (SSR) and insertion-deletion (Indel) markers targeting a particular substitution segment and background segments. Then, a total of 87 heterozygous and 88 homozygous individuals that contained only a single substitution segment of corresponding chromosomal regions were obtained. For the 87 plants heterozygous at a particular substitution segment, an F2 progeny of 20 individuals was planted to select a desirable homozygous genotype at the target region (Supplementary Fig. S1). Finally, the updated CSSL population comprising 175 new lines and 46 original lines was developed, in which all substitution segments together almost covered the entire japonica genome. To define the genotypes of the introduced segments more precisely, this newly developed CSSL population (named as DNZ) was subjected to genotyping with the Rice 8 K SNP array.
The SNP genotyping of the DNZ population revealed 3,610 polymorphic SNPs between NIP and ZS97 evenly distributed across the 12 rice chromosomes, with a median density of 50.3 kb per adjacent SNP (Supplementary Table S1). The population comprises 142 (64%) lines with only a single substitution segment, 50 (23%) lines with two substitution segments, and 29 lines with three or more introduced segments (Supplementary Fig. S2), which in total contain 408 substitution segments with an average length of 2.3 Mb, covering 96% of the NIP genome (Supplementary Table S2). Based on the recombination breakpoints caused by the 408 substituted segments, a bin map contains 418 bins ranging from 1.8 kb to 8.2 Mb with a median size of 473 kb across the whole genome was constructed for the DNZ population (Supplementary Table S2).
Phenotypic variation of panicle and grain traits
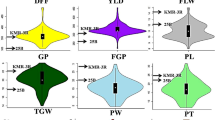
Two parents (NIP and ZS97) exhibited significant differences in panicle and grain traits, with lower values in NIP than ZS97 for all seven assayed traits, except for primary branch (Supplementary Table S3). The frequency distribution of these traits all showed a large continuous variation in the DNZ population (Supplementary Fig. S3), indicating that the panicle and grain-related traits were in quantitative inheritance controlled by polygenes or QTLs. Most of the lines were similar to the parent ZS97 in all assayed traits (Fig. 1a; Supplementary Table S3); however, several lines exhibited phenotypic values surpassing ZS97 at various degree in both directions (Fig. 1a), which suggests that some QTLs or genes located in the corresponding substitution segments had either negative or positive effects on the panicle and grain traits.
Phenotypic and QTL analysis of the DNZ CSSL population. (a) Variation of the phenotypic traits in the DNZ CSSL population. In the box plots, the horizontal line within the box indicates the median value; the bars of the box indicate the limits as 1.5 times the interquartile range from the box; the dots outside the bars indicate the most extreme data points or possible outliers. The red dot indicates the phenotype value of ZS97. (b) Overviews of QTL results for seven traits in the DNZ CSSL population. Rectangle color density indicates the magnitude of -log10 (P) values in the ridge regression significance test. The horizontal position of the rectangle indicates the relative physical position of QTL support interval on the chromosome. The triangle indicates approximately physical location of a known gene for grain size. GL, GW, TGW, SN, PB, PL and PW represent grain length, grain width, thousand-grain weight, spikelet number, primary branch, panicle length and panicle weight, respectively. The sub-figures (a, b) were prepared using R software (version 3.5.2) (http://www.R-project.org/).
Detection of QTLs for the panicle and grain traits
The ridge regression analyses for the QTL detection were performed in the DNZ population and the original population with the bin genotypes. In total, 43 bins containing 89 QTLs were identified for seven panicle and grain traits (Supplementary Table S4), and 37 bins containing 82 loci detected for these traits in the original population. The same or overlapping bin carrying several QTLs for multiple traits was considered as only one QTL. Hence, 43 bin QTLs were identified in the DNZ population. Most (24/43) of these loci were repeatedly detected in the two populations (Supplementary Table S5), suggesting their stable effects in different environments. Therefore, the QTLs identified in the DNZ population are present below in details.
Among the loci in the DNZ population, 18 were located in bins with sizes of less than 500 kb, and 7 loci were mapped in much smaller bins (less than 200 kb) (Supplementary Table S4). The QTLs detected in small bins were significant for further fine mapping and candidate gene pinpointing. The phenotypic variation explained (PVE) by each QTL ranged from 0.7 to 16.9%, in which only three bin QTLs had major-effect (PVE ≥ 10%), while other bins containing 70 loci exhibited minor effects (PVE < 5%) (Supplementary Table S4). These results confirm that the panicle and grain traits are complex quantitative traits controlled by multiple genes. The majority of the QTLs exhibited a positive effect, with the NIP alleles increasing panicle and grain size (Supplementary Table S4).
Regarding grain size, 32 bins containing 49 QTLs were identified distributed on all chromosomes except chromosome 12 in the DZN population (Supplementary Table S4). Several QTLs were identified in common for grain length, grain width and thousand-grain weight, which is consistent with the significant correlation between these three traits (Supplementary Fig. S3). For example, three QTLs, qGL1-3, qGW1-3 and qTGW1-2, were colocalized in Bin029 on chromosome 1. The loci qGL3-2, qGW3-5 and qTGW3-4 mapped in Bin147 on chromosome 3 were colocalized in the same region surrounding TGW35,25,26 (Fig. 1b). Three loci, qGL8 and qTGW8-2 with the largest effect on grain length or thousand-grain weight, along with qGW8, were mapped in Bin329 (approximately 354 kb) on chromosome 8, which may be the same QTL for all three grain traits (Fig. 1b; Supplementary Table S4). The loci qGL11 for grain length and qTGW11 for thousand-grain weight were mapped in Bin390 on chromosome 11.
Similarly, there were 21 bins harboring 40 QTLs for panicle architecture (Supplementary Table S4). Four loci (qPL4-2, qSN4-2, qPW4-2, and qPB4-2) were detected for panicle architecture in the same region surrounding NAL1, which was previously reported as affecting panicle length, spikelet number and panicle weight27. Two QTLs on chromosomes 5 and 7 were identified in common for four panicle-related traits (Supplementary Table S4). Four loci (qPB7-1, qPL7, qSN7-1 and qPW7-1), detected for primary branch, panicle length, spikelet number and panicle weight, respectively, were colocalized in the same region (Bin295) encompassing Ghd7, a cloned gene regulating multiple yield-related traits28. Three QTLs (qPW7-2, qSN7-2 and qPB7-2) were co-localized in Bin309 (appropriately 110 kb) containing SGDP7, a gene for small grain and dense panicle29. The colocalization of these QTLs is a genetic refection of the significant correlation between the panicle-related traits (Supplementary Fig. S3).
Fine mapping of qGL11 for grain length
To verify a newly identified locus (qGL11 or qTGW11), a CSSL (named N232) carrying a single substitution segment covering qGL11 was crossed with ZS97 to generate a CSSL-derived F2 population. N232 had a longer grain length and larger grain weight than ZS97 (Fig. 2a,b). First, a small F2 population comprising 107 individuals was genotyped using 12 polymorphic markers in this segment (Supplementary Table S6). The QTL analysis in this population revealed that qGL11 was located in the interval between markers M7 and M9, explaining 52% of the phenotypic variance for grain length. The additive effect of the NIP alleles was 0.15, and the dominant effect was 0.03 (Fig. 2c). Then, a large CSSL-derived segregating population comprising 3500 individuals was used to select recombinants between M7 and M9 by using ten additional Indel markers (Supplementary Table S6). Based on the progeny phenotyping of grain size for the informative recombinants, qGL11 was delimited to a 25-kb region flanked by markers ID2 and ID5 (Fig. 2d). This region contains five predicted genes according to the RAP database30 (https://rapdb.dna.affrc.go.jp/). Among the five annotated genes, only Os11g0528700 (OsGH3.13), encoding an indole-3-acetic acid (IAA)-amido synthetase, exhibited a significant expression difference in the young panicle between a pair of NILs (NIL-NIP and NIL-ZS97), which carry a single NIP segment (less than 2 Mb) covering the contrasting NIP and ZS97 alleles at qGL11 within the ZS97 background (Fig. 2e). Sequence comparison revealed that OsGH3.13 had substantial variation among rice varieties. In its promoter region (3-kb upstream of the predicted start codon), 14 SNPs and three Indels were found between ZS97 and NIP. In particular, the A/G variant at the -2458 site upstream of the start codon may cause auxin-response element (GATACA) defects in OsGH3.13NIP (Fig. 2f). In the coding region, there were three nonsynonymous SNPs resulting in amino acid changes (Supplementary Table S7). Thus, OsGH3.13 is a possible candidate gene for qGL11.
Fine mapping of qGL11. (a) Graphic genotype of N232 showing a single substitution segment encompassing qGL11. (b) Comparison of the grain size in ZS97 and N232. Differences between ZS97 and N232 in grain length, grain width, grain thickness and thousand-grain weight. Data are given as the mean and SE (n = 6 individuals). (c) The QTL was detected in a CSSL-derived F2 population. PVE, Add and Dom respectively represent phenotypic variation explained, additive effect and dominant effect. (d) Fine mapping of qGL11 in a 25-kb region using a large segregating population (n = 3500). Five predicted genes are indicated, including Os11g0528700 (700) which was selected as the candidate gene. L3-L10 represent recombinant plants. Phenotype data for GL (grain length) and TGW (thousand-grain weight) are provided as the mean and SE (n = 10). (e) Quantitative RT-PCR analysis of gene expression in the 8-cm panicle. Transcription levels relative to NIL-ZS97, which was set to 1, are presented as the mean and SE of triplicates. LOC_Os03g13170 (Ubiquitin) is the control gene. (f) Sequence comparison of Os11g0528700 among four cultivars. The triangle indicates a nucleotide deletion. **, * and NS indicate significant differences at P < 0.01, P < 0.05, and no significance, respectively, by Student’s t-test.
Validation of the qGL11 effect in NILs
To confirm the qGL11 effect, NIL-NIP and NIL-ZS97 were evaluated in grain size and other yield-related traits (Fig. 3a). Phenotypic comparison of NILs showed that the average grain length of NIL-NIP was 4.3% longer than that of NIL-ZS97, and the thousand-grain weight was 3.7% greater than that of NIL-ZS97, while grain width and grain thickness was not significantly different between NILs (Fig. 3b,c). Notably, the grain yield of NIL-NIP was increased by approximately 20% compared to that of NIL-ZS97 (Fig. 3b). The effective panicle number was slightly increased, but not significantly, between NILs (Supplementary Fig. S4). Other agronomic traits, such as heading date, plant height, panicle length, primary branch, spikelet number, and seed setting ratio, also showed no significant difference between the two NILs (Supplementary Fig. S4). To investigate whether cell expansion was involved in NILs, scanning electron microscopy analysis was conducted and revealed that NIL-NIP had an average mastoid spacing of 95.9 ± 4.4 µm, significantly larger than those of NIL-ZS97 (average 83.5 ± 5.6 µm). The NIL-NIP epidermal cells were approximately 19.2% longer than their counterparts (Fig. 3d–f), suggesting that the longer cells of NIL-NIP largely accounted for the differences in grain size. It has been reported that OsGH3.13 is an IAA-amido synthetase, specifically catalyzing the conversion of active IAA into its inactive form via the conjugation of IAA with amino acids21,22. Liquid chromatography-mass spectrometry analyses were thus performed to measure the IAA and its conjugates in NILs. The results revealed higher free IAA concentration in NIL-NIP than in NIL-ZS97. Consistent with this observation, the relative contents of two IAA conjugates IAA-Glu and IAA-Asp were lower in NIL-NIP than in NIL-ZS97 (Fig. 3g).
Differences between NIL-ZS97 and NIL-NIP with the contrasting alleles at qGL11. (a) Graphic genotype representation of NIL-NIP. (b) Differences between NIL-ZS97 and NIL-NIP in seed morphology. Scale bar: 5 mm. (c) Comparison of the grain size and grain yield in NIL-ZS97 and NIL-NIP. Data are given as the mean and SE (n = 10). (d–f) Scanning electron microscopy analysis of the lemma. Differences between NIL-ZS97 and NIL-NIP in spikelet morphology (d), cell morphology (e), longitudinal cell length (f) in the lemmas of spikelet hulls. Values provided as the mean and SE (n = 10). (g) IAA and IAA-amino acid conjugate species in NIL-ZS97 and NIL-NIP. Content relative to NIL-ZS97, which was set to 1, is presented as the mean and SE of triplicates. **, * indicate a significant difference at P < 0.01 and P < 0.05, respectively, by Student’s t-test. NS, no significance.
Sequence comparison of OsGH3.13 among four varieties, NIP, ZS97, 9311 and ZH11, revealed that ZH11 and NIP had the same sequence, while 9311 and ZS97 shared the same sequence (Fig. 2f). Therefore, the other NIL (R108) that harbors a single NIP segment (less than 5 Mb) encompassing qGL11 in the 9311 background (Fig. 4a) was also evaluated. Similarly, OsGH3.13 exhibited a lower expression in R108 than in 9311 (Fig. 4b). Consistent with the grain size difference between NIL-NIP and NIL-ZS97, R108 revealed longer grain length and larger thousand-grain weight than 9311, with no significant difference in grain width and grain thickness (Fig. 4c–g). Taken together, these results indicate that qGL11NIP is a positive factor, increasing grain length and grain weight in both the ZS97 and 9311 genetic backgrounds.
Comparative analysis of NILs containing qGL11 in the background of 9311. (a) Graphic representation of the genotype of R108 that carries a single substituted segment encompassing qGL11. (b) Relative expression of OsGH3.13 of 9311 and R108 in the 8-cm panicle. Transcription levels relative to 9311, which was set to 1, are presented as the mean and SE (n = 3). Ubiquitin is the control gene. (c–g) Differences between 9311 and R108 in seed morphology (c), scale = 5 mm, grain length (d), grain width (e), grain thickness (f) and thousand-grain weight (g). (d–g) Data are given as the mean and SE (n = 6). ** and NS indicate statistically significant differences by Student’s t-test at P < 0.01 and no significance, respectively.
OsGH3.13 mutants increased grain size
To determine whether OsGH3.13 affects grain size, several mutants of OsGH3.13 were generated separately in 9311 and ZH11 by using the CRISPR/Cas9 strategy (Fig. 5a–c). A total of 22 mutants were first verified as positive transgenic plants by PCR using specific Cas9 primers, and sequence analysis identified the nucleotide mutations produced in the target exon of the gene in the positive plants. Finally, six homozygous independent mutants of OsGH3.13 with various variants were obtained and characterized in the corresponding genetic background of ZH11 or 9311 (Fig. 5b,c). Three independent mutants in 9311 (MT9311-1, MT9311-2, MT9311-3), which carried a 28-bp deletion, 6-bp deletion and 1-bp insert in the target region, respectively. The 28-bp deletion and 1-bp insert resulted in frameshifts causing a premature termination of transcription of the gene, while the 6-bp deletion might cause an encoded incomplete peptide. As expected, these mutants exhibited increased grain length and thousand-grain weight compared to 9311 (Fig. 5d–h). Similarly, three independent mutations in ZH11 (MTZH11-1, MTZH11-2, MTZH11-3) exhibited longer grain length and larger grain weight compared to ZH11 (Fig. 5i–m). All six mutants significantly increased average grain yield per plant by 14–20% compared to the counterpart 9311 or ZH11 while not influencing the other agronomic traits, such as plant height, effective panicle number, primary branch, panicle length, spikelet number, and seed setting ratio (Supplementary Fig. S5). These results indicate that OsGH3.13 could functionally regulate grain length, thousand-grain weight, and final grain yield in rice.
CRISPR/Cas9-induced mutants of the gene and their phenotypes. (a) Schematic gene model of OsGH3.13 showing the sgRNA target site and sequence. The protospacer-adjacent motif (CGG) is shown in red font. (b, c) Homozygous mutations of OsGH3.13 in 9311 (b), and ZH11 (c). Scale bars: 10 mm. (d–m) Differences between the mutants and corresponding wild type in grain size and grain yield per plant. Data are given as the mean and SE (n = 6). Asterisks (**) indicate significant difference from the wild type by Student’s t-test at P < 0.01. NS, no significance.
Discussion
It has been reported that CSSLs as an advanced backcross population can effectively reduce the interaction between QTLs and minimize the genetic background interference, thus improving the power to detect, fine-map and clone QTLs31,32,33,34,35,36. However, most CSSLs carry multiple and long substituted chromosomal segments with a low marker density, which could decrease the detection power, especially for those minor QTLs that are much more easily masked by genetic interactions. In this study, we developed NIP/ZS97 CSSLs with high-density SNPs and identified the role of OsGH3.13 in affecting grain size. This updated set of CSSLs includes 192 (87%) lines containing only one or two substitution segments (Supplementary Fig. S2), facilitating the identification of the QTLs of complex traits. In this study, a large number of minor QTLs were detected for panicle and grain traits. Many QTLs are mapped in the same or overlapping regions of known genes or QTLs previously reported in rice (Supplementary Table S4). Moreover, a minor QTL, qGL11 for grain length, was identified and finely mapped using the CSSL population and a CSSL-derived secondary segregating population (Fig. 2). These data indicate that CSSLs with high-density SNPs can provide an efficient tool for the identification of minor-effect QTLs associated with yield-related traits in rice, which will be of great value for rice breeding.
