Abstract
Previous studies showed that mutation of folC caused decreased expression of the dihydropteroate synthase encoding gene folP2 in Mycobacterium tuberculosis (M. tuberculosis). We speculated that mutation of folC in M. tuberculosis might affect the susceptibility to sulfamethoxazole (SMX). To prove this, 53 clinical isolates with folC mutations were selected and two folC mutants (I43A, I43T) were constructed based on M. tuberculosis H37Ra. The results showed that 42 of the 53 clinical isolates (79.2%) and the two lab-constructed folC mutants were more sensitive to SMX. To probe the mechanism by which folC mutations make M. tuberculosis more sensitive to SMX, folP2 was deleted in H37Ra, and expression levels of folP2 were compared between H37Ra and the two folC mutants. Although deletion of folP2 resulted in increased susceptibility to SMX, no difference in folP2 expression was observed. Furthermore, production levels of para-aminobenzoic acid (pABA) were compared between the folC mutants and the wild-type strain, and results showed that folC mutation resulted in decreased production of pABA. Taken together, we show that folC mutation leads to decreased production of pABA in M. tuberculosis and thus affects its susceptibility to SMX, which broadens our understanding of mechanisms of susceptibilities to antifolates in this bacterium.
Similar content being viewed by others
Introduction
Tuberculosis (TB), which is caused by the single infectious agent Mycobacterium tuberculosis (M. tuberculosis), was known as the “white plague” in the preantibiotics era1. Today, even with various available antimicrobials, TB is still one of the deadliest diseases, claiming an estimated 1.5 million deaths worldwide in 20182. The occurrence of multidrug-resistant (MDR) M. tuberculosis, which is co-resistant to rifampicin (RIF) and isoniazid (INH), complicates the control of TB3,4. It was estimated that there were globally 484,000 MDR/RIF-resistant (RR) TB cases in 20182. The frequent increases in drug resistance reduce the number of drugs available to treat TB and renew the interest in old anti-TB drugs. Increasing our understanding of these old drugs with respect to action and resistance mechanisms is a faster and cheaper strategy compared with the research and development of new drugs.
Folate antagonists are one kind of renewed drugs that impose negative effects on different components of folate synthesis and metabolic pathways. The bacterial folate pathway is a valuable target for antimicrobial drug design due to the following two reasons. First, most of the microorganisms need to synthesize folates de novo, but mammals lack such a pathway and thus acquire folates from the diet; second, folate serves as a major one-carbon donor for multiple vital physiological processes, such as the biosynthesis of thymine, purines, and methionine5. Sulfonamides, the first antimicrobials used by humans to cure bacterial infections, compete with para-aminobenzoic acid (pABA) and thus disrupt bacterial dihydropteroate biosynthesis by blocking the substrate required for dihydropteroate synthase6. Sulfamethoxazole (SMX) is one of the commonly used sulfonamides and is typically used in combination with trimethoprim (TMP), also known as cotrimoxazole (SXT), to treat respiratory and urinary tract infections7,8, or to prevent infection of Pneumocystis carinii in HIV-infected patients9. Afterward, clinical M. tuberculosis isolates were found to be susceptible to SXT regardless of their resistance to first-line anti-TB drugs10. However, other studies showed that SMX was active against M. tuberculosis, but TMP was not11,12,13. In addition, SMX was shown to have a synergistic effect with rifampicin and an additive effect with ethambutol in the treatment of M. tuberculosis isolates12. Recently, several reports showed that prophylaxis of HIV-positive patients with SXT reduced the risk of TB, indicating a potential role for SXT in the treatment of latent M. tuberculosis infection14,15,16,17.
Para-aminosalicylic acid (PAS), another folate antagonist, is the second antimicrobial used for the treatment of TB after introduction of streptomycin (STR)18. The combined usage of PAS and STR laid the foundation for the modern 6- to 9-month TB therapy19. Subsequently, because of gastrointestinal disturbance, PAS was replaced by ethambutol, a more tolerated drug. Although PAS has been used for more than 70 years, its mechanisms of action and resistance are still not fully understood. PAS is a structural analog of pABA, and the addition of pABA antagonizes the activity of PAS20. As a prodrug, PAS needs to be sequentially bio-activated by two enzymes involved in M. tuberculosis folate biosynthesis (dihydropteroate synthase and dihydrofolate synthase), and then the bio-activated metabolite of PAS inhibits dihydrofolate reductase (DHFR) and, eventually, bacterial growth19. Consequently, mutations of folC (the DHFR coding gene) impair PAS bio-activation and result in PAS resistance in M. tuberculosis clinical isolates21,22. Except for mutations in folC, mutations in the thymidylate synthase coding gene thyA23 and the riboflavin biosynthesis-related gene ribD also confer PAS resistance in M. tuberculosis clinical isolates24. Mutations in the above three genes could be identified in about two-thirds of the PAS-resistant clinical isolates25.
