Abstract
Ligusticum L., one of the largest members in Apiaceae, encompasses medicinally important plants, the taxonomic statuses of which have been proved to be difficult to resolve. In the current study, the complete chloroplast genomes of seven crucial plants of the best-known herbs in Ligusticum were presented. The seven genomes ranged from 148,275 to 148,564 bp in length with a highly conserved gene content, gene order and genomic arrangement. A shared dramatic decrease in genome size resulted from a lineage-specific inverted repeat (IR) contraction, which could potentially be a promising diagnostic character for taxonomic investigation of Ligusticum, was discovered, without affecting the synonymous rate. Although a higher variability was uncovered in hotspot divergence regions that were unevenly distributed across the chloroplast genome, a concatenated strategy for rapid species identification was proposed because separate fragments inadequately provided variation for fine resolution. Phylogenetic inference using plastid genome-scale data produced a concordant topology receiving a robust support value, which revealed that L. chuanxiong had a closer relationship with L. jeholense than L. sinense, and L. sinense cv. Fuxiong had a closer relationship to L. sinense than L. chuanxiong, for the first time. Our results not only furnish concrete evidence for clarifying Ligusticum taxonomy but also provide a solid foundation for further pharmaphylogenetic investigation.
Similar content being viewed by others
Introduction
Comprising numerous economically important species, Apiaceae has attracted increasing attention1. Over the past decades, extensively valuable progress has been made in evolution, phylogeny and systematics based on the investigation of morphology2, molecular barcode3,4,5, and whole genome sequence1, among others. Encompassing ca. 60 species6, Ligusticum L. one of the largest genera in Apiaceae, belongs to the subtribe Seselinae, tribe Ammineae, and subfamily Apioideae of Apiaceae and is widely distributed in alpine belts, meadows and forests of the Eurasian continent and North America, especially the Himalayas and North America, the two diversity centers7,8. However, the evolutionary scheme and the circumscription to putatively allied genera of Ligusticum have been a long-standing debate3,9,10,11,12,13,14. Based on the preceding research, a strongly supported phylogenetic implication and unambiguous classification framework of Ligusticum, with paramount importance in deciphering the evolutionary history of Apioideae, is predominantly hampered by the diversity of various diagnostic characters and the insufficiency of effective molecular data sets7,9.
Ligusticum comprises famous traditional oriental medicinal herbs, the bulk of which contain high amounts of natural active compounds15, such as alkaloids (ligustrazine)15,16,17,18, phenolic acids (ferulic acid)15,17,18, phthalide lactones (ligustilide)19 and volatile oils15,17,18, having prominent pharmaceutical values, represented by L. chuanxiong17,18, one of most pivotal medicinal plants. Owing to the clinical efficacy in the treatment of headaches, dysmenorrhea, menstrual disturbance, stroke, and cardiovascular and cerebrovascular diseases15,17,18, the dry rhizomes of L. chuanxiong (known as Chuanxiong Rhizoma or Chuan-Xiong) are extensively utilized in China, Korea and Japan. However, it is worth noting that the original plant of Chuan-Xiong was first defined by Qiu20 as Ligusticum chuanxiong Hort, a horticultural scientific name, because hitherto, exclusively extant plants were cultispecies, and they were mainly cultivated in Sichuan, China, following the extinction of wild species. The original plant of Japanese Chuan-Xiong (called “Senkyu” in Japanese) is Cnidium officinale Makino21, which was initially placed in the genus Cnidium but was then clustered into Ligusticum by Kenji Kondo et al., based on the rbcL sequence21 and was further revised into Ligusticum and named as L. officinale22 according to universal proof based on molecular evidence23,24 (L. officinale is used hereafter). Moreover, according to the ancient records that L. officinale was first introduced from China to Japan in the Edo era, and based on the sequence analysis of ITS and 18S rRNA23, recent studies proposed that L. officinale was basically synonymous to L. chuanxiong but incongruent with trnK24. Likewise, our recent research revealed that L. officinale is not closer to L. chuanxiong than to L. jeholense25, which also belongs to Ligusticum, has the trivial name LiaoGaoBen or HuoGaoBen, and is widely distributed in northern China and Korea15,26. In addition, L. jeholense together with L. sinense, are the original plants of another well-known traditional medicine, GaoBen15,27. Although numerous studies based on morphology28, karyotype29, and mini-barcodes stated that L. sinense is the wild species of L. chuanxiong26, in Oriental Medicine practices, the explicitly specific property differentiation in channel tropism and therapeutic effect were indicated30. In herbal markets, L. sinense is frequently mistaken in folk medicine and is even deliberately mixed in commercial products of Chuan-Xiong due to its indistinguishable flavor and features of L. chuanxiong using traditional identification methods. Therefore, the issue about whether L. chuanxiong and L. officinale have a close relationship to L. sinense urgently deserves further exploration. Unfortunately, three additional Chinese endemic herbs are also locally known as Chuan-Xiong increasing challenges to authentication29. Even though their rhizome is highly similar to that of L. chuanxiong but with a lower quality compared to L. chuanxiong, according to the records of ancient Chinese medicine classics and experiences from practical application31. Two of those three, original plants heretofore are recognized as different cultivated accessions of L. chuanxiong, and one is named L. sinense cv. Fuxiong, which has been reported to be a triploid plant derived from L. chuanxiong. Nevertheless, the phylogenetic relationship among them is still unknown as incompatible frameworks were presented using data derived from karyotypes29, pollen morphology28, chemical components and DNA fragments32. Similarly, the subtle distinction of L. jeholense and L. tenuissimum (Korean name, Go-Bon)33 resulted in improper utilization and counterfeit medicines being sold frequently in China and Korea. So far, a few complicated methods have been proposed, including multiplex PCR24,33 and high-performance liquid chromatography34 for precise identification, which depends on stringently experimental conditions. Generally, the reliable phylogeny implications and accurate and effective plant identification for those species, in this context, has become progressively imperative. Not only is it critical for promoting market supervision and improving the safety and quality of TCM (traditional Chinese medicine) but it is also of great benefit in elucidating the evolutionary event of Ligusticum.
Chloroplasts (CP), one of the most crucial organelles for photosynthesis in plants35 excluding a few algae, saprophytes and parasitical species, show a semiautonomous proliferation-deduced origin from cyanobacterium in a universally accepted endosymbiotic event36 and contain almost all necessary components regarding autosynthesis. In angiosperms, the chloroplast genome (CP genome) is predominately uniparental inheritance, amplified mainly through ameiosis replication with infrequent recombination37, and exhibits a quadripartite structure within a molecular framework ranging from 115 to 165 kb38. In general, the CP genome contain 110–130 unique genes of which the gene contents, gene order and genome structure are conserved39. Recently, the CP genome has been increasingly demonstrated able to provide sufficient variations, whereby showing high resolution in plant classification superior to that of mini-barcode fragments40. In past decades, phylogenomic approaches to clarify contentious phylogenetic relationships have been successfully employed: for instance, at high taxonomic levels, the phylogenetic relationship of basal lineages of angiosperms41 and the controversy over tree topology of the extant four orders in gymnosperms42 have been settled; at low taxonomic levels, the relationship of wild and domesticated rice has been resolved43. Moreover, utilizing the CP genome promotes the authentication in a variety of TCM that seem indistinguishable via traditional methods and have now well-discerned based on the CP genome22,44.
Hence, in present study, to address foregoing issues, CP genomes of the aforementioned seven species, which are valuable herbal plants in the Ligusticum genus, were sequenced and utilized. Our principal aims herein were to: (1) scrutinize the evolutionary dynamics of the seven plastomes within Ligusticum by examining the genome organization, gene content and sequence divergence to shed light on evolutionary patterns among plastomes of Ligusitcum; (2) identify highly variable candidate regions for species discrimination and population genetic study of Ligusitcum; (3) infer phylogenies to better understand the relationships of Ligusticum species as well as contribute to pharmaphylogenetic investigation.
Results
Plastome features
Seven assembled plastomes of Ligusticum exhibited a typical quadripartite structure with one large single-copy region (LSC) and one small single-copy region (SSC) which were separated by a pair of IRs amenable to the nature of most CP genomes in angiosperms. The CP genomes ranged in size from 148,275 bp for L. sinense to 148,564 bp for L. chuanxiong cv. Gansu and resulted from the associated length difference in four parts: LSC, from 93,682 bp for L. sinense to 94,012 bp for L. chuanxiong cv. Gansu; SSC, from 17,607 bp for L. officinale to 17,629 bp for L. jeholense; IRs, from 18,463 bp for L. jeholense to 18,484 bp for L. sinense (Supplementary Table S2). Although a slight size difference occurred among these CP genomes, two huge deletions were specifically observed in the LSC of L. sinense, prominently responsible for its genome contraction (they were verified together with other Indels of a size greater than 20 bp based on PCR amplification and Sanger sequencing, Supplementary Table S3). The overall GC content of these species was almost equivalent and showed an uneven distribution across the whole CP genome with an average GC content of approximate 37.60% that was most enriched in IRs from 47.78 to 47.80% followed by LSC, about 35.99%, and lowest in SSC, from 31.09 to 31.14%, which imputed the location of transfer RNA (tRNA) and ribosomal RNA (rRNA), of which the GC content reached 55% (Supplementary Table S4). In addition, a similar GC percentage ~ 46.79% in homologous protein-coding regions (CDS) among these species was discovered to be consistent with the identity of the overall GC content. Within CDS, the AT content of each position in the triplet codon displayed the canonical bias of the CP genome, distinguished from nuclear and mitochondrial DNA45 that a higher AT percentage was observed at the third position in involved species, up to 70.30%, along with a sharp decrease in the second and first positions (Supplementary Table S4).
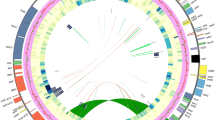
Compared to noncoding regions, coding sequences accounted for ~ 56.90%, showing more conserved features that encoded an identical set of 126 functional genes (Fig. 1), of which 113 were unique, harboring 79 protein-coding genes, 30 tRNA and 4 rRNA, with a coincidence of genomic organization in terms of the gene order and orientation (Table 1). Of these, 13 genes were duplicated, including three protein-coding genes, four rRNA genes and six tRNA genes, all of which resulted from IR duplication. Similar to many angiosperms, introns were discovered in 19 genes comprising 12 protein-coding genes and 6 tRNA genes. Among them, clpP, ycf3 and rps12 contained two introns, especially rps12, a trans-spliced gene, of which the 5′ end was located in the LSC region, whereas two replicated 3′ ends were contained within IRa and IRb regions, respectively. In addition, trnK-UUU possessed the longest intron that contained matK.
Codon usage and RNA editing sites
Since usage bias of synonymous codons is widespread in organisms, it plays a vital role in evolution. Knowledge of codon preference could greatly help in understanding the selection pressure on gene expression46,47 and improve the translation efficiency using major codons48. Here, beyond the major initiator codon, in these seven species, alternative start codons were discovered in two distinct genes where ACG was used as a start codon for ndhD and GTG for rps19. Using alternative codon initiating is a ubiquitous phenomenon in eudicot plants, while previous reports also pointed out that RNA editing could restore ACG to the conventional start codon49,50. Overall, the 79 distinct protein-coding genes in each of the seven species were composed of 23,446–23,482 triplet codons. Of those encoded amino acids, in all presented species, the most abundant was leucine (4.14–4.17%), and the least abundant was cysteine (0.24%), which is similar to most of the reported CP genomes of angiosperm plants. The relative synonymous codon usage (RSCU) value analysis demonstrated that almost every amino acid with a synonymous codon showed a usage bias (Supplementary Table S5, Supplementary Fig. S1). Interestingly, A- or T-ended codons accounted for nearly half of the synonymous codons with commonly higher RSCU values in contrast to the other half that ended with C or G. Possibly, those reported preferences are driven by the mutational pressure in the A/T composition bias of the CP genome51,52,53.
RNA editing events have been proved universally in CP genomes since first reported54. Regarding the current CP genomes, a total of 56 potential RNA editing sites from 32 genes were predicted in each species (Supplementary Table S6). In all seven CP genomes, the event of S converting to L occurred with predominant frequency; by contrast, R converting to C occurred with the lowest frequency, which is in accordance with a previous investigation that the change of S to L becomes more frequent as the number of amino acids increases55.
Repeat structure and simple sequence repeat analyses
Simple sequence repeats (SSRs), known as microsatellite sequences, consist of tandem short repeat units, ubiquitously distributed across the CP genome, mostly with the nature of uniparental inheritance and non-recombination56. Owing to the high degree of polymorphism, co-dominance and efficiency of amplification, SSRs are valuable molecular markers for mining population genetics and phylogenetic studies57. On average, 46 SSRs (from 42 to 49) with two motif types were identified in each species (Supplementary Table S7, Supplementary Fig. S2). SSR motifs presenting a heterogeneity frequency were predominantly rich in A/T bases. Of these SSR repeat units, 13% were detected in protein-coding regions. To capture the dynamic evolution of CP genomes within Ligusticum and Apioideae, the SSR characteristics of available CP genomes of representative plants in Apioideae were also investigated. Interestingly, C/G units had higher variability within Ligusticum, and from early-diverging lineage of Apioideae (Daucus carota) to Peucedaneae (Angelica gigas), SSR characters exhibited an increasing and prolonged tendency of SSRs, primarily in mononucleotide A/T rather than other motifs. Furthermore, the differences in SSR motif numbers among those species further demonstrated the potential of using cpSSR markers in genetic analysis among genera of Apioideae.
On average, 39 long repeats accounting for ~ 0.8% of CP genomes were detected in presented species (Supplementary Table S8, Supplementary Fig. S2), with an apparent species-specific distribution, and none were located in protein-coding regions. In contrast to SSRs, the number of four kinds of long repeats (forward, reverse, complementary and palindromic repeats) in Ligusticum and Apioideae displayed a significant change in certain species, ranging from 31 to 89 without a constant pattern. For instance, forward repeats were significantly enriched in L. chuanxiong cv. Gansu compared to its relatives, the proportion of large-sized repeats and palindromic repeats sharply increased in L. tenuissimum, and complementary repeats occasionally disappeared in Ligusticum. Moreover, repeats of L. tenuissimum were wholly distributed in adjacent regions of IR boundaries, which were likely to lead to its IR expansion as that in previous reports, many evidences showed the repeat sequence contributing to plastome structural variation.