One of the notable findings of this study is that OsGH3.13 affects grain length and thousand-grain weight in rice through regulating the IAA level. IAA is deactivated through either conjugation to amino acids or chemical oxidation, which has critical roles in maintaining local auxin homeostasis and normal plant development37. OsGH3.13 is an IAA-amido synthetase, which catalyzes the conjugation of IAA to amino acids. Previously, TLD1 was reported as OsGH3.13, downregulating IAA content, resulting in IAA deficiency and dramatic changes in plant architecture and enhancing rice drought tolerance22. A gain-of-function rice mutant (tld1-D) increased number of tillers, enlarged leaf angles, and dwarfism in rice. However, the loss-of-function mutant tld1/OsGH3.13 did not show visible differences in plant growth as well as grain size from wild-type plants. The OsGH3.13 homologous genes OsGH3.8 and OsGH3.1 were described as participating in disease resistance to rice pathogens38,39. Overexpression of OsGH3.8 suppressed cell wall relaxation and inhibited plant growth38. In this study, we used a CSSL-derived segregating population to narrow down qGL11 to a 25-kb region that contains the gene OsGH3.13 (Fig. 2). We find that OsGH3.13NIP has IAA-conjugating activity, leading to an increase of IAA and a decrease of IAA-Asp and IAA-Glu in NIL-NIP relative to NIL-ZS97 (Fig. 3). Sequence analysis shows several allelic variations, namely, OsGH3.13NIP, OsGH3.13ZS, and OsGH3.139311. In particular, the A/G variant at the site -2458 upstream of the gene start codon results in a defective motif of GATACA40, which may cause a significant difference in expression level. In agreement with the expression variation, NIL-NIP (or R108), in which OsGH3.13 was expressed at a low level, exhibits an increased grain length and grain weight compared to NIL-ZS97 (or 9311), in which the gene is expressed at a high level (Figs. 2e, 4b). Moreover, the CRISPR-induced OsGH3.13 mutants in two genetic backgrounds (9311 and ZH11) confirmed that the function of OsGH3.13 increased grain length and grain weight, and even the final grain yield per plant (Fig. 5). As OsGH3.13 underlying qGL11 could not affect grain width and grain thickness, we assume that the QTL/gene affects grain weight mainly by regulating grain length, although the regulation mechanism is required further investigation. In addition, OsGH3.13, like TGW6 (an IAA-glucose hydrolase gene), is involved in auxin homeostasis influencing grain size. However, OsGH3.13 influences grain length and weight with an effect on husk size (Fig. 3). It is different from TGW6, which regulates grain weight with no effect on husk size7. Our data establish a connection between IAA-amido synthetase and grain size and present the feasibility of modulating IAA homeostasis to influence grain size. Taken together, these results indicate that OsGH3.13 corresponds to qGL11 for grain size, with the OsGH3.13NIP alleles exhibiting a positive effect on grain length and grain weight. These naturally favorable alleles can increase grain yield without affecting other agronomic traits and have great potential in rice breeding for yield improvement in rice.
Materials and methods
Plant materials
Previously, a CSSL population consisting of 143 lines was generated from a cross of the genome-sequenced cultivars ZS97 and NIP23,24. As some lines of the original population carried multiple introgressed NIP segments, it was necessary to develop the lines each with only single NIP segment of a particular chromosomal region in the ZS97 background. Therefore, a new CSSL population (named DNZ) comprising 221 lines was developed by a backcrossing scheme with MAS (Supplementary Fig. S1) and used for further study.
A CSSL-derived F2 population segregating at the target region was generated for fine mapping. In the process of fine mapping, the pairwise NILs (e.g. NIL-NIP and NIL-ZS97) that carry the contrasting NIP and ZS97 alleles at the QTL of interest within the common background of ZS97 were generated from the heterozygous recombinant line that contains the smallest introgression segment covering the QTL. The line R108 was also selected from the CSSL population derived from a cross of NIP as the donor and the cultivar 9311 as the recurrent parent for the QTL confirmation41.
SNP genotyping and bin mapping of QTL
The DNZ population was genotyped using the rice 8 K array, which contains 7,519 SNP, by the China Golden Marker Co. (Beijing, China). Based on the SNP genotyping, a bin map was constructed for the CSSL population. The QTL analyses with the bins as markers were performed using linear ridge regression in R software with the package “ridge” (http://www.R-project.org/)42, as described previously24. A significance level of P < 0.01 was set as the threshold for the QTL declaration. The phenotypic variance explained by each QTL was decomposed by using ‘lmg’ from the ‘relaimpo’ package42. QTL nomenclature followed the principles as suggested43. The QTL IciMapping V4.2 (http://www.isbreeding.net/) was applied for the QTL validation in the CSSL-derived segregating population.
DNA extraction and marker analysis
Genomic DNA from fresh young leaves of each line was extracted using the CTAB method44. Polymorphic SSR and Indel markers were used for MAS, The SSRs were selected from Gramene Database45, and Indel markers were developed according to the sequence variation between NIP and ZS97 (http://ricevarmap.ncpgr.cn)46. The primers were designed according to the Nipponbare reference genome by Primer3 (http://redb.ncpgr.cn/modules/redbtools/primer3.php). SNP markers in a given region were detected by sequencing. The sequences were analyzed using Sequencer 5.0 (Gene Codes Corporation). All primers were synthesized at Sangon Biotech (Shanghai) and are listed in Supplementary Table S6.
Quantitative real-time PCR analysis
Total RNAs were isolated with the TRIzol kit (Invitrogen, CA) according to the manufacturer’s instructions. The RNA was treated with DNase I (Invitrogen), and approximately 3 μg of total RNA was used to synthesize first-strand cDNA using oligo (dT)18 as primer (Promega, Shanghai). Quantitative real-time (qRT) PCR was performed using gene-specific primers (Supplementary Table S6) and the FastStart Universal SYBR Green Master (Roche) on a real-time PCR ViiA7 system (Applied Biosystems). The rice Ubiquitin gene was used as the internal control. The relative quantification method was used to evaluate gene expression level47. At least three biological replicates were performed for each experiment.