Recently, Liu et al. found that deletion of the folP2 gene in Mycobacterium smegmatis, which encodes an ortholog of the dihydropteroate synthase27, resulted in increased susceptibility to SMX26. Recently, Wei et al. found that mutation of folC in M. tuberculosis clinical isolates led to a decreased expression of folP228. We thus speculated that mutation of folC in M. tuberculosis might lead to decreased expression of folP2 and hence affect the susceptibility to SMX. To test this hypothesis, we collected PAS-resistant clinical isolates and determined their susceptibilities to SMX. We constructed lab strains with folC point mutations and folP2 deletion, and expression levels of folP2 were compared between the folC mutants and their parental strain. In addition, pABA production levels were compared between the folC mutants and their parental strain. In the present paper, we report our results.
Results
Characteristics of the selected clinical isolates
The characteristics of the isolates used in this study are summarized in Table 1. A total of 95 strains were selected. One of these is M. tuberculosis H37Rv (ATCC 27294), and the other 94 strains are randomly selected clinical isolates from different patients. Among the 94 clinical isolates, 80 (85.1%) were isolated from sputum. Concerning drug resistance, 68 (72.3%) and 19 (20.2%) isolates were defined as MDR and extensively drug-resistant (XDR), respectively. Isolates from male patients (63, 67%) were twofold more than those from female patients (31, 33%). The dominating age group was 25–44 years old, which accounted for 48 (51.1%) patients from whom M. tuberculosis strains were isolated. The detailed information of all isolates is listed in Table S2.
ThyA, ribD, and folC mutational types in PAS-resistant isolates
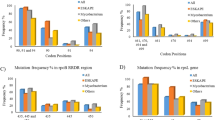
Among the 95 strains selected in this study, five clinical isolates plus H37Rv were sensitive to PAS (< 1 μg/ml), and the remaining 89 (94.7%) clinical isolates were resistant to PAS (> 1 μg/ml) on Löwenstein–Jensen (L–J) solid medium (Table 2). The resistance to PAS on L–J medium was subsequently confirmed by the minimum inhibitory concentration (MIC) method on 7H10 medium (> 2 μg/ml) (Table 3). The thyA, ribD, and folC genes of all 95 isolates were sequenced. No mutations were observed in all three genes in H37Rv, five PAS-sensitive isolates, and two PAS-resistant isolates. The remaining 87 PAS-resistant isolates harbored mutations in at least one of these three genes. Most mutations are nucleotide substitutions, and only one base insertion was found (in folC) (Table 3). For mutations in thyA, nucleotide alterations occurred at positions 64, 127, 290, 329, 399, 440, 505, and 704. Mutation at position 399 (one isolate) resulted in a premature stop codon. The dominating mutation in thyA occurred at position 704 (15 of 23 isolates), changing arginine to proline. Mutations in ribD were uncommon, and only 10 isolates with ribD mutations were identified. Among them, seven isolates had mutations at the putative regulatory zone (nucleotide position − 12), and the remaining three isolates had mutations in the coding region (nucleotide position 745). For mutations in folC, the leading mutation in this gene was at nucleotide position 128 (33 of 53 isolates). Two isolates with mutations at nucleotide position 128 switched T to G, and the other 31 isolates converted T to C (Table 3).
Most PAS-resistant clinical isolates with folC mutations were sensitive to SMX
To test SMX susceptibilities, H37Rv, five PAS-sensitive strains, and two PAS-resistant strains with no mutation in thyA, ribD, and folC were used as control strains. SMX MICs of these eight strains were all determined to be 50 μg/ml (Table 3). PAS-resistant strains with either thyA or ribD mutations were not more sensitive to SMX. On the contrary, among the 53 PAS-resistant strains with folC mutations, 42 (79.2%) were more sensitive to SMX compared with the eight control strains (Tables 2 and 3).
M. tuberculosis H37Ra ΔfolP2, M. tuberculosis H37Ra ΔfolC pMV361::folC(I43T), and M. tuberculosis H37Ra ΔfolC pMV361::folC(I43A) were more sensitive to SMX than the parental strain
SMX susceptibilities were tested in two lab-constructed folC mutants21 and M. tuberculosis H37Ra ΔfolP2. The results showed that, compared with their parental strain, all three mutants were more sensitive to SMX (Table 4). In addition, the results of the killing curve assay showed that the two folC mutants were more susceptible to SMX than their parental strain (Fig. 1).
FolC mutations did not affect the expression of folP2
Results of the quantitative real-time PCR assay showed that no difference in the folP2 expression level could be observed between M. tuberculosis H37Ra and the two folC mutants, which was confirmed by subsequent western blot analysis (Figs. 2 and 3). To determine production levels of pABA in the three different H37Ra strains, an Escherichia coli (E. coli) W3110 ΔpabB mutant was used as previously described29. When E. coli W3110 ΔpabB was cultured in E minimal medium plus culture filtrates from different H37Ra strains, there was obviously less growth when culture filtrates from the two folC mutants were used (Fig. 4), suggesting a decreased production of pABA.
Comparison of the protein levels of FolP2 during the exponential phase in H37Ra, H37Ra folC(I43A), and H37Ra folC(I43T) strains, as indicated by western blot. Experiments were repeated at least three times, and representative results are shown. (a) Total protein (20 μg per lane) was separated by SDS-PAGE and stained by Coomassie brilliant blue. Lane M, the prestained protein marker. Lane 1, purified FolP2 protein. Lanes 2 and 3, total protein of H37Ra. Lanes 4 and 5, total protein of H37Ra folC(I43A). Lanes 6 and 7, total protein of H37Ra folC(I43T). (b) Western blot analysis of total protein immunoblotted with mouse antiserum with anti-FolP2. Lane 1, anti-FolP2 visualizing the purified FolP2 protein. Lanes 2 and 3, anti-FolP2 visualizing the total protein of H37Ra. Lanes 4 and 5, anti-FolP2 visualizing the total protein of H37Ra folC(I43A). Lanes 6 and 7, anti-FolP2 visualizing the total protein of H37Ra folC(I43T).
Comparison of the transcription levels of the gene folP2 during the exponential phase in wild-type H37Ra and H37Ra folC point mutant strains (I43A, I43T) by quantitative real-time PCR. mRNA expression levels were normalized against sigA as an endogenous control. Data are representative of three experiments. Standard deviations are indicated by error bars.
Mutation of folC caused decreased production of pABA. Growth curves of E. coli W3110 ΔpabB were measured in the presence of pABA and culture filtrates from wild-type H37Ra (WT-filtrate) and the two H37Ra folC mutant strains (MT-filtrates). Experiments were performed in triplicate. Standard deviations are indicated by error bars.
Discussion
A huge obstacle in defeating TB is the increasing resistance to scanty anti-TB drugs. This predicament reinforced our interest in exploring the usable medicines in the approved drugs treasure chest. Antifolates, due to their toxicity in folate biosynthesis, which is vital and greatly different between human cells and bacteria, are potentially important choices to treat MDR TB. In fact, after being put on the shelf for several decades, PAS was reintroduced in the 1990s for treating MDR TB30. However, resistance to PAS appeared in clinical M. tuberculosis isolates in the early 2000s. Although SXT has not been tried to treat TB clinically, attempts have been made to make it an option for treating drug-resistant TB31,32,33,34. In this context, better use of antifolates against TB has gained importance, and further in-depth studies on the mechanisms of susceptibility and resistance of antifolates are required.
Very recently, researchers found that in PAS-resistant clinical isolates of M. tuberculosis with folC mutation, protein expression levels of folP2 were significantly lower than in strains with no folC mutation28. Though FolP2 of M. tuberculosis has been predicted to be defective in dihydropteroate synthesis activity27, deletion of the folP2 gene in M. smegmatis resulted in increased susceptibility to SMX26. We also knocked out the folP2 gene in M. tuberculosis H37Ra, and found that the resulting ΔfolP2 mutant was more sensitive to SMX. Therefore, we speculated that decreased expression of folP2 caused by folC mutation might lead to increased susceptibility to SMX.
As expected, most of the PAS-resistant isolates with mutations in the folC gene showed increased susceptibility to SMX. In addition, two lab-constructed folC mutants were also more sensitive to SMX. These data suggest that mutation of folC did lead to increased susceptibility to SMX in M. tuberculosis.