IR contraction and expansion
The absence of one copy of three genes, commonly duplicated and situated in the vicinity of junction sites of IR and LSC, was detected, deserving a more thorough examination. Subsequently, the evolutionary trajectories of the contraction and expansion of IR within Ligusticum (Supplementary Fig. S3) and Apioideae (Fig. 2) were investigated. The border of SSC/IRa crossed by ycf1 maintained a relatively conserved state, in which a nearly constant fluctuation of ca. 100 bp was observed throughout the evolution of Apioideae. Symmetrically, the SSC/IRb boundary, located in the pseudogene fragment ψycf1 and neighboring ndhF, showed an erratic shift, resulting in inconsistent deviation of the border to ndhF and fluctuation in ψycf1 length. Notably, multiple dynamic expansions and contractions indicated that the junction of the LSC/IRb endpoint moved from spanning rps19 in D. carota (putative ancestral IRb/LSC boundary) to rpl2 in Anethum graveolens, followed by lineage-specific IR contraction to ycf2, causing Seselinae and Peucedaneae to lack one copy of ycf2, but both copies were present in L. tenuissimum. Considering the phylogenetic topology that L. tenuissimum was a sister to L. chuanxiong, belonging to the clade comprising species of the tribes Seselinae and Peucedaneae, and an enhanced expansion footprint in L. tenuissimum compared to the ancestor extending rps19 into IR, we remain parsimonious in proposing an independent IR expansion reoccurring in L. tenuissimum. In addition, within the seven presented Ligusticum spp., an on-going shift of the LSC-IR junction and an alleviated dislocation of SSC were demonstrated whereas severe reduction hardly occurred.
Comparison of the border position of LSC, SSC and IRs across the Apioideae. JLB (IRb /LSC), JSB (IRb/SSC), JSA (SSC/IRa) and JLA (IRa/LSC) denote the junction between each corresponding region. Genes and their locations were showed using boxes with corresponding names and with ψ representing the pseudogenes. Genes transcribed clockwise and counter clockwise are presented above and below of components, respectively. The distance between the end or start coordinate of a given gene and the border sites are indicated. The black box in phylogenetic tree denotes the branch-specific IR contraction and gray box denotes branch-specific IR expansion. These features are not to scale.
Furthermore, we investigated the synonymous substitution rate of genes, ycf2 and rpl2 which are duplicated and de-duplicated because of IR contraction and expansion. Exceeding our expectation, the Ks value was highly variable among lineages (Supplementary Fig. S4) but without significant correlation with the copy number variation that resulted from IR contraction and expansion, which usually accelerates synonymous substitution58,59.
Comparative genomic divergence and structure arrangement
In the view of subsequently taking full advantage of hidden mutation information in CP genomes for assisting phylogenetic inference and species identification, an in-depth investigation of the genomic structure and sequence divergence among Ligusticum and Apioideae was performed. Exceptionally, the CP genome sequences of L. chuanxiong cv. Yunnan and L. sinense cv. Fuxiong were identical, and the two were definitely two different species: one diploid with flowers and seeds and one triploid reproduced solely by means of vegetative propagation29. This result was double checked via subsequent conventional Sanger amplicon sequencing of variation hotspot regions. Synteny and sequence divergence analyses demonstrated that the seven Ligusticum species exhibited a high degree of colinearity (Supplementary Fig. S5) and sequence identity at the genome-scale level. The nucleotide diversity value (Pi) across the genomes ranged from 0 to 2.5% and nearly 45.6% of the compared regions showed 100% identity (Fig. 3). A higher divergence in the LSC region and lower divergence in IR were demonstrated, implying general conservatism of IR in contrast to other regions, which is in congruence with characteristics for the majority of angiosperms. Furthermore, the most divergent loci were located in petA-psbJ-psbL with mean Pi = 0.023, and six additionally different hypervariable regions (Pi > 0.004) were determined, including pebH-petB, trnL-trnH-psbA, accD-psaI-ycf4, ycf4-cemA, psbH-petB and ycf1, providing potential candidate regions for genus-specific barcode marker mining. Recently, increasing researches revealed wide prospects of using Indel marker derived from CP genomes for species identification and authentication of herbs22,25,60. Here, the insertion and deletion were detected to be located mainly in noncoding regions especially marked in psbB-petB and trnM-psbD. Intriguingly, Indels have also been found frequently occurring in ndhB and ycf2 coding regions next to the boundary of IRb/LSC (Supplementary Fig. S6), which indicates intensive changes around corresponding junctions; this is compatible with common phenomena found in closely related species61. Despite conserved synteny in gene order and orientation of CP genomes among Apioideae (Supplementary Fig. S7), a higher sequence divergence and length variation, specifically in trnL-psbA and the regions between rpoB and psbD, where protein-coding genes are rarely situated, were disclosed. In contrast, the IRb/LSC boundary of Apioideae was more conserved (Supplementary Fig. S8) compared to the Ligusticum genus in consideration of the comparison among Apioideae using genetically distant related species. If the same underlying mechanisms within Apioideae were involved, accordingly, a high divergence would be detected.
Phylogenetic analyses
To address the relationship among the seven medicinal species, phylogenetic analysis was carried out using entire CP genome sequences because of an extreme synteny among those CP genome and limited parsimony informative characters in the CDS of 79 shared protein-coding genes where scarcely 129 sites were found. Three inference methods, including Bayesian inference (BI), maximum likelihood (ML) and maximum parsimony (MP), were employed along with Seseli montanum as an outgroup. The topologies generated by whole genomes were highly concordant, and almost every node was highly supported, regardless of the different methods used. Among all constructed trees (Fig. 4), Ligusticum comprised two separate subclades where L. tenuissimum presented as a sister clade to the remaining seven taxa. L. chuanxiong cv. Gansu and L. chuaxiong demonstrated a closer relationship, together forming a clade sister to L. officinale, receiving a robust supporting value (~ 100%) for all methods, and this entire group was clustered as the sister clade to L. jeholense following the clade of L. sinense cv. Fuxiong and L. chuanxiong cv. Yunnan. To further verify the phylogenetic relationship obtained, the phylogenetic signal across the genome was measured. As expected, this highly consistent tree was supported by phylogenetic signal analysis based on the value of delta site-wise log-likelihood scores (ΔSLS) with all four strong sites (absolute ΔSLS > 0.5) and 81.3% weak sites (absolute ΔSLS ≤ 0.5) favouring, however, here a lower highest value reaching at most 2.3 was observed (Supplementary Fig. S9). Additionally, extremely short branch lengths within those seven species were observed.
As yet, the increasing availability of CP genomes for Apiaceae provides us unprecedented resources to precisely clarify phylogenetic relationships and investigate the taxonomic status of Ligusticum within Apioideae via phylogenomic analyses. Here, based on 70 shared protein-coding sequences involving 37 species that contain 4196 parsimony informative characters and L. chuanxiong were used to represent those seven species that were attributed to limited parsimony information in CDS. Phylogenetic trees were constructed using maximum parsimony, maximum likelihood and Bayesian inference, with Panax ginseng as an outgroup. The phylogeny produced from each analysis was topologically identical, and most nodes agreed well with previous relevant plastid genome analyses within Apioideae22,62 (Fig. 5). Overall, 29 of 33 nodes received a maximally supported value of 100% bootstrap (BS) and 1.0 posterior probability (PP). Notably, Ligusticum formed a monophyletic clade that was placed within Seselinae and allied with a group comprised of S. montanum and Coriandrum sativum, with weak support in ML and MP but moderate support in BI. An ambiguous circumscription between Seselinae and Peucedaneae was depicted in the present cladogram, which is similar to earlier studies. In accordance with previous phylogeny results based on CP genomes, Glehnia littoralis, positioned within the genus Angelica, was moderately supported22. Moreover, two Prangos plants were weakly clustered as a sister clade to the group consisting of Seselinae and Peucedaneae, which requires further reexamination.
Phylogenetic topology of 36 species within Apiaceae constructed based on the CDS of 70 protein-coding gene sequences using MP, ML, and BI methods with P. ginseng as an outgroup. The supported value of MP, ML, and BI were shown along with each node, and stars represent a bootstrap value ≥ 99% and posterior probability = 0.99.