Trait measurement
All plant materials except the original CSSLs were planted in an randomized block design with two replications at the experimental station of Huazhong Agricultural University at Wuhan (114°30′ E, 30°60′ N) in 2018 for phenotype investigation. Each line was grown in a two-row plot with 10 plants per row, with a spacing of 16.7 cm between plants and 26.6 cm between rows. The original CSSLs were grown in the same design in 2013. At the mature stage, panicle weight (PW), panicle length (PL) and primary branch (PB) were evaluated following the methods described previously48. Spikelet number (SN), seed setting ratio (SS), grain length (GL), grain width (GW) and thousand-grain weight (TGW) were measured using a yield traits scorer in a high-throughput phenotyping facility49.
IAA and IAA-amino acid conjugate measurement
Extraction of IAA and IAA-amino acid conjugates from rice leaves and their analysis followed the method described previously50. Briefly, leaf samples were collected and frozen in liquid nitrogen. Approximately 100 mg powder of each sample was extracted overnight at 4 °C with 1 mL pure methanol containing 0.1 mg/L lidocaine. Following centrifugation at 10,000×g for 10 min, the supernatant was absorbed and filtrated using a filter (SCAA-104, 0.22-µm pore size; Shanghai ANPEL Scientific Instrument). The extracts were analyzed using liquid chromatography-mass spectrometry (ABI 4000 Q-Trap, Applied Biosystems). The measurements were repeated three times for each sample.
Scanning electron microscopy
Young spikelets (4 days before heading) were collected for scanning electron microscopy analyses. The samples were prepared following the procedure as previously described51. The mastoid spacing of the lemmas of outer spikelet hulls was measured under a scanning electron microscope (JSM-6390LV, JEOL) at an accelerating voltage of 10 kV and a spot size of 30 nm. Scanning electron microscopy analysis was carried out on at least ten biological replications of mounted sample.
Vector construction and rice transformation
A clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 vector targeting the given gene was constructed following a previously described method15. Briefly, a 23-nucleotide fragment (5′-ACTACAAGAGCCGCCACTTCCGG-3′) including the protospacer-adjacent motif (CGG) was designed as the target sequence using the CRISPRP tool (http://rice.hzau.edu.cn/cgi-bin/rice/CRISPR_rice). The resultant construct was introduced into Agrobacterium tumefaciens strain EHA105 and transferred into rice variety Zhonghua 11 (ZH11) and 9311 using the Agrobacterium-mediated transformation method52. All primers used for the transgenic tests are listed in Supplementary Table S6.
Data availability
All data generated or analysed during this study are included in this published article and its Supplementary Information files.
References
Zuo, J. & Li, J. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 48, 99–118. https://doi.org/10.1146/annurev-genet-120213-092138 (2014).
Li, N. & Li, Y. Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 33, 23–32. https://doi.org/10.1016/j.pbi.2016.05.008 (2016).
Li, N., Xu, R., Duan, P. & Li, Y. Control of grain size in rice. Plant Reprod. 31, 237–251. https://doi.org/10.1007/s00497-018-0333-6 (2018).
Li, N., Xu, R. & Li, Y. Molecular networks of seed size control in plants. Annu. Rev. Plant Biol. 70, 435–463. https://doi.org/10.1146/annurev-arplant-050718-095851 (2019).
Hu, Z. et al. A novel QTL qTGW3 encodes the GSK3/SHAGGY-like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol. Plant 11, 736–749. https://doi.org/10.1016/j.molp.2018.03.005 (2018).
Liu, L. et al. Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Acad. Sci. USA 112, 11102–11107. https://doi.org/10.1073/pnas.1512748112 (2015).
Ishimaru, K. et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 45, 707–713. https://doi.org/10.1038/ng.2612 (2013).
Eshed, Y. & Zamir, D. An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141, 1147–1162 (1995).
Koumproglou, R. et al. STAIRS: A new genetic resource for functional genomic studies of Arabidopsis. Plant J. 31, 335–364. https://doi.org/10.1046/j.1365-313x.2002.01353.x (2002).
Yamamoto, T., Yonemaru, J. & Yano, M. Towards the understanding of complex traits in rice: Substantially or superficially?. DNA Res. 16, 141–154. https://doi.org/10.1093/dnares/dsp006 (2009).
Subudhi, P. K. et al. A chromosome segment substitution library of weedy rice for genetic dissection of complex agronomic and domestication traits. PLoS ONE 10, e0130650. https://doi.org/10.1371/journal.pone.0130650 (2015).
Wang, X. & Roy, I. Using advanced intercross lines for high-resolution mapping of HDL cholesterol quantitative trait loci. Genome Res. 13, 1654–1664. https://doi.org/10.1101/gr.1185803 (2003).