We noted that 11/53 (20.8%) PAS-resistant isolates with folC mutations were not more sensitive to SMX, including isolates with I43A or I43T mutations. All PAS-resistant isolates used in this study were MDR/XDR clinical isolates obtained from different patients, suggesting the complexity of the genetic backgrounds of those isolates. A previous study using transmission electron microscopy showed that the cell walls of MDR and XDR strains were thicker than those of the susceptible M. tuberculosis isolates35, indicating another possible explanation.
However, subsequent quantitative real-time PCR and western blot analysis showed that folC mutations did not affect the expression of folP2, suggesting that the increased susceptibilities of the folC mutants to SMX were not caused by decreased expression of folP2.
With respect to the mechanisms of action, SMX shares one thing with PAS: both drugs compete with pABA. As a result, deficiency in pABA biosynthesis usually leads to increased susceptibility to both drugs29,36. We thus speculated that mutation of folC might lead to decreased production of pABA and hence affect the susceptibility to SMX. Subsequent comparison of pABA production levels between the two lab-constructed folC mutants and their parental strain confirmed this hypothesis.
To the best of our knowledge, this is the first study on the interaction of the two antifolates in the treatment of MDR or XDR TB clinical isolates. We found that the folC mutation in M. tuberculosis leads to decreased production of pABA and hence increases sensitivity to SMX. Since our previous data showed that a small proportion (~ 9%) of the MDR strains had a mutation in folC21, it would be interesting to test the efficacy of SMX or SXT against those MDR strains in vivo. The regulation of folate metabolism is still obscure in M. tuberculosis, and our pioneering observations provide new evidence to guide future research.
Materials and methods
M. tuberculosis clinical isolates and drug susceptibility testing
Clinical samples (Table 1), such as sputum and pleural fluid, were pretreated and cultured in a BACTEC MGIT 960 system (Becton Dickinson, Sparks, MD, USA), according to the manufacturer’s instructions37. Positive cultures were subjected to (i) mycobacterium species identification and (ii) drug susceptibility testing (DST) using the proportion method on L–J solid medium (Encode, Zhuhai, China), where MDR was defined as resistance to at least isoniazid and rifampicin, and XDR was defined as MDR plus resistance to any fluoroquinolones (ofloxacin, levofloxacin or moxifloxacin) and at least one injectable drug (amikacin or capreomycin). Mycobacterium species identification was performed based on (i) sequence polymorphisms in 16S rRNA, hsp65, and rpoB38 and (ii) the results of bacterial growth on L–J solid medium containing 5 μg/ml TCH and 500 μg/ml PNB. MDR and XDR isolates were chosen for preliminary screening of PAS resistance using the same method and medium as described above. For DSTs of PAS or SMX on Middlebrook 7H10 solid medium (Difco, Becton Dickinson, Sparks, MD, USA), the two drugs (purchased from Merck, Darmstadt, Germany) were dissolved in deionized water and dimethyl sulfoxide (Merck) at concentrations of 10 mg/ml and 60 mg/ml, respectively. The drugs were frozen at − 20 °C after sterilization. The critical concentration of PAS was 1 μg/ml on L–J medium and 2 μg/ml on 7H10 medium according to policy guidelines on DST for second-line anti-TB drugs39. Isolates were frozen in 25% glycerol at − 70 °C until use. Our study was conducted in accordance with the Declaration of Helsinki. The institutional review board of Chongqing Public Health Medical Center approved this study and waived the requirement for written informed consent. The institutional review board waived the need for informed consent because all patients’ data were analyzed in anonymity and no additional informed consent was required.
ThyA, ribD, and folC amplification and sequence analysis of PAS-resistant clinical isolates
The entire open reading frame flanking 150 bp of the upstream putative regulatory sequence of each of the three genes was amplified using the following primers: for folC amplification, the forward primer was folC-F (5′-CGGTCAGCAGTATCAACAGCACGGC-3′) and the reverse primer was folC-R (5′-CGCCGCCTGGAAAAGGAGTTGG-3′); for thyA amplification, the forward primer was thyA-F (5′-TGATCTCCCGGAAATGCGCCTGGT-3′) and the reverse primer was thyA-R (5′-GGTTTTCGGCATGGCCTCCGTTGTA-3′); and for ribD amplification, the forward primer was ribD-F (5′-CCGGCAAAAGTCCTGGCACGCCACG-3′) and the reverse primer was ribD-R (5′-GTTCTTGGGTGCGGCGAGCGGTGGT-3′). The primers were designed according to the M. tuberculosis reference sequence (Gene Bank accession number AL123456.3). For thyA amplification, an S1000 Thermal cycler (BIO-RAD, Hercules, CA, USA) was used with the following PCR program: denaturation at 98 °C for 10 min, followed by thirty cycles of denaturation at 98 °C for 15 s, annealing at 63.2 °C for 15 s, and extension at 72 °C for 30 s, and a final extension at 72 °C for 5 min. Amplification of ribD and folC was carried out following the same protocol, except the annealing temperatures were 66.5 °C for ribD and 64.3 °C for folC.
Agarose gel electrophoresis was performed, and the DNA was purified with an EZNA bacterial DNA kit (Omega Bio-Tek, Norcross, GA, USA) according to the manufacturer’s instructions. The purified DNA products were sequenced in an automatic DNA sequencer (model 3730XL, ABI, Foster City, CA, USA) with the same primers used for the PCR amplifications. Sequencing results were compared with reference sequences using nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and polymorphisms were depicted according to Sequence Variant Nomenclature (http://varnomen.hgvs.org/).
Construction of H37Ra ΔfolP2
A modified strategy for specialized transduction was used to construct the M. tuberculosis H37Ra ΔfolP2 mutant40. Genomic regions flanking folP2, 824 bp upstream (a region containing MRA_1215) and 827 bp downstream (a region containing MRA_1217), were amplified by PCR. The primers used for the amplification of the upstream region of folP2 were folP2koLFP and folP2koLRP, and those for the downstream region were folP2koRFP and folP2koRRP. The recombinant plasmid p0004s-L + R was constructed by inserting the Van91I-digested PCR products into Van91I-digested plasmid p0004s. Then, p0004s-L + R was digested with PacI and ligated into the PacI-digested shuttle phasmid vector phAE159. After ligation, the recombinant cosmid phAE159-p0004s-L + R was transduced into E. coli HB101 in an in vitro λ-packaging reaction (MaxPlax Lambda Packaging Extracts, Epicentre Biotechnologies, Madison, WI, USA). The phasmid DNA prepared from confirmed selected hygromycin-resistant transductant was electroporated into M. smegmatis mc2155 to generate the specialized transducing phage. As described in a previous study40, the transducing phage at the most efficient titer was used to infect H37Ra at a multiplicity of infection of 10. Successful transduction of H37Ra was confirmed by comparing the size of the PCR-amplified products of hygromycin-resistant colonies with the wild-type H37Ra using primers folP2LYZ and folP2RYZ (Table S1).
Drug susceptibility testing of M. tuberculosis H37Ra ΔfolP2, M. tuberculosis H37Ra ΔfolC pMV361::folC(I43T), and M. tuberculosis H37Ra ΔfolC pMV361::folC(I43A)
DST was performed as previously described41. Briefly, M. tuberculosis H37Ra ΔfolP2, M. tuberculosis H37Ra ΔfolC pMV361::folC(I43T), and M. tuberculosis H37Ra ΔfolC pMV361::folC(I43A) were grown at 37 °C in Middlebrook 7H9 medium plus 10% oleic acid–albumin–dextrose–catalase (OADC) to the mid-log phase (OD600 0.4–0.8). Bacterial cultures were centrifuged, and cell pellets were collected and washed twice with 7H9. After tenfold serial dilution, 10 μL of diluted bacterial cells (approximately 105 CFU/ml) were plated on Middlebrook 7H10 medium plus 10% OADC containing various concentrations of PAS (0, 0.0025, 0.005, 0.01, 0.02, 0.04, 0.08, 0.16, 0.32, 0.64, 1.28, 2.56, 5.12, and 10.24 μg/ml) and SMX (0, 0.78125, 1.5625, 3.125, 6.25, 12.5, 25, 50, 100, and 200 μg/ml). The plates were incubated at 37 °C for 3 weeks to determine the MIC values. The MIC was defined as the lowest required concentration of antibiotics to inhibit the growth of 99% of the bacterial CFUs.
Quantitative real-time PCR assays
An RNeasy Mini kit (Qiagen, Germany) was used to extract total RNA, and a ReverTra Ace qPCR kit (TOYOBO, Osaka, Japan) was used to synthesize cDNA. All kits were used according to the manufacturers’ instructions. Gene expression levels were quantified using quantitative real-time PCR analysis on a 7900 HT Sequence Detection System (ABI, Foster City, CA, USA) with ABI Power SYBR Green PCR Master Mix (ABI). mRNA expression levels were normalized to sigA as an endogenous control. Primers for folP2 amplification (folP2-qRT-L and folP2-qRT-R) were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China) (Table S1).
Purification of recombinant histidine-tagged FolP2
The folP2 gene of M. tuberculosis H37Ra was amplified using primers M folP2-L and M folP2-R (Table S1) and subsequently cloned into pET28a to obtain the pET28a::folP2 recombinant plasmid. The recombinant plasmid was verified and transformed into E. coli BL21 (DE3). The recombinant strain E. coli BL21 pET28a::folP2 was grown at 37 °C in liquid LB medium to the mid-log phase (OD600 0.4–0.8), 0.2 mM isopropyl-β-D-thiogalactopyranoside was added, and the cells were incubated at 16 °C for another 20 h. After induction, the bacterial cells were centrifuged and resuspended in lysis buffer [50 mM Tris–HCl, 500 mM NaCl, and 20 mM imidazole (pH 8.0)]. Bacterial cell suspensions were lysed by ultrasonication and centrifuged. Supernatants were collected and mixed with prewashed nickel-nitrilotriacetic acid HisTrap HP affinity resin (GE Healthcare, USA) at 4 °C overnight for target protein binding, and non-specific binding proteins were washed away with wash buffer (50 mM Tris–HCl, 0.5 M NaCl, and 60 mM imidazole [pH 8.0]). The affinity resins were washed with elution buffer (50 mM Tris–HCl, 0.5 M NaCl, and 250 mM imidazole [pH 8.0]) to collect recombinant histidine-tagged FolP2. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was used to verify the target protein.
Antibody preparation of FolP2
Antibody preparation of FolP2 protein of M. tuberculosis H37Ra was performed as previously described29.
Western blot analysis
Western blot analysis was performed as previously described29, except that in this case antiserum against the recombinant FolP2 was used.
Measurement of pABA production levels in different M. tuberculosis H37Ra strains using the E. coli W3110 ΔpabB mutant
M. tuberculosis H37Ra, M. tuberculosis H37Ra ΔfolC pMV361::folC(I43T), and M. tuberculosis H37Ra ΔfolC pMV361::folC(I43A) were cultured in Middlebrook 7H9 with OADC to log-phase and culture filtrates were collected with 0.22 μm filters (Millipore, Merck). Measurement of pABA production levels in these three strains was performed as previously described29.
Data availability
All data generated or analyzed during this study are included in this article and its Supplementary Information files.
References
Spekker, O. et al. Tracking down the White Plague: The skeletal evidence of tuberculous meningitis in the Robert J. Terry Anatomical Skeletal Collection. PLoS ONE 15, e0230418. https://doi.org/10.1371/journal.pone.0230418 (2020).
World Health Organization (WHO). Global Tuberculosis Report 2019 (WHO, Geneva, 2019).
Islam, M. M. et al. Drug resistance mechanisms and novel drug targets for tuberculosis therapy. J. Genet. 44, 21–37. https://doi.org/10.1016/j.jgg.2016.10.002 (2017).
Hameed, H. M. A. et al. Molecular targets related drug resistance mechanisms in MDR-, XDR-, and TDR-Mycobacteriumtuberculosis strains. Front. Cell. Infect. Microbiol. 8, 114. https://doi.org/10.3389/fcimb.2018.00114 (2018).
Bermingham, A. & Derrick, J. P. The folic acid biosynthesis pathway in bacteria: Evaluation of potential for antibacterial drug discovery. BioEssays 24, 637–648 (2002).
Minato, Y. et al. Mutual potentiation drives synergy between trimethoprim and sulfamethoxazole. Nat. Commun. 9, 1003. https://doi.org/10.1038/s41467-018-03447-x (2018).
Hughes, W. T., McNABB, P. C., Makres, T. D. & Feldman, S. Efficacy of trimethoprim and sulfamethoxazole in the prevention and treatment of Pneumocystiscarinii pneumonitis. Antimicrob. Agents Chemother. 5, 289–293 (1974).
Laursen, H. & Nielsen, M. L. Urinary tract pathogens-sensitivity to trimethoprim/sulphamethoxazole. Scand. J. Infect. Dis. 2, 205–209. https://doi.org/10.3109/inf.1970.2.issue-3.09 (1970).
Smilack, J. D. Trimethoprim-sulfamethoxazole. Mayo Clin. Proc. 74, 730–734. https://doi.org/10.4065/74.7.730 (1999).
Forgacs, P. et al. Tuberculosis and trimethoprim–sulfamethoxazole. Antimicrob. Agents Chemother. 53, 4789–4793. https://doi.org/10.1128/AAC.01658-08 (2009).
Huang, T. S. et al. Susceptibility of Mycobacteriumtuberculosis to sulfamethoxazole, trimethoprim and their combination over a 12 year period in Taiwan. J. Antimicrob. Chemother. 67, 633–637. https://doi.org/10.1093/jac/dkr501 (2012).
Macingwana, L. et al. Sulfamethoxazole enhances the antimycobacterial activity of rifampicin. J. Antimicrob. Chemother. 67, 2908–2911. https://doi.org/10.1093/jac/dks306 (2012).
Vilcheze, C. & Jacobs, W. R. Jr. The combination of sulfamethoxazole, trimethoprim, and isoniazid or rifampin is bactericidal and prevents the emergence of drug resistance in Mycobacteriumtuberculosis. Antimicrob. Agents Chemother. 56, 5142–5148. https://doi.org/10.1128/aac.00832-12 (2012).
Nunn, A. J. et al. Role of co-trimoxazole prophylaxis in reducing mortality in HIV infected adults being treated for tuberculosis: Randomised clinical trial. BMJ (Clin. Res. Ed.) 337, a257. https://doi.org/10.1136/bmj.a257 (2008).
Hasse, B. et al. Co-trimoxazole prophylaxis is associated with reduced risk of incident tuberculosis in participants in the Swiss HIV Cohort Study. Antimicrob. Agents Chemother. 58, 2363–2368. https://doi.org/10.1128/aac.01868-13 (2014).
Hakim, J. et al. Enhanced prophylaxis plus antiretroviral therapy for advanced HIV infection in Africa. N. Engl. J. Med. 377, 233–245. https://doi.org/10.1056/NEJMoa1615822 (2017).
Ku, S. W. et al. Cotrimoxazole prophylaxis decreases tuberculosis risk among Asian patients with HIV. J. Int. AIDS Soc. 22, e25264. https://doi.org/10.1002/jia2.25264 (2019).
Lehmann, J. Para-aminosalicylic acid in the treatment of tuberculosis. Lancet 247, 15–16 (1946).
Chakraborty, S., Gruber, T., Barry, C. E., Boshoff, H. I. & Rhee, K. Y. Para-aminosalicylic acid acts as an alternative substrate of folate metabolism in Mycobacteriumtuberculosis. Science 339, 88–91. https://doi.org/10.1126/science.1228980 (2013).
Youmans, G. P., Raleigh, G. W. & Youmans, A. S. The tuberculostatic action of para-aminosalicylic acid. J. Bacteriol. 54, 409 (1947).
Zhao, F. et al. binding pocket alterations in dihydrofolate synthase confer resistance to para-aminosalicylic acid in clinical isolates of Mycobacteriumtuberculosis. Antimicrob. Agents Chemother. 58, 1479–1487. https://doi.org/10.1128/AAC.01775-13 (2014).
Zheng, J. et al. Para-aminosalicylic acid is a prodrug targeting dihydrofolate reductase in Mycobacteriumtuberculosis. J. Biol. Chem. 288, 23447–23456 (2013).
Rengarajan, J. et al. The folate pathway is a target for resistance to the drug para-aminosalicylic acid (PAS) in mycobacteria. Mol. Microbiol. 53, 275–282 (2004).
Minato, Y. et al. Mycobacteriumtuberculosis folate metabolism and the mechanistic basis for para-aminosalicylic acid susceptibility and resistance. Antimicrob. Agents Chemother. 59, 5097–5106. https://doi.org/10.1128/aac.00647-15 (2015).
Zhang, X. et al. Genetic determinants involved in p-aminosalicylic acid resistance in clinical isolates from tuberculosis patients in northern China from 2006 to 2012. Antimicrob. Agents Chemother. 59, 1320–1324. https://doi.org/10.1128/aac.03695-14 (2015).
Liu, T. et al. Role of folP1 and folP2 genes in the action of sulfamethoxazole and trimethoprim against mycobacteria. J. Microbiol. Biotechnol. 25, 1559–1567. https://doi.org/10.4014/jmb.1503.03053 (2015).
Gengenbacher, M., Xu, T., Niyomrattanakit, P., Spraggon, G. & Dick, T. Biochemical and structural characterization of the putative dihydropteroate synthase ortholog Rv1207 of Mycobacteriumtuberculosis. FEMS Microbiol. Lett. 287, 128–135. https://doi.org/10.1111/j.1574-6968.2008.01302.x (2008).
Wei, W. et al. Multi-omics comparisons of p-aminosalicylic acid (PAS) resistance in folC mutated and un-mutated Mycobacteriumtuberculosis strains. Emerg. Microbes Infect. 8, 248–261. https://doi.org/10.1080/22221751.2019.1568179 (2019).
Yang, S. S. et al. Deletion of sigB causes increased sensitivity to para-aminosalicylic acid and sulfamethoxazole in Mycobacteriumtuberculosis. Agents Chemother. 61, 1. https://doi.org/10.1128/aac.00551-17 (2017).
Peloquin, C. A. et al. Pharmacokinetic evaluation of para-aminosalicylic acid granules. Pharmacotherapy 14, 40–46. https://doi.org/10.1002/j.1875-9114.1994.tb02787.x (1994).
Palomino, J. C. & Martin, A. The potential role of trimethoprim–sulfamethoxazole in the treatment of drug-resistant tuberculosis. Future Microbiol. 11, 539–547. https://doi.org/10.2217/fmb.16.2 (2016).
Alsaad, N. et al. Pharmacokinetic evaluation of sulfamethoxazole at 800 milligrams once daily in the treatment of tuberculosis. Antimicrob. Agents Chemother. 60, 3942–3947. https://doi.org/10.1128/aac.02175-15 (2016).
Davies Forsman, L. et al. Intra- and extracellular activities of trimethoprim–sulfamethoxazole against susceptible and multidrug-resistant Mycobacteriumtuberculosis. Antimicrob. Agents Chemother. 58, 7557–7559. https://doi.org/10.1128/aac.02995-14 (2014).
Alsaad, N. et al. Evaluation of co-trimoxazole in the treatment of multidrug-resistant tuberculosis. Eur. Respir. J. 42, 504–512. https://doi.org/10.1183/09031936.00114812 (2013).
Velayati, A. A. et al. Differences in cell wall thickness between resistant and nonresistant strains of Mycobacteriumtuberculosis: Using transmission electron microscopy. Chemotherapy 55, 303–307. https://doi.org/10.1159/000226425 (2009).
Thiede, J. M. et al. Targeting intracellular p-aminobenzoic acid production potentiates the anti-tubercular action of antifolates. Sci. Rep. U.K. https://doi.org/10.1038/Srep38083 (2016).
Kruuner, A., Yates, M. D. & Drobniewski, F. A. Evaluation of MGIT 960-based antimicrobial testing and determination of critical concentrations of first- and second-line antimicrobial drugs with drug-resistant clinical strains of Mycobacteriumtuberculosis. J. Clin. Microbiol. 44, 811–818. https://doi.org/10.1128/jcm.44.3.811-818.2006 (2006).
Kim, S. H. & Shin, J. H. Identification of nontuberculous mycobacteria using multilocous sequence analysis of 16S rRNA, hsp65, and rpoB. J. Clin. Lab. Anal. https://doi.org/10.1002/jcla.22184 (2018).
World Health Organization (WHO). Policy Guidance on Drug-Susceptibility Testing (DST) of Second-Line Antituberculosis Drugs (WHO, Geneva, 2008).
Bardarov, S. et al. Specialized transduction: An efficient method for generating marked and unmarked targeted gene disruptions in Mycobacteriumtuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148, 3007 (2002).
Li, K. et al. Deletion of nudB causes increased susceptibility to antifolates in Escherichiacoli and Salmonellaenterica. Antimicrob Agents Chemother https://doi.org/10.1128/AAC.02378-16 (2017).
Acknowledgements
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB29020000), the National Science and Technology Major Project of China during the 13th Five-Year Plan period (Grant No. 2018ZX10302104), the Natural Science Foundation of Chongqing (Grant No. cstc2018jcyjAX0428), and the Joint Medical Research Project of the Science and Technology Bureau of Chongqing and the Health Commission of Chongqing (Grant No. 2018QNXM020).
Author information
Authors and Affiliations
Contributions
J.Y.D. and Y.K.C. designed the research; R.Q.W., K.L., and J.F.Y. performed the experiments with technical support from J.Y.D.; J.Y.D. analyzed the data and wrote the manuscript; all authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, R., Li, K., Yu, J. et al. Mutations of folC cause increased susceptibility to sulfamethoxazole in Mycobacterium tuberculosis. Sci Rep 11, 1352 (2021). https://doi.org/10.1038/s41598-020-80213-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-80213-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.