Discussion
Since the first CP genome of Apiaceae was reported63, along with the rapid development of sequencing technology in past decades, approximately 50 CP genomes were figured out within Apiaceae. However, plastid sequences for Ligusticum, the taxonomic scheme and the placement of which is one of the most difficult genera to clarify in Apiaceae, are scarcely available. The seven newly established CP genomic sequences significantly enrich the molecular resources for Ligusticum. The conserved features of gene content and organization, gene orientation, and intron number among those CP genomes were revealed to be similar to the variability within previously reported species of Camellia64, Panax65 and Epimedium66. Despite the fact that a long time has passed since its divergence from D. carota, those genomes share an identical gene set, with a similar organization further indicating the structural conservation in contrast to Circaeasteraceae67. Although a higher nucleotide variability value was presented in certain divergence hotspot regions based on the diversity investigation and complete genome pairwise alignment, the entire variation exhibited a conserved tendency. Strikingly, in our study, the plastid genome sequences of two species were completely identical, which has barely been reported and may be caused by recent divergence. Overall, a moderate divergence among sequences was demonstrated, compared to several genera recently reported44,61,68,69,70. Of note, petA-psbJ-psbL, the most divergent region revealed in the present study, has also been demonstrated to be highly divergent in many genera68,69,71,72. Even though high diversity was observed in trnH-psbA and ycf1, which have extensively suggested to be taken as universal barcodes73 and depicted has a capacity for sufficient variation information for taxon discrimination in angiosperms72,74, no one hotspot alone was enough to distinguish these seven herbs. Thus, with plastid scale-level analyses, we proposed a combination strategy of those hotspot regions to enable us to definitively distinguish these species and elucidate a comprehensive resolution, which prior studies were not able to achieve based on a single fragment.
Intriguingly, in the present study, we noticed the dramatic branch-specific CP size reduction in the sub-clade consisting of Peucedaneae and Seselinae, including a considerable number of famous oriental medicinal plants. Subsequently, the lack of one copy of the gene clusters located at the boundary of IRa/LSC attributed to IR contraction was observed. Compared with the IR type of D. carota, rpl2, rpl23, trnL-CAU and ycf2 were lost in A. gigas, L. chuanxiong and S. montanum, implying a branch-specific IR shift. For a certain species within this branch, multiple rounds of contraction and expansion occurred resulting in CP genome re-expansion and of which the IR is even larger than that of the ancestor type; for instance, the IR border of L. tenuissimum re-extended to rpl22, which led to a duplication of rps19 that primitively spanned the junction in D. carota, and could potentially be a crucial character in Ligusticum taxonomy. As early as the last century, the frequency fluctuation and large size shift of IR within Apioideae, especially, within the apioid superclade, were noticed and used to reconstruct the phylogenetic relationship75, which re-placed L. officinale in the Angelica group based on the restriction map. Likewise, IR variations in Berberidaceae76,77, conifers78,79, legumes59, and ferns80 were found and used to reconstruct their phylogeny. The rapid development of sequencing technology has allowed us to establish the CP genome, which enables us to further precisely confirm and define the endpoint of IR. Indeed, the examination of IR shifts within Ligusitcum at nucleotide level were reported for the first time and to the best of our knowledge, such a large-scale shift within one genus has not been reported in apioid superclade. Large-scale expansion and contraction (over 1 kb) of IR across Apioideae were revealed to be primarily confined at the boundary of LSC/IRb displaying a lineage-specific flux whereas the situation of SSC/IR exhibited a constant character similar to most non-monocot angiosperms81,82. Recently, as the availability of plastid genomes increases, IR border shifts become more frequently reported, yet large-scale expansion and contraction of IR are considered uncommon phenomena81, principally observed in heterotrophs83 and a few autotrophs84. However, unlike the case of rearrangement that frequently occurred in the CP genome of which IR remarkably shifted, for instance, Pelargonium85, conifers79,86, Clemati87, legumes59, Asarum88, etc., the genome structure and gene order within the apioid superclade are significantly conserved81. Hence, the large-scale alteration of IR within this clade has great benefit for the study of the underlying mechanism causing large-scale expansion and contraction of IR81. In comparison to small alterations of IR which are supposed to result from gene conversion89, large-scale IR alterations are attributed to the double-strand break along with illegitimate recombination accounted by repetitive sequence possibly75,89,90. But herein, at the boundary of LSC/IRb of seven new plastomes, without a directly supporting evidence of those hypotheses, the repetitive motif, was observed in accordance with some previously reported plastomes in Apiaceae81. Nevertheless, around the junction of LSC/IRb in L. tenuissimum, ploy(A) and ploy(T) tracts were discovered. Furthermore, according to previous reports, a novel fragment, the derivation of which remains unsettled, was detected concomitantly residing between LSC/IRa in the CP genome of the apioid superclade and might be contributing to IR shift in Apiaceae81 while a ~ 500 bp insertion was recovered in our seven plastomes but was absent in L. tenuissimum. In Petroselinum, the corresponding homologous regions are highly similar to the intergenic sequence of cob-atp4 of the mitochondrial genome81 and were postulated to be transferred from the mitochondrial genome; however, the insertion fragment of seven Ligusticum plant plastomes did not have a specific homologous region in mitochondrial DNA. To be prudent here, those aforementioned mechanisms that were presumed responsible for the IR shift should not be precluded without further convincing evidence. Previously, a decelerated synonymous rate of duplicated genes led by IR fluxes was uncovered58,59 while herein, no significant changes of the synonymous rate in Ligusticum was manifested, similar to the ycf2 retention event in gingko91.
Combining 37 available CP genomes, a phylogenetic analysis of Apiaceae was carried out, of which the phylogenetic trees were highly similar to the topological structure that was recently reported based on CP genomes except for the node with a weakly supported value. Considering a limited variation in protein-coding regions within Ligusticum, phylogenetic inference of eight Ligusticum species was performed based on the entire genome. L. tenuissimum was a sister to the clade comprising the remainder, with a highly supported value, which is in line with the relationship deduced from morphological cladistic analysis using 40 characters9. Here, L. chuanxiong cv. Gansu and L. chuanxiong were clustered as a sister clade to L. officinale, verifying a closer relationship of L. chuanxiong cv. Gansu and L. chuanxiong, coincident with the origin investigation by herbal textual research, i.e., they belong to the western type of Chuan-Xiong30. In addition, we further confirmed the former revision and repositioning of L. officinale in the Ligusticum genus based on molecular cytogenetic92 and barcoding21,23 analyses, providing the molecular evidence for the ancient record that L. officinale was introduced from China. Above all, a closer relationship of L. chuanxiong to L. jeholense rather than L. sinense was revealed for the first time. Therefore, here, we approved the original nomenclature of L. chuanxiong presented by Qiu20 instead of the revision by Fu26. Previous studies based on karyotype suggested L. sinense cv. Fuxiong was a triploid of L. Chuanxiong29, while, as another foremost discovery, we provided the distinct result that L. sinense cv. Fuxiong had a closer relationship with L. chuanxiong cv. Yunnan and with affinity to L. sinense rather than to L. chuanxiong. We purely presume that the sequence identity of L. sinense cv. Fuxiong and L. chuanxiong cv. Yunnan resulted from incomplete lineage sorting or a recent divergence event, owing to the triploid event of L. sinense cv. Fuxiong, of which the ancestor probably derived from Yunnan that was demonstrated as the most diverse center of Ligusticum in China7. Furthermore, L. sinense cv. Fuxiong or the ancestor was introduced and domesticated in Jiangxi. Our present analyses simultaneously encompassed different original plants with nomenclatural types of L. chuanxiong, which are actually distinct, whereas previous researchers did not realize this. In light of the present findings, we strongly support the hypotheses that ancient Chuan-Xiong was independently derived from two regional groups of original plants with different distributions and cultivation centers, one in the north of China, including L. chuanxiong and L. chuanxiong cv. Gansu, and the other mainly in the south, including L. sinense cv. Fuxiong and L. chuanxiong Yunnan. Our data also elucidate a relatively distant relationship between L. chuanxiong cv. Yunnan and L. chuanxiong suggesting the scientific name should be revised for L. chuanxiong cv. Yunnan. Furthermore, we obtained phylogenetic trees based on CP inheritance and our assumption could be further scrutinized by integrating nuclear and mitochondrial data.
Materials and methods
Plant materials, DNA extraction and sequencing
The seven Liguisticum species used in this study were collected from different places. The detailed collection and identification information of each sample is listed in Supplementary Table S1. The voucher specimens were deposited in the herbarium of the traditional Chinese medicine planting center of Sichuan, Industrial Crop Research Institute, Sichuan Academy of Agricultural Sciences, China, and living plants were permanently planted in germplasm nurseries of traditional Chinese medicinal plants, at the scientific research base of the Industrial Crop Research Institute, Sichuan Academy of Agricultural Sciences, China. Fresh leaves of each plant were collected and frozen in liquid nitrogen and then surrounded by dry ice. Leaves were divided into two parts: one was used for sequencing, and the other underwent long-term storage at − 80 °C for later use. Total genomic DNA was extracted using the modified CTAB method93 and then the concentration and purity were examined using spectrophotometric methods by a Nanodrop-2000 spectrometer (Nanodrop Technologies, Wilmington, DE, USA) and DNA agarose gel electrophoresis by comparison with marker and reference DNA samples. A 350 bp sequencing library was prepared strictly according to the manufacturer’s instruction using highly pure DNA samples and was subsequently subjected to the HiSeq X Ten platform. At least 10 Gb data of 150 bp pair-end reads for each sample were obtained.
Genome assembly, annotation, and sequence features
Using publicly available CP genome sources of relatives as a reference, CP genomes of seven species were assembled using NOVOPlasty 3.094 with default kmer value, and the junction regions were confirmed by Sanger sequencing. The assemblies were annotated via the online website OGDRAW 1.3.1 (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html)95 with default parameters along with tRNA identification. The annotations were manually examined and revised by comparison with homologous genes and reference genomes using several types of software: Geneious v10.2.2, MEGA 7.096 and Apollo v1.11.897. The linear genome map was drawn using OGDRAW. The assembled and annotated results were submitted to NCBI with corresponding accession numbers (Supplementary Table S1). The following sequence features were analyzed: (1) GC content was calculated using an in-house Perl script; (2) SSRs were detected using MISA98 with the setting file as follows: mononucleotide-ten, dinucleotide-six, trinucleotide-five, tetranucleotide-five, pentanucleotide-five, and hexanucleotide-five, interruptions-one hundred; (3) Codon usage bias was calculated by MEGA 7.0, and the RSCU (Relative synonymous codon usage) ratio with a threshold value of 1 was applied to estimate the usage preference of synonymous codons; (4) Direct (forward), reverse, complement and inverted (palindromic) dispersed repeats were examined via the online program REPuter99 with parameters as follows: hamming distance was set to 3, minimum and maximum sizes of repeats were 30 bp and 500 bp, respectively, and redundant repeats were manually removed; (5) RNA editing sites were predicted through the online program Predictive RNA Editor for Plants (PREP-Cp)100 with a cutoff value of 0.8.
Genome comparison
Prank101 and MAFFT version 7102 were used for multiple sequence alignment with default parameters. The sequence identity of CP genomes was intercompared and visualized using mVISTA103. The annotation of L. chuanxiong was taken as a reference. Colinearity and rearrangement of the CP genome were determined by Mauve104 with default parameters. When running Mauve, L. chuanxiong cv Ganshu and A. graveolens were taken as reference for detecting within Ligusticm and Apiaceae, respectively. The nucleotide diversity value was calculated by DnaSP v6105 using a sliding window length of 600 bp and a 200 bp step size. Pairwise synonymous substitution values were examined based on bioperl and were normalized following Chaw’s method91. The Wilcoxon test was implemented to determine the significance level.
Phylogenetic analysis
In total, 43 CP genomes were used for phylogenetic relationship inference, of which 36 were downloaded from NCBI (Supplementary Table S9). Two data sets were used: (1) for phylogenetic analysis among Ligusticum, the entire CP genome sequence was used, and (2) for the intergeneric phylogenetic analysis within Apiaceae, the CDS of common protein-coding gene were used. The whole genome was aligned using MAFFT version 7. The CDS of seventy common protein-coding genes were detected and extracted using an in-house Perl script, and multiple sequence alignments of each gene were executed separately via two programs, Clustal W 2.1106 and MAFFT version 7. Afterwards, the aligned CDS sequences were concatenated into one data set for further analysis. Phylogenetic analysis was performed based on three different algorithms, MP, ML and BI. An optimal nucleotide substitution model was implemented via the analysis of jModelTest 2107 and the model with the best corrected Akaike Information Criterion (AICc) value was selected. RAxML version 8108 was used for ML tree construction with 1000 bootstrapping replicates and the nucleotide substitution model GTR+G was token based on the results of jModelTest 2. Using PAUP version 4109 with a heuristic search method (repeat 1000), the MP tree was constructed and tested by the bootstrap method as well. Mrbayes v3.2.6110 was used for BI tree construction with at least, 2,000,000 iterations of the Markov Chain Monte Carlo method. When p values converged, the majority-rule consensus tree was constructed based on the remaining 75% of the sample. The unconstrained ML tree stated above was set as T1 for phylogenetic signal exploration while the alternative tree (T2) was obtained based on the ML constrained method referring to the framework described by Shen111, the method of which site-wise log-likelihood and ΔSLS was calculated. The threshold for strong and weak sites was set based on Shen’s method111.
Data availability
All the scripts and commands used can be found at https://github.com/can11sichuan/test/tree/master.
References
Iorizzo, M. et al. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat. Genet. 48, 657–666 (2016).
Sun, F. J. & Downie, S. R. Phylogenetic analyses of morphological and molecular data reveal major clades within the perennial, endemic western North American Apiaceae subfamily Apioideae. J. Torrey Bot. Soc. 137, 133–156 (2010).
Spalik, K., Reduron, J. P. & Downie, S. R. The phylogenetic position of Peucedanum sensu lato and allied genera and their placement in tribe Selineae (Apiaceae, subfamily Apioideae). Plant Syst. Evol. 243, 189–210 (2004).
Zhou, J., Peng, H., Downie, S. R., Liu, Z. W. & Gong, X. A molecular phylogeny of Chinese Apiaceae subfamily Apioideae inferred from nuclear ribosomal DNA internal transcribed spacer sequences. Taxon 57, 402–416 (2008).
Zhou, J., Gong, X., Downie, S. R. & Peng, H. Towards a more robust molecular phylogeny of Chinese Apiaceae subfamily Apioideae: Additional evidence from nrDNA ITS and cpDNA intron (rpl16 and rps16) sequences. Mol. Phylogenet. Evol. 53, 56–68 (2009).
Pu, F. T. A revision of the genus Ligusticum (Umbelliferae) in China. Acta Phytotaxonomica Sinica 29, 385–393 (1991).
Zhou, J., Pu, F. D., Peng, H. J., Pan, Y. Z. & Gong, X. Karyological studies of ten Ligusticum species (Apiaceae) from the Hengduan Mountains Region of China. Caryologia 61, 333–341 (2008).
Pu, F. T. & Watson, M. F. Ligusticum L. in Flora of China (eds. Sheh, M. L. et al.) 140–151 (Missouri Botanical Garden, 2005).
Sun, N., He, X. J. & Zhou, S. D. Morphological cladistic analysis of Ligusticum (Umbelliferae) in China. Nord. J. Bot. 26, 118–128 (2008).
Mathews, S., Clements, M. D. & Beilstein, M. A. A duplicate gene rooting of seed plants and the phylogenetic position of flowering plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 383–395. https://doi.org/10.1098/rstb.2009.0233 (2010).
Pimenov, M. G., Kljuykov, E. V. & Ostroumova, T. A. Towards a clarification in the taxonomy of Sino-Himalayan species of Selinum L. s. l. (Umbelliferae). The genus Oreocome Edgew. Willdenowia 31, 101–124 (2001).
Pimenov, M. G., Kljuykov, E. V. & Ostroumova, T. A. A revision of Conioselinum Hoffm. (Umbelliferae) in the Old World. Willdenowia 33, 353–377 (2003).
Downie, S. R., Katz-Downie, D. S. & Watson, M. F. A phylogeny of the flowering plant family Apiaceae based on chloroplast DNA rpl16 and rpoC1 intron sequences: Towards a suprageneric classification of subfamily apioideae. Am. J. Bot. 87, 273–292 (2000).
Downie, S. R., Watson, M. F., Spalik, K. & Katz-Downie, D. S. Molecular systematics of Old World Apioideae (Apiaceae): relationships among some members of tribe Peucedaneae sensu lato, the placement of several island-endemic species, and resolution within the apioid superclade. Can. J. Bot. 78, 506–528 (2000).
Donkor, P. O., Chen, Y., Ding, L. Q. & Qiu, F. Locally and traditionally used Ligusticum species—A review of their phytochemistry, pharmacology and pharmacokinetics. J. Ethnopharmacol. 194, 530–548. https://doi.org/10.1016/j.jep.2016.10.012 (2016).
Xiao, Y. Q., Li, L. I., You, X. L., Taniguchi, M. & Baba, K. Studies on chemical constituents of the rhizomae of Ligusticum chuanxiong. China J. Chin. Mater. Med. 27, 519–522 (2002) (in Chinese).
Ran, X., Ma, L., Peng, C., Zhang, H. & Qin, L. P. Ligusticum chuanxiong Hort: A review of chemistry and pharmacology. Pharm. Biol. 49, 1180–1189 (2011).
Li, W. X., Tang, Y. P., Chen, Y. Y. & Duan J. A. Advances in the chemical analysis and biological activities of Chuanxiong. Molecules 17, 10614–10651 (2012).
Yan, R., Ko, N. L., Li, S. L., Tam, Y. K. & Lin, G. Pharmacokinetics and metabolism of ligustilide, a major bioactive component in Rhizoma Chuanxiong, in the rat. Drug Metab. Dispos. 36, 400–408 (2008).
Qiu, S. H., Zeng, Y. Q., Pan, K. Y., Tang, Y. C. & Xu, J. M. On the nomenclature of the Chinese plant drug “Chuanxiong”. Acta Phytotaxonomica Sinica 17, 101–103 (1979) (in Chinese).
Kondo, K., Terabayashi, S., Okada, M., Yuan, C. & He, S. Phylogenetic relationship of medicinally important Cnidium officinale and Japanese Apiaceae based on rbcL sequences. J. Plant. Res. 109, 21–27 (1996).
Park, I. et al. Sequencing and comparative analysis of the chloroplast genome of Angelica polymorpha and the development of a novel Indel marker for species identification. Molecules 24, 1038. https://doi.org/10.3390/molecules24061038 (2019).
Liu, Y., Cao, H., Han, G., Fushimi, H. & Komatsu, K. MatK and ITS nucleotide sequencing of crude drug Chuanxiong and phylogenetic relationship between their species from China and Japan. Acta Pharmaceutica Sinica 37, 63 (2002) (in Chinese).
Zhu, S. et al. Molecular identification of “Chuanxiong” by nucleotide sequence and multiplex single base extension analysis on chloroplast trnK gene. Biol. Pharmaceutical Bull. 30, 527–531 (2007).
Xiong, M. et al. Development of germplasm identification markers and phylogenetics analysis of Ligusticum chuanxiong. Chin. Traditional Herbal Drugs 51, 169–181 (2020) (in Chinese).
Pu, F. T. A revision of the genus Ligusticum (Umbelliferae) in China (Cont.). Acta Phytotaxonomica Sinica 29, 525–548 (1991).
Kim, B. et al. Endothelium-independent vasorelaxant effect of Ligusticum jeholense root and rhizoma on rat thoracic aorta. Molecules 20, 10721–10733 (2015).
Pingli, W. Pollen morphology and its relationship of Ligusticum sinense cv. Chuanxiong, L. sinense cv. Fuxiong and L. sinense. Acta Botanica Yunnanica 12, 173–178 (1990) (in Chinese).
Shumin, F. & Haidao, Z. Studies on the origin of the traditional Chinese drug Fuxiong and its relationships with Ligusticum chuanxiong and L. sinense. Acta Phytotaxonomica Sinica 22, 38–42 (1984) (in Chinese).
Shan, F. & Hao, J. Herbal textual research on origin and development of Chuanxiong. China J. Chin. Mater. Med. 36, 2306 (2011) (in Chinese).
Chen, L., Peng, C., Liu, Y., Chen, H. & Xiang, C. Discussion on forming pattern of Dao-di Herbs Ligusticum chuanxiong. China J. Chin. Mater. Med. 36, 2303 (2011) (in Chinese).
Wang, S. T. L., Sun, S., Huang, N., Sun, X. & Wang, H. ITS2 sequence analysis of germplasms of Ligusticum sinense cv. Chuanxiong, L. sinense Oliv. and L. jeholense Nakai. J. Agric. Univ. Hebei 40, 25–31 (2017) (in Chinese).
Jigden, B. et al. Authentication of the oriental medicinal plant Ligusticum tenuissimum (Nakai) Kitagawa (Korean Go-Bon) by multiplex PCR. Planta Med. 76, 648–651 (2010).
Wang, J. H., Xu, L., Yang, L., Liu, Z. L. & Zhou, L. G. Composition, antibacterial and antioxidant activities of essential oils from Ligusticum sinense and L. jeholense (Umbelliferae) from China. Rec. Nat. Prod. 5, 314–318 (2011).
Neuhaus, H. E. & Emes, M. J. Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Phys. 51, 111–140 (2000).
Sanchez-Baracaldo, P., Raven, J. A., Pisani, D. & Knoll, A. H. Early photosynthetic eukaryotes inhabited low-salinity habitats. Proc. Natl. Acad. Sci. USA 114, E7737–E7745 (2017).
Birky, C. W. The inheritance of genes in mitochondria and chloroplasts: Laws, mechanisms, and models. Annu. Rev. Genet. 35, 125–148 (2001).
Tanvi, K. et al. Chloroplast genome sequence of Pigeonpea (Cajanus cajan (L.) Millspaugh) and Cajanus scarabaeoides (L.) Thouars: Genome organization and comparison with other legumes. Front. Plant Sci. 7, 1847. https://doi.org/10.3389/fpls.2016.01847 (2016).
Daniell, H., Lin, C. S., Yu, M. & Chang, W. J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 17, 134. https://doi.org/10.1186/s13059-016-1004-2 (2016).
Krawczyk, K., Nobis, M., Myszczynski, K., Klichowska, E. & Sawicki, J. Plastid super-barcodes as a tool for species discrimination in feather grasses (Poaceae: Stipa). Sci. Rep. 8, 1924. https://doi.org/10.1038/s41598-018-20399-w (2018).
Moore, M. J., Bell, C. D., Soltis, P. S. & Soltis, D. E. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc. Natl. Acad. Sci. USA 104, 19363–19368 (2007).
Wu, C. S., Wang, Y. N., Liu, S. M. & Chaw, S. M. Chloroplast genome (cpDNA) of Cycas taitungensis and 56 cp protein-coding genes of Gnetum parvifolium: Insights into cpDNA evolution and phylogeny of extant seed plants. Mol. Biol. Evol. 24, 1366–1379 (2007).
Wambugu, P. W., Brozynska, M., Furtado, A., Waters, D. L. & Henry, R. J. Relationships of wild and domesticated rices (Oryza AA genome species) based upon whole chloroplast genome sequences. Sci. Rep. 5, 13957. https://doi.org/10.1038/srep13957 (2015).
Li, X. X. et al. Comparison of four complete chloroplast genomes of medicinal and ornamental Meconopsis species: genome organization and species discrimination. Sci. Rep. 9, 10567. https://doi.org/10.1038/s41598-019-47008-8 (2019).
Clegg, M. T., Gaut, B. S., Learn, G. H. & Morton, B. R. Rates and patterns of chloroplast DNA evolution. Proc. Natl. Acad. Sci. USA 91, 6795–6801 (1994).
Iannacone, R., Grieco, P. D. & Cellini, F. Specific sequence modifications of a cry3B endotoxin gene result in high levels of expression and insect resistance. Plant Mol. Biol. 34, 485–496 (1997).
Rouwendal, G. J. A., Mendes, O., Wolbert, E. J. H. & de Boer, A. D. Enhanced expression in tobacco of the gene encoding green fluorescent protein by modification of its codon usage. Plant Mol. Biol. 33, 989–999. https://doi.org/10.1023/A:1005740823703 (1997).
Bulmer, M. Are codon usage patterns in unicellular organisms determined by selection-mutation balance?. J. Evolut. Biol. 1, 15–26 (1988).
Takenaka, M., Zehrmann, A., Verbitskiy, D., Hartel, B. & Brennicke, A. RNA editing in plants and its evolution. Annu. Rev. Genet. 47, 335–352 (2013).
Hoch, B., Maier, R. M., Appel, K., Igloi, G. L. & Kossel, H. Editing of a chloroplast mRNA by creation of an initiation codon. Nature 353, 178–180. https://doi.org/10.1038/353178a0 (1991).
Morton, B. R. The role of context-dependent mutations in generating compositional and codon usage bias in grass chloroplast DNA. J. Mol. Evol. 56, 616–629 (2003).
Williams, A. V., Boykin, L. M., Howell, K. A., Nevill, P. G. & Small, I. The complete sequence of the Acacia ligulata chloroplast genome reveals a highly divergent clpP1 gene. PLoS ONE 10, e0125768. https://doi.org/10.1371/journal.pone.0125768 (2015).
Wang, Y. et al. Complete chloroplast genome sequence of Aquilaria sinensis (Lour.) Gilg and evolution analysis within the Malvales order. Front. Plant Sci. 7, 280. https://doi.org/10.3389/fpls.2016.00280 (2016).
Lenz, H., Hein, A. & Knoop, V. Plant organelle RNA editing and its specificity factors: Enhancements of analyses and new database features in PREPACT 3.0. BMC Bioinform. 19, 255. https://doi.org/10.1186/s12859-018-2244-9 (2018).
Luo, J. et al. Comparative chloroplast genomes of photosynthetic orchids: Insights into evolution of the Orchidaceae and development of molecular markers for phylogenetic applications. Plos One 9, e99016. https://doi.org/10.1371/journal.pone.0099016 (2014).
Olmstead, R. G. & Palmer, J. D. Chloroplast DNA systematics: A review of methods and data-analysis. Am. J. Bot. 81, 1205–1224 (1994).
Cavalier-Smith, T. Chloroplast evolution: Secondary symbiogenesis and multiple losses. Curr. Biol. 12, R62–R64 (2002).
Perry, A. S. & Wolfe, K. H. Nucleotide substitution rates in legume chloroplast DNA depend on the presence of the inverted repeat. J. Mol. Evol. 55, 501–508 (2002).
Wang, Y. H., Qu, X. J., Chen, S. Y., Li, D. Z. & Yi, T. S. Plastomes of Mimosoideae: Structural and size variation, sequence divergence, and phylogenetic implication. Tree Genet. Genomes 13, 41. https://doi.org/10.1007/s11295-017-1124-1 (2017).
Kim, Y. et al. Molecular discrimination of Cynanchum wilfordii and Cynanchum auriculatum by InDel markers of chloroplast DNA. Molecules 23, 1337. https://doi.org/10.3390/molecules23061337 (2018).
Liu, L. X. et al. Chloroplast genome analyses and genomic resource development for epilithic sister genera Oresitrophe and Mukdenia (Saxifragaceae), using genome skimming data. BMC Genomics 19, 235. https://doi.org/10.1186/s12864-018-4633-x (2018).
Samigullin, T. H., Logacheva, M. D., Degtjareva, G. V., Terentieva, E. I. & Vallejo-Roman, C. M. Complete plastid genome of critically endangered plant Prangos trifida (Apiaceae: Apioideae). Conserv. Genet. Resources 10, 847–849. https://doi.org/10.1007/s12686-017-0945-4 (2018).
Ruhlman, T. et al. Complete plastid genome sequence of Daucus carota: implications for biotechnology and phylogeny of angiosperms. BMC Genom. 7, 222. https://doi.org/10.1186/1471-2164-7-222 (2006).
Huang, H., Shi, C., Liu, Y., Mao, S. Y. & Gao, L. Z. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evolut. Biol. 141, 51. https://doi.org/10.1186/1471-2148-14-151 (2014).
Zhao, Y. B. et al. The complete chloroplast genome provides insight into the evolution and polymorphism of Panax ginseng. Front. Plant Sci. 5, 696. https://doi.org/10.3389/fpls.2014.00696 (2015).
Zhang, Y. J. et al. The complete chloroplast genome sequences of five Epimedium species: lights into phylogenetic and taxonomic analyses. Front. Plant Sci. 7 (2016).
Sun, Y. et al. Complete plastome sequencing of both living species of Circaeasteraceae (Ranunculales) reveals unusual rearrangements and the loss of the ndh gene family. BMC Genom. 18, 592. https://doi.org/10.1186/s12864-017-3956-3 (2017).
Zhao, M. L. et al. Comparative chloroplast genomics and phylogenetics of nine Lindera species (Lauraceae). Sci. Rep. 8, 8844. https://doi.org/10.1038/s41598-018-27090-0 (2018).
Li, X., Zuo, Y., Zhu, X., Liao, S. & Ma, J. Complete chloroplast genomes and comparative analysis of sequences evolution among seven Aristolochia (Aristolochiaceae) medicinal species. Int. J. Mol. Sci. https://doi.org/10.3390/ijms20051045 (2019).
Zhang, X. et al. Comparative analyses of chloroplast genomes of Cucurbitaceae species: Lights into selective pressures and phylogenetic relationships. Molecules 23, 2165. https://doi.org/10.3390/molecules23092165 (2018).
Xu, F. et al. Comparative analysis of two sugarcane ancestors Saccharum officinarum and S. spontaneum based on complete chloroplast genome sequences and photosynthetic ability in cold stress. Int. J. Mol. Sci. https://doi.org/10.3390/ijms20153828 (2019).
Dong, W., Liu, J., Yu, J., Wang, L. & Zhou, S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE 7, e35071. https://doi.org/10.1371/journal.pone.0035071 (2012).
Kress, W. J. & Erickson, D. L. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. Plos One 2, e508. https://doi.org/10.1371/journal.pone.0000508 (2007).
Dong, W. et al. ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 5, 8348. https://doi.org/10.1038/srep08348 (2015).
Plunkett, G. M. & Downie, S. R. Expansion and contraction of the chloroplast inverted repeat in Apiaceae subfamily Apioideae. Syst. Bot. 25, 648–667 (2000).
Ma, J. et al. The complete chloroplast genome sequence of Mahonia bealei (Berberidaceae) reveals a significant expansion of the inverted repeat and phylogenetic relationship with other angiosperms. Gene 528, 120–131. https://doi.org/10.1016/j.gene.2013.07.037 (2013).
Kim, Y. D. & Jansen, R. K. Chloroplast DNA restriction site variation and phylogeny of the Berberidaceae. Am. J. Bot. 85, 1766–1778 (1998).
Guo, W. H. et al. Predominant and substoichiometric isomers of the plastid genome coexist within Juniperus plants and have shifted multiple times during cupressophyte evolution. Genome Biol. Evolut. 6, 580–590 (2014).
Wu, C. S. & Chaw, S. M. Highly rearranged and size-variable chloroplast genomes in conifers II clade (cupressophytes): evolution towards shorter intergenic spacers. Plant Biotechnol. J. 12, 344–353 (2014).
Stein, D. B., Palmer, J. D. & Thompson, W. F. Structural evolution and flip-flop recombination of chloroplast DNA in the fern genus Osmunda. Curr. Genet. 10, 835–841 (1986).
Downie, S. R. & Jansen, R. K. A comparative analysis of whole plastid genomes from the Apiales: expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst Bot 40, 336–351 (2015) (316).
Samigullin, T. H., Logacheva, M. D., Terenteva, E. I., Degtjareva, G. V. & Vallejo-Roman, C. M. Plastid genome of Seseli montanum: complete sequence and comparison with plastomes of other members of the Apiaceae family. Biochemistry (Mosc) 81, 981–985. https://doi.org/10.1134/S0006297916090078 (2016).
Naumann, J. et al. Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visseri (Hydnoraceae). Genome Biol. Evol. 8, 345–363. https://doi.org/10.1093/gbe/evv256 (2016).
Zhu, A., Guo, W., Gupta, S., Fan, W. & Mower, J. P. Evolutionary dynamics of the plastid inverted repeat: The effects of expansion, contraction, and loss on substitution rates. New Phytol. 209, 1747–1756. https://doi.org/10.1111/nph.13743 (2016).
Chumley, T. W. et al. The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 23, 2175–2190 (2006).
Strauss, S. H., Palmer, J. D., Howe, G. T. & Doerksen, A. H. Chloroplast genomes of two conifers lack a large inverted repeat and are extensively rearranged. Proc. Natl. Acad. Sci. 85, 3898–3902. https://doi.org/10.1073/pnas.85.11.3898 (1988).
Choi, K. S., Jeong, K. S., Ha, Y.-H. & Choi, K. Complete chloroplast genome sequences of Clematis: IR expansion and relative rates of synonymous substitutions. Preprints https://doi.org/10.20944/preprints201804.0106.v1 (2018).
Sinn, B. T., Sedmak, D. D., Kelly, L. M. & Freudenstein, J. V. Total duplication of the small single copy region in the angiosperm plastome: Rearrangement and inverted repeat instability in Asarum. Am. J. Bot. 105, 71–84. https://doi.org/10.1002/ajb2.1001 (2018).
Goulding, S. E., Olmstead, R. G., Morden, C. W. & Wolfe, K. H. Ebb and flow of the chloroplast inverted repeat. Mol. Gen. Genet. 252, 195–206. https://doi.org/10.1007/BF02173220 (1996).
Wang, R. J. et al. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol. 8, 36. https://doi.org/10.1186/1471-2148-8-36 (2008).
Lin, C. P., Wu, C. S., Huang, Y. Y. & Chaw, S. M. The complete chloroplast genome of Ginkgo biloba reveals the mechanism of inverted repeat contraction. Genome Biol. Evolut. 4, 1201–1201 (2012).
Lee, S. H., Choi, H. W., Sung, J. S. & Bang, J. W. Inter-genomic relationships among three medicinal herbs: Cnidium officinale, Ligusticum chuanxiong and Angelica polymorpha. Genes Genom 32, 95–101 (2010).
Attitalla, I. H. Modified CTAB method for high quality genomic DNA extraction from medicinal plants. Pak. J. Biol. Sci. 14, 998–999 (2011).
Dierckxsens, N., Mardulyn, P. & Smits, G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18. https://doi.org/10.1093/nar/gkw955 (2016).
Greiner, S., Lehwark, P. & Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. https://doi.org/10.1093/nar/gkz238 (2019).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Lewis, S. E. et al. Apollo: A sequence annotation editor. Genome Biol. 3, RESEARCH0082. https://doi.org/10.1186/gb-2002-3-12-research0082 (2002).
Thiel, T., Michalek, W., Varshney, R. K. & Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 106, 411–422. https://doi.org/10.1007/s00122-002-1031-0 (2003).
Kurtz, S. & Schleiermacher, C. REPuter: Fast computation of maximal repeats in complete genomes. Bioinformatics 15, 426–427 (1999).
Mower, J. P. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 37, W253–W259 (2009).
Löytynoja, A. & Goldman, N. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science 320, 1632–1635 (2008).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. https://doi.org/10.1093/molbev/mst010 (2013).
Mayor, C. et al. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16, 1046–1047 (2000).
Darling, A. C. E., Mau, B., Blattner, F. R. & Perna, N. T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403 (2004).
Librado, P. & Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. https://doi.org/10.1093/bioinformatics/btm404 (2007).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 9, 772 (2012).
Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Wilgenbusch, J. C. & Swofford, D. Inferring evolutionary trees with PAUP. Curr. Protocols Bioinform, Chapter 6, Unit 6.4. https://doi.org/10.1002/0471250953.bi0604s00 (2003).
Ronquist, F. et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012).
Shen, X. X., Hittinger, C. T. & Rokas, A. Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nat. Ecol. Evol. 1, 126. https://doi.org/10.1038/s41559-017-0126 (2017).
Acknowledgements
This research was supported by the Financial Innovation Ability Promotion Project of Sichuan province (Grant numbers: 2017QNJJ-004, 2016TSCY-001 and 2018LWJJ-019), Application Foundation Projects of Sichuan province (Grant number: 2018JY0633), Breeding Project of Sichuan Province (Grant number: 2016NYZ0036-4-1) and Scientific and Technological Program of Zhejiang province (Grant number: 2017F10024).
Author information
Authors and Affiliations
Contributions
C.Z., F.-S.M. and F.P. designed this experiment; S.T., M.X. and W.-J.Z. collected the plant materials and validated the analysis results; C.Y., Y.W., M.-Q.L., X.-F.S., and M.X. performed the data analysis, C.Y., Y.W., and M.-Q.L. wrote the manuscript; C.Z., F.-S.M., F.P., Y.W., and M.-Q.L. revised the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yuan, C., Sha, X., Xiong, M. et al. Uncovering dynamic evolution in the plastid genome of seven Ligusticum species provides insights into species discrimination and phylogenetic implications. Sci Rep 11, 988 (2021). https://doi.org/10.1038/s41598-020-80225-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-80225-0
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.