Ali, M. L., Sanchez, P. L., Yu, S., Lorieux, M. & Eizenga, G. C. Chromosome segment substitution lines: A powerful tool for the introgression of valuable genes from oryza wild species into cultivated rice (O. sativa). Rice 3, 218–234. https://doi.org/10.1007/s12284-010-9058-3 (2010).
Xu, J. et al. Developing high throughput genotyped chromosome segment substitution lines based on population whole-genome re-sequencing in rice (Oryza sativa L.). BMC Genom. 11, 656–669. https://doi.org/10.1186/1471-2164-11-656 (2010).
Tang, X., Gong, R., Sun, W., Zhang, C. & Yu, S. Genetic dissection and validation of candidate genes for flag leaf size in rice (Oryza sativa L.). Theor. Appl. Genet. 131, 801–815. https://doi.org/10.1007/s00122-017-3036-8 (2017).
Balakrishnan, D., Surapaneni, M., Mesapogu, S. & Neelamraju, S. Development and use of chromosome segment substitution lines as a genetic resource for crop improvement. Theor. Appl. Genet. 132, 1–25. https://doi.org/10.1007/s00122-018-3219-y (2019).
Ma, X. et al. Genomic structure analysis of a set of oryza nivara introgression lines and identification of yield-associated QTLs using whole-genome resequencing. Sci. Rep. 6, 27425–27436. https://doi.org/10.1038/srep27425 (2016).
Qiao, W. et al. Development and characterization of chromosome segment substitution lines derived from oryza rufipogon in the genetic background of O. sativa spp. indica cultivar 9311. BMC Genom. 17, 580–591. https://doi.org/10.1186/s12864-016-2987-5 (2016).
International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature 436, 793–800. https://doi.org/10.1038/nature03895 (2005).
Zhang, J. et al. Extensive sequence divergence between the reference genomes of two elite indica rice varieties Zhenshan 97 and Minghui 63. Proc. Natl. Acad. Sci. USA 113, E5163–E5171. https://doi.org/10.1073/pnas.1611012113 (2016).
Staswick, P. E. et al. Characterization of an arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17, 616–627. https://doi.org/10.1105/tpc.104.026690 (2005).
Zhang, S. et al. Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 151, 1889–1901. https://doi.org/10.1104/pp.109.146803 (2009).
Chen, Q., Mu, J., Zhou, H. & Yu, S. Genetic effect of japonica alleles detected in indica candidate introgression lines. Sci. Agric. Sin. 40, 2387–2394 (2007).
Sun, W. et al. Identification of genomic regions and the isoamylase gene for reduced grain chalkiness in rice. PLoS ONE 10, e0122013. https://doi.org/10.1371/journal.pone.0122013 (2015).
Xia, D. et al. GL3.3, a novel QTL encoding a GSK3/SHAGGY-like kinase, epistatically interacts with GS3 to produce extra-long grains in rice. Mol. Plant 11, 754–756. https://doi.org/10.1016/j.molp.2018.03.006 (2018).
Ying, J. et al. TGW3, a major QTL that negatively modulates grain length and weight in rice. Mol. Plant 11, 750–753. https://doi.org/10.1016/j.molp.2018.03.007 (2018).
Zhang, G. et al. LSCHL4 from japonica cultivar, which is allelic to NAL1, increases yield of indica super rice 93–11. Mol. Plant 7, 1350–1364. https://doi.org/10.1093/mp/ssu055 (2014).
Xue, W. et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. https://doi.org/10.1038/ng.143 (2008).
Bai, X. et al. Duplication of an upstream silencer of FZP increases grain yield in rice. Nat. Plants 3, 885–893. https://doi.org/10.1038/s41477-017-0042-4 (2017).
Sakai, H. et al. Rice annotation project database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 54(2), e6. https://doi.org/10.1093/pcp/pcs183 (2013).
Yano, M., Kojima, S., Takahashi, Y., Lin, H. & Sasaki, T. Genetic control of flowering time in rice, a short-day plant. Plant Physiol. 127, 1425–1429. https://doi.org/10.1104/pp.127.4.1425 (2001).
Qiu, X., Gong, R., Tan, Y. & Yu, S. Mapping and characterization of the major quantitative trait locus qSS7 associated with increased length and decreased width of rice seeds. Theor. Appl. Genet. 125, 1717–1726. https://doi.org/10.1007/s00122-012-1948-x (2012).
Zhu, S. et al. Mapping quantitative trait loci for heat tolerance at the booting stage using chromosomal segment substitution lines in rice. Physiol. Mol. Biol. Plants 23, 817–825. https://doi.org/10.1007/s12298-017-0465-4 (2017).
Mulsanti, I. W. et al. Finding the superior allele of japonica-type for increasing stem lodging resistance in indica rice varieties using chromosome segment substitution lines. Rice 11, 25–25. https://doi.org/10.1186/s12284-018-0216-3 (2018).
Zhao, D. et al. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commu. 9, 1240–1253. https://doi.org/10.1038/s41467-018-03616-y (2018).
Yuan, Z. et al. Genetic dissection of seed storability and validation of candidate gene associated with antioxidant capability in rice (Oryza sativa L.). Int. J. Mol. Sci. 20, 4442–4457. https://doi.org/10.3390/ijms20184442 (2019).
Zhao, Y. Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu. Rev. Plant Biol. 69, 417–435. https://doi.org/10.1146/annurev-arplant-042817-040226 (2018).
Ding, X. et al. Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate and jasmonate independent basal immunity in rice. Plant Cell 20, 228–240. https://doi.org/10.1105/tpc.107.055657 (2008).
Domingo, C., Andres, F., Tharreau, D., Iglesias, D. J. & Talon, M. Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol. Plant-Microbe Interact. 22, 201–210. https://doi.org/10.1094/MPMI-22-2-0201 (2009).
Li, P. & Xue, H. Structural characterization and expression pattern of the rice PLT gene family. Acta Biochim. Biophys. Sin. 43, 688–697. https://doi.org/10.1093/abbs/gmr068 (2011).
Xu, H., Sun, Y., Zhou, H. & Yu, S. Development and characterization of contiguous segment substitution lines with background of an elite restorer line. Acta Agron. Sin. 33, 979–986 (2007).
R Core Team. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, Vienna, 2018). http://www.R-project.org.
McCouch, S. R. Gene nomenclature system for rice. Rice 1, 72–84. https://doi.org/10.1007/s12284-008-9004-9 (2008).
Murry, M. G. & Thompson, W. F. Rapid isolation of high molecular-weight plant DNA. Nucleic Acids Res. 8, 4321–4326. https://doi.org/10.1093/nar/8.19.4321 (1980).
McCouch, S. R. et al. Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9, 257–279. https://doi.org/10.1093/dnares/9.6.199 (2002).
Zhao, H. et al. RiceVarMap: A comprehensive database of rice genomic variations. Nucleic Acids Res. 43, D1018–D1022. https://doi.org/10.1093/nar/gku894 (2015).
Livak, K. & Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△Ct method. Methods 25, 402–408. https://doi.org/10.1006/meth.2001.1262 (2001).
Wang, P., Zhou, G., Cui, K., Li, Z. & Yu, S. Clustered QTL for source leaf size and yield traits in rice (Oryza sativa L.). Mol. Breed. 29, 99–113. https://doi.org/10.1007/s11032-010-9529-7 (2012).
Yang, W. et al. Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat. Commun. 5, 5087–5095. https://doi.org/10.1038/ncomms6087 (2014).
Chen, W. et al. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant 6, 1769–1780. https://doi.org/10.1093/mp/sst080 (2013).
Zhu, K. et al. ERECT PANICLE2 encodes a novel protein that regulates panicle erectness in indica rice. Genetics 184, 343–350. https://doi.org/10.1534/genetics.109.112045 (2010).
Hiei, Y., Ohta, S., Komari, T. & Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. https://doi.org/10.1046/j.1365-313x.1994.6020271.x (1994).
Acknowledgements
This research was funded by grants from the National Natural Science Foundation of China (31671656, 31971864), the Fundamental Research Funds for the Central Universities (2662018YJ025), and the National Key Research and Development Program of China (2016YFD0100301). We thank Shaowen Yuan, Fangzhou Liu, Xiaohai Zheng and Li Tian for assistance with phenotypic evaluation and sample preparation. We are grateful to Kun Xie for assistance in date analysis.
Author information
Authors and Affiliations
Contributions
S.Y. designed and conceived research; D.W. and Q.S. performed phenotyping and fine mapping; Z.Y. and K.F. conducted the transgenic experiment; D.W., Q.S. and W.S. developed the population. D.W., Q.S. and C.Z. analyzed the data; D.W. and S.Y. wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, D., Sun, W., Yuan, Z. et al. Identification of a novel QTL and candidate gene associated with grain size using chromosome segment substitution lines in rice. Sci Rep 11, 189 (2021). https://doi.org/10.1038/s41598-020-80667-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-80667-6
This article is cited by
-
GS6.1 controls kernel size and plant architecture in rice
Planta (2023)
-
KASP mapping of QTLs for yield components using a RIL population in Basmati rice (Oryza sativa L.)
Euphytica (2023)
-
QTL mapping of drought-related traits in the hybrids of Populus deltoides ‘Danhong’×Populus simonii ‘Tongliao1’
BMC Plant Biology (2022)
-
Identification, pyramid, and candidate gene of QTL for yield-related traits based on rice CSSLs in indica Xihui18 background
Molecular Breeding (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.