Abstract
Persistent HPV infection associated with immune modulation may result in high-grade squamous intraepithelial lesions (CIN)2/3. Currently, there is little information on the cervicovaginal microbiome, local cytokine levels and HPV infection related to CIN. Follow-up of patients after local surgery provides an opportunity to monitor changes in the cervicovaginal environment. Accordingly, we undertook this longitudinal retrospective study to determine associations between HPV genotypes, cervicovaginal microbiome and local cytokine profiles in 41 Japanese patients with CIN. Cervicovaginal microbiota were identified using universal 16S rRNA gene (rDNA) bacterial primers for the V3/4 region by PCR of genomic DNA, followed by MiSeq sequencing. We found that Atopobium vaginae was significantly decreased (p < 0.047), whereas A. ureaplasma (p < 0.022) increased after surgery. Cytokine levels in cervical mucus were measured by multiplexed bead-based immunoassays, revealing that IL-1β (p < 0.006), TNF-α (p < 0.004), MIP-1α (p < 0.045) and eotaxin (p < 0.003) were significantly decreased after surgery. Notably, the level of eotaxin decreased in parallel with HPV clearance after surgery (p < 0.028). Thus, local surgery affected the cervicovaginal microbiome, status of HPV infection and immune response. Changes to the cervicovaginal microbiota and cervical cytokine profile following surgery for cervical intraepithelial neoplasia may be important for understanding the pathogenesis of CIN in future.
Similar content being viewed by others
Introduction
Infection of the cervix by human papillomaviruses (HPVs) can cause cervical intraepithelial neoplasia (CIN). Transient HPV infections cause low-grade squamous intraepithelial lesions (CIN1), whereas persistent HPV infection presumably associated with accumulating genetic changes and immune modulation can result in high-grade squamous intraepithelial lesions (CIN2–3) or cervical cancer. Although some viral infections elicit strong host immune responses and are subsequently eliminated by the host, HPV effectively evades such immune rejection, allowing the establishment of persistent viral infection1.
The microbiome has a variety of functions including regulation of the immune system and metabolism of the host. The potential role of commensal cervicovaginal bacteria in modulating immune responses is largely unknown at the present time2. However, there is emerging evidence suggesting that the cervicovaginal microbiota is important for HPV persistence and eventually for the development of premalignant lesions3. Thus, we aimed to identify relationships between local host immune responses, the cervicovaginal microbiota and HPV infection in patients with CIN. However, there was a great deal of heterogeneity in the cervicovaginal microbiota among patients with CIN, possibly associated with confounding factors including smoking, number of sexual partners, sexual behavior, food intake, oral contraceptive use, and other factors. It was thus difficult to compare the relationships between HPV infections, microbiota and local host immune response in the cohort of patients as a whole, but monitoring the same individual patients with CIN over time after surgery avoid this problem of heterogeneity.
Laser cone resection, diathermy and Loop Electrosurgical Excision Procedure (LEEP) are commonly-applied local surgical interventions for CIN in developed countries. Surgery may change not only the microbial diversity4 but also the cervicovaginal environment, including immune responses5 and the status of HPV infection6. Thus, we undertook this retrospective longitudinal study to elucidate associations between cervicovaginal microbiota, HPV infection and cytokine profiles in individual premenopausal women with CIN before and after surgery.
Results
Characteristics of patients with CIN who received surgery and those under observation only
We compared the cervicovaginal microbiota in women with CIN before and after surgery to assess whether there were any correlations between bacterial composition and status of HPV infection (Figure S1). Demographic features of the enrolled patients with CIN treated by surgery or the group under observation without surgery are provided in Table 1. There were no significant differences between the two groups in multiple factors including parity and smoking. The number of different HPV genotypes was significantly decreased after surgery (p = 0.000), but there was no significant change over the same time period in the observation group (p = 0.414). In Table S1, it can be seen that 24 patients (85.7%) converted to cytology-negative (NILM) after surgery, and that 16 of 28 (57.1%) with HPV infections tested negative after surgery. The number of patients with multiple infections decreased from nine to two. In contrast, the number of patients with multiple infections did not change in the observation group.
Characteristics of the cervicovaginal microbiota from patients with CIN
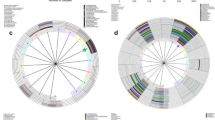
Cervicovaginal microbiota were investigated using PCR on extracted genomic DNA with universal 16S rRNA gene (rDNA) bacterial primers for the V3/4 region followed by MiSeq sequencing. In total 2,968,275 reads were obtained from 82 specimens with an average number of reads per specimen of 36,198.5 and an average of 18.7 operational taxonomic units (OTUs) per specimen, as shown in Table 2. A total of 147 taxa was found in these specimens. At the phylum level, Firmicutes (65.1%), Actinobacteria (23.7%), Bacteroidetes (6.6%) and Fusobacteria (2.2%) were the most dominant. At the genus level, a total of 27 genera were present at an abundance > 0.1%; of these, Lactobacillus (56.1%), Gardnerella (11.3%), Bifidobacterium (5.9%), Atopobium (5.6%), and Prevotella (4.6%) were the dominant taxa according to the previous classification7. The relative abundance of the representative microbiota in the first collection and the second is depicted in Fig. 1. The most dominant species was L. iners in 63 specimens (76.8%). At the genus level, Lactobacillus and Gardnerella were the most abundant in both the first and second collections (Fig. 1, Table 2).
Heatmap of the relative abundance of the representative microbiota at first and second collections. Cervicovaginal microbiota were collected from 41 patients with CIN and identified by 16S rRNA V3/4 sequencing. First collection included before surgery and observation 1 described in figure. Color gradation indicates the relative abundance of microbiota. The scale of color gradation is indicated at the bottom. The number of microbial species qualified by the observed species richness (Sobs) is indicated as α-diversity in each specimen. Heatmap was drawn by Microsoft Excel.
Relationships between the microbiota in the patients
There was an inverse relationship between the presence of L. cirspatus and anaerobes including Dialister, Atopobium vaginae, Adlercreutzia, Parimonas and Clostridium throughout the first and second collections, as assessed by QIIME2.0 and shown in Fig. 2. In contrast, the presence of anaerobic bacteria including Prevotella, Dialister, Atopobium vaginae, Sneathia, Adlercreutzia, Peptoniphilus, Megashpaera, Parvimonas and Clostridium were positively correlated with each other, with no difference after surgery relative to before surgery. There was a strong correlation between L. crispatus and L. jensenii in the first collection and after surgery as determined by SpeciateIT.
Symbiotic relationship among microbiota. Spearman’s rank correlation for multiple comparisons was estimated for each taxon as the relative abundance of each symbiont. Color and shade indicate the extent of positive and negative correlation. Dark yellow blocks indicate strong positive correlations (correlation coefficient 0.6–1.0). Pale yellow blocks indicate weak positive correlations (correlation coefficient 0.2–0.6). Pale blue indicates weak negative correlations (correlation coefficient − 0.2 to − 0.6) and dark blue strong negative correlations (correlation coefficient − 0.6 to − 1.0). Correlations were examined by QIIME2.0 (a–c). Lactobacillus species correlations were examined by SpeciateIT (d–f). Correlation significance: *p = 0.01–0.05, **p = 0.001–0.01, ***p < 0.001. Correlation tables were determined using Excel.
Changes in the relative abundance of microbial phyla after surgery
We compared the relative abundance of microbial phyla from patients after surgery with patients in the observation-only group. Proteobacteria were significantly decreased whereas Tenericutes were increased after surgery, as shown in Table 3 and Figure S2. There was no change over time in the observation group. At the genus level, Atopobium vaginae and Methylobacteriaceae were significantly decreased, whereas Ureaplasma increased after surgery.
Correlation between microbiota and cytokines in the first collection specimen
Levels of proinflammatory cytokines including IL-1β and TNF-α were significantly increased with the presence of anaerobics microbiota, whereas inversely correlated with Lactobacillus in Table 4. Levels of TNF-α, IL-10 and RANTES were inversely correlated with L. crispatus.
Changed cytokine profile after surgery
The levels of IL-1β, TNF-α, MIP-1α and eotaxin were significantly decreased after surgery but not in patients without surgery over the same time period (Table 5). Because the number of HPV genotypes detected was significantly decreased after surgery, we examined the association between the HPV infection status and cytokine profiles (Table 6). We focused on cytokine levels in patients who were positive for 7 or 13 high risk HPV genotypes before surgery but who were negative after surgery. We found that the level of eotaxin was significantly decreased in parallel with HPV negativity after surgery.
Discussion
Local interplay between the microbiome and the immune response may be important for understanding the pathogenesis of the sequence of events from HPV infection, CIN development and progression to cervical cancer8. As one approach to investigating this issue, we examined how removal of the neoplastic lesions by surgery impacted on associations between microbiome diversity, local immune responses and the number of HPV genotypes in patients with CIN. This showed that Atopobium was significantly decreased after surgery (Table 3 and Figure S2), in parallel with the decreased number of HPV genotypes detected after surgery. Atopobium was positively correlated with the presence of anaerobic bacteria (Fig. 2) and with HPV persistence9. Both Atopobium and Gardnerella were associated with CIN10. Decreased HPV infections and removal of neoplastic lesions might be associated with decreased diversity of microbiota10,11,12,13. Thus, the presence of CIN lesions could contribute to the maintenance of microbiome diversity3.
Ureaplasma was found in the vagina or cervix of 40–80% of premenopausal asymptomatic women14 but were also reported to be associated with CIN15. Our data showed that Ureaplasma increased after surgery. One explanation for this could be that surgical intervention increases the opportunity for Ureaplasma growth or that the environment before surgery was not appropriate for this species due to competition from other microbes. Prevotella may provide nutrients such as ammonia and amino acids to other members of the microbial community such as Gardnerella and Peptostreptococcus16 and assume a role as the hub for vaginal microbiota17. Indeed, there was a positive correlation between the presence of Prevotella and other anaerobics regardless of surgery. Prevotella could therefore be critical for maintenance of a dysbiome in the vagina (Fig. 2).
Using QIIME2.0, we separated Lactobacillus into L. crispatus, L. iners and unclassified L. spp and employed SpeciateIT for species of Lactobacillus classification. L. crispatus is the most common vaginal H2O2-producing Lactobacillus species, followed by L. jensenii, whereas L. iners does not produce H2O2 (which has been associated with increased risk of abnormal vaginal microbiota)18. In vitro experiments demonstrated that L. iners and Gardnerella disrupt the cervical epithelial barrier by regulating adherens junction proteins, cervical immune responses and miRNA expression, whereas L. crispatus has a protective effect19. In our analysis, the presence of L. crispatus was positively correlated with L. jensenii, and L. crispatus and others including L. jensenii were negatively correlated with anaerobics (Fig. 2).
High levels of proinflammatory cytokines were associated with the presence of anaerobic bacteria and inversely correlated with the presence of Lactobacillus (Table 4). High levels of multiple proinflammatory cytokines were strongly associated with highly diverse bacterial communities in patients, suggesting that specific genital bacteria induced a robust local immune response2. Atopobium was reported to induce strong expression of IL-1β and TNF-α in cultured cells20. In other studies, high levels of IL-1β were associated with bacterial vaginosis21 or cervical dysplasia22. In contrast, TNF-α, IL-10, and RANTES were down-regulated in bacterial vaginosis in patients where L. crispatus dominated. The level of TNF-α was decreased post-LEEP compared with patients without LEEP5. In addition to TNF-α, we found that IL-1β, MIP-1α and eotaxin were decreased after surgery (Table 5). MIP-1α was reported to be a biomarker for precursor lesions in cervical cancer22,23,24. HPV clearance after surgery was inversely correlated with the level of eotaxin. This is consistent with a report that eotaxin was measurable in cultured cervical cells with integrated HPV16/18 genomes25. Marks observed high levels of eotaxin in the cervical mucus of patients with HPV infections26. One possible explanation for this finding is that precursor lesions infected with HPVs produce eotaxin. High expression of IFN-γ was correlated with L. crispatus for unknown reasons. However, the cause of the high expression of proinflammatory cytokines remains controversial because cervical neoplasia, bacterial vaginosis and HPV infections are known to be factors influencing each other.
The present study was rigorous in the diagnosis, recruitment, treatment and management of patients, and specimens were all taken by a single colposcopist. Clinical data including histology, cytology and HPV genotypes were precisely recorded (Table S1). Yin reported on the effects of surgery on TNF-α expression, as a biomarker of inflammation27. The level of TNF-α was increased one month after surgery, and decreased again thereafter. It takes approximately 6 months to heal the wound of the LEEP surgery5. The patients received limited oral antibiotics and anti-inflammatory drugs for two days after surgery. To avoid interference caused by the surgery per se, specimens were taken at a mean of 280 days, and the interval of IQR was 129–364 days, as shown in Table 1. Therefore, we believe that inflammation or drugs did not affect the microbial environments and cytokine profiles. The rate of residual HPV DNA 7–9 months after conization was < 10% as reported by Kim et al6. Therefore, specimens were taken 7–9 months after surgery in this aspect. Of note, some patients remained HPV-positive whatever neoplastic lesion was removed (Table S1). It is therefore necessary to be aware of the issue of late recurrence. The etiology of multiple infection in terms of cervical carcinogenesis is unknown. Multiple infections result from many different factors including an immunocompromised state or increased chances of infection from multiple sex partners. Multiple infections are often observed in LSIL, but thereafter monoclonal cells infected with a certain high-risk HPV genotype grow rapidly, whereas cells infected with other HPV genotypes decrease, possibly as a result of immune responses. Consequently, infection with a single strain is usually observed in HSIL or cancer. This is likely the reason why the number of different HPV genotypes was decreased or reduced to zero by surgery. The elimination or diminished HPV genotypes after surgery is possibly due to the resection of the infected lesion. However, HPV infections might be present beyond the surgical area. Whether or not these are eliminated would depend on the individual immune response.
Pre- or post-menopausal status is a critical factor influencing diversity of the cervicomicrobiome28. Young healthy women had dominant L. crispatus or L. iners communities29, whereas postmenopausal women had a paucity of Lactobacillus and dominant Streptococcus, Prevotella17 and Atopobium28. Cervical mucus is more abundant in young women; the frequency of CIN peaks in women in their 30’s. LEEP, diathermy and laser cone treatments are the most appropriate options for fertility-sparing surgery in women of childbearing age. Taking these factors together, we enrolled premenopausal women, mainly in their 30′s in order to exclude an age-associated effect on the reduction of cervical mucus. Ravel reported that there are differences in the vaginal microbiome according to race29. Human immunodeficiency virus-infected individuals represent a unique cohort of patients with HPV infections at increased risk of developing cervical cancer. We therefore recruited immunocompetent Japanese patients for the present study. We also fixed the anatomical site of the sampling lesion and the sampling devices because of differences resulting from using different methods30,31.
There are some limitations to this study. We showed sequencing results in Table 2, but these could be different if the target region of 16s rRNA genes analyzed or methods themselves were different32. There may also be some selection bias for the enrolled patients. Patients with CIN2 and tiny lesions were apt to be assigned to observation only, whereas patients with CIN3 and larger lesions were assigned to surgery. However, there was in fact little difference between them, because the HSIL category in pathology includes both CIN2 and CIN3. Another limitation is the lack of adjustment for risk factors and possible confounders between groups. Tobacco smoking is a risk factor for bacterial vaginosis, and Peptostreptococcus and Veillonella are associated with smoking33,34. Smoking is associated with a lower proportion of Lactobacillus than observed in non-smokers35. However, there was no difference between the surgery and observation groups regarding smoking (Table 1). The effect of smoking was not seen here, possibly due to the small number of patients. A further limitation is that the time course of sample taking was limited. Brotman reported an average of 29 samples per participant to examine the association between HPV infections and the vaginal microbiome12. Fluctuations over time of the cervicovaginal microbiota from the same individual were observed36. We did not determine the status of bacterial vaginosis including pH and Nugent Score. We have no patients with recurrence nor progression of CIN. Therefore, we have not examined the association between CIN pathogenesis and vaginal environment possibly due to the short observation period. A larger cohort and multiple longitudinal clinical studies will be needed to expand and validate our findings. Moreover, mechanistic studies using in vitro and in vivo models will be required to fully understand the complex relationships among the cervicomicrobiome, HPV infections, local immune responses and regression-vs-persistence-vs-progression of neoplastic disease of the cervix.
In summary, we have undertaken a longitudinal retrospective study to determine associations between the cervicovaginal microbiota, HPV infections and cytokine profiles in premenopausal patients with CIN, comparing patients receiving surgery with those under observation only. The dominant microbiota in Japanese premenopausal patients with CIN was L. iners, the abundance of which was unchanged by surgery. There was an inverse relationship between L. crispatus and the presence of anaerobic bacteria. At the genus level, Atopobium vaginae was significantly decreased, whereas Ureaplasma increased after surgery. We found that high levels of proinflammatory cytokines including IL-1β and TNF-α were significantly increased in parallel with the presence of anaerobics, and inversely correlated with Lactobacillus dominance. Levels of IL-1β, TNF-α, MIP-1α and eotaxin were significantly decreased after surgery. Of note, the expression of eotaxin in parallel with HPV clearance after surgery was significantly decreased. In conclusion, we found that surgical intervention dramatically changed the cervicovaginal microbiome and local immune responses. We could find the association among microbiota, HPV and local immune response with the recurrence or progression of CIN in future.
Materials and methods
Study subjects
Specimens were collected from 41 Japanese patients with CIN, aged 24–48 years (median 35), who attended the outpatient clinic at Fujita Health University Hospital, Aichi Prefecture, Japan, for routine gynecological examinations from April 2016 to March 2019. Histology results were classified according to the Richart classification. Cytological interpretation was classified according to the Bethesda 2001 system.
The patients were divided into a group requiring surgery (n = 28) and observation only (n = 13) according to the clinical decision-making process (Figure S1). Of the former, five underwent laser cone resection and 23 LEEP with diathermy. First collection of specimens designated “before surgery” or “observation 1” was from all patients. The second collection designated “after surgery” or “observation 2” was taken from each group. At first collection, disease as classified by histology into chronic cervicitis (n = 1), five CIN1, 23 CIN2 and 12 CIN3. Cytology results were classified as negative for intraepithelial lesions or malignancy (NILM) (n = 1), five atypical squamous cells of undetermined significance (ASC-US), eight low-grade squamous intraepithelial lesions (LSIL), 24 high-grade squamous intraepithelial lesions (HSIL), and three atypical squamous cells, cannot exclude high-grade squamous intraepithelial lesions (ASC-H). In the “after surgery” group, there were 24 NILM, one ASC-US, one LSIL, and two HSIL. In the “observation 2” group, histological classification was chronic cervicitis (n = 1), three CIN1, three CIN2, five CIN3 and one not determined. Cytology results were one NILM, one LSIL, and two HSIL.
We excluded patients who (a) were younger than 20 years or older than 49 years; (b) were pregnant or postmenopausal; (c) had undergone previous treatment with chemotherapy, radiation, or surgery for CIN; (d) had cancer; or (e) took medication for sexually transmitted diseases. Specimens taken during menstrual period were excluded. The study protocol was approved by the Ethics Committees of Fujita Health University and the National Institute of Infectious Diseases. Written informed consent was obtained from each patient. All the methods were performed in accordance with the relevant guidelines and regulations. The anatomy site of the sample taking was fixed. Cervical mucus specimens were collected using BD BBL Culture Swab (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) for microbial analysis and using Merocel cervical sponges (Medtronic Xomed, Inc., Jacksonville, FL, USA) for cytokine analysis and stored at − 80 °C. The cervical brush was inserted into the cervical canal to collect ectocervical and endocervical cells for HPV genotypes and stored at − 80 °C.
HPV genotyping
For HPV genotyping assays, total DNA was extracted with the QIAamp DNA Mini Kit (QIAGEN GmbH, Hilden, Germany). Each cytobrush was soaked in 400 μl of phosphate-buffered saline, according to the manufacturer’s protocol for swabs. The extracted DNA was eluted with 120 μl Buffer AE. Quality of extracted DNA was determined spectrophotometrically using the NanoDrop ND-1000 (NanoDrop Technologies Inc., Wilmington, USA). HPV genotyping assays were performed by polymerase chain reaction (PCR) with PGMY primers followed by reverse line blot hybridization37. This assay can detect the 31 HPV genotypes, HPV 6, 11, 16, 18, 26, 31, 33, 34, 35, 39, 40, 42, 44, 45, 51, 52, 53, 54, 55, 56, 57, 58, 59, 66, 68, 69, 70, 73, 82, 83 and 84. Details of the pathological results and HPV genotypes are shown in Table S1. Two groups of HPV genotypes were selected for further analysis: seven most common high-risk HPV genotypes in Japan (HPV 16, 18, 31, 33, 45, 52, and 58), and 13 genotypes (HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68) found worldwide.
DNA extraction for microbial analysis
DNA was extracted from cervicovaginal mucus, collected using a BD BBL Culture Swab Plus with a ChargeSwitch Forensic DNA Purification Kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. DNA concentrations were measured with a Synergy H1 microplate reader (BioTek, Winooski, VT) and a QuantiFluor dsDNA system (Promega, Madison, WI, USA) following the manufacturer’s instructions.
Library preparation and sequencing
Cervicovaginal microbiota were determined using extracted genomic DNA by PCR with universal 16S rRNA gene (rDNA) bacterial primers for the V3/4 region followed by MiSeq sequencing. Libraries were prepared by a two-step tailed PCR method. First, two PCR analyses were conducted, the first with Bakt_341F and Bakt_805R primers38, the second with index primers. Library concentrations were measured using a Synergy H1 microplate reader (BioTek) and a QuantiFluor dsDNA System (Promega), and library quality was assessed with a Fragment Analyzer (Advanced Analytical Technologies, Ankeny, IA, USA) and a dsDNA 915 Reagent Kit (Agilent, Santa Clara, CA, USA), following the manufacturer’s instructions. Paired-end sequencing (2 × 300 bp) was carried out on the Illumina MiSeq platform (Illumina, San Diego, CA, USA) with the MiSeq Reagent Kit v3 (Illumina).
Microbial data analysis
Reads that began with a sequence that completely matched the primer used were extracted by using the fastx_barcode_splitter tool in the FASTX-Toolkit, and then the primer sequence was trimmed. Next, the Sickle tool39 with a quality value of 20 was used to trim and filter the reads; trimmed reads and paired-end reads with fewer than 150 bases were discarded. The FLASH paired-end merge script40 was used to merge the remaining reads under the following conditions: fragment length after merge, 420 bases; read fragment length, 280 bases; and minimum overlap length, 10 bases. All merged sequences were used for further analysis.
QIIME2.0 (2019.4), with the default parameter values, was used for sequence denoising using the DADA2 method, for chimera checking and then for taxonomic assignments with the Greengenes database (13_8) clustered at 97% identity41,42.
RDP classifier was used for taxonomic assignments for the genus Lactobacillus; the merged sequences (reads) were used as the input to the RDP classifier. Because the region of the gene to be analyzed was different and a new database had to be created, the 16S rRNA gene sequences of 12 species of the genus Lactobacillus (L. coleohominis, L. crispatus, L. gasseri, L. iners, L. jensenii, L. mucosae, L. paracasei, L. paraplantarum, L. plantarum, L. reuteri, L. rhamnosus, and L. vaginalis) included in the database attached to SpeciateIT (https://sourceforge.net/projects/speciateit/) were downloaded from the Ribosomal Database Project website (http://rdp.cme.msu.edu/hierarchy/hb_intro.jsp) using the following options: strain = both; source = isolates, size ≥ 1,200 bases; quality = good, and taxonomy = nomenclatural. Following SpeciateIT instructions, a database for species discrimination analysis was created from the 16S rRNA gene sequences, and then a species discrimination analysis was performed using SpeciateIT, with the created database and the output sequences of RDP classifier classified as the genus Lactobacillus. Alpha diversity estimators observed species richness (Sobs)—the observed OTUs was calculated for the overall bacterial community using QIIME2.0.
Protein extraction from cervical sponges
Protein for cytokine analysis was extracted from Merocel cervical sponges, using previously described methods25. First, the wet weight of each sponge was recorded, and each was then placed in a 2-ml Spin-X centrifuge filter tube (Corning Inc., Corning, NY, USA), and 300 μl of extraction buffer containing PBS(Sigma-Aldrich, St. Louis, MO, USA), with the addition of 256 mM NaCl and 100 μg/ml aprotinin (Wako, Amagasaki, Japan) were slowly added. The sponges were incubated at 4 °C for 2 h and then centrifuged at 14,000 rpm for 15 min at 4 °C, followed by the addition of 30 µl of fetal bovine serum to the 270 µl of extract. The sample was then vortexed briefly, aliquoted, and frozen at − 80 °C until further testing. The remaining extracts were stored at − 80 °C until the time of total protein measurement.
Cytokine measurements using cytometric bead array
Measurements of cytokine levels were as reported previously25. The following cytokines, chemokines, and growth factors were measured using multiplexed bead-based immunoassays (Cytometric Bead Array; CBA) according to the manufacturer’s protocol (BD Biosciences): interleukin (IL)-1α (Cat# 560153), IL-1β (Cat# 558279), IL-6 (Cat# 558276), IFN-γ (Cat# 558269), tumor necrosis factor (TNF)-α (Cat# 558273), granulocyte–macrophage colony-stimulating factor (GM-CSF) (Cat# 558335), granulocyte colony-stimulating factor (G-CSF) (Cat# 558326), IL-10 (Cat# 558274), IL-8 (Cat# 558277), IL-17A (Cat# 560383), monocyte chemoattractant protein (MCP)-1 (Cat# 558287), macrophage inflammatory protein (MIP)-1α (Cat# 558325), RANTES (Cat# 558324), and eotaxin (Cat# 558329). Cervical extracts were thawed and diluted 1:1 to 1:1000 in extraction buffer depending on the cytokine levels. Briefly, a 10-point standard curve ranging from 0 to 2500 pg/ml for each cytokine was prepared using the cytokine standard provided in each kit. Samples and cytokine standards were incubated in the capture bead mixture for 1 h and phycoerythrin-conjugated antibodies against each cytokine were added to the sample-bead mixture for 2 h of incubation at room temperature. All buffers used were from the CBA human soluble protein master buffer kit (Cat# 558265, BD Biosciences). Beads were washed and analyzed using a BD FACSCalibur flow cytometer (BD Biosciences). Mean fluorescence intensity for each bead cluster was converted into cytokine concentrations based on the 10-point standard curve, using FCAP Array software (BD version 3.0.1).
Adjustment of cytokine levels
Cytokine levels were adjusted by weighted volume, according to a previous report25,43. To compare differences in sponge weights after specimen collection, the dilution factor was calculated as [(x − y) + 300 mg of buffer]/(x − y), where x equals the weight of the sponge after collection and y is the weight of the dry sponge. Each cytokine measured was multiplied by this dilution factor to obtain weight-normalized values.
Statistical analysis
All statistical analyses were performed using SPSS for Windows (ver. 22.0.0.0; IBM Corp, Armonk, NY, USA). Mann–Whitney U tests after Bonferroni correction were used to compare continuous data between groups. To compare before with after intervention, the Wilcoxon signed-rank test was used. Spearman’s rank correlation for multiple comparisons was estimated for 1) the presence of each taxon and 2) the association between cytokine levels and microbiota. We defined p < 0.05 as significant.
References
Stanley, M. A. & Sterling, J. C. Host responses to infection with human papillomavirus. Curr. Probl. Dermatol. 45, 58–74. https://doi.org/10.1159/000355964 (2014).
Anahtar, M. N. et al. Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract. Immunity 42, 965–976. https://doi.org/10.1016/j.immuni.2015.04.019 (2015).
Wiik, J. et al. Cervical microbiota in women with cervical intra-epithelial neoplasia, prior to and after local excisional treatment, a Norwegian cohort study. BMC Womens Health 19, 30. https://doi.org/10.1186/s12905-019-0727-0 (2019).
Zhang, H. et al. Cervical microbiome is altered in cervical intraepithelial neoplasia after loop electrosurgical excision procedure in china. Sci. Rep. 8, 4923. https://doi.org/10.1038/s41598-018-23389-0 (2018).
Saftlas, A. F. et al. Influence of a loop electrosurgical excision procedure (LEEP) on levels of cytokines in cervical secretions. J. Reprod. Immunol. 109, 74–83. https://doi.org/10.1016/j.jri.2015.01.002 (2015).
Kim, Y. T. et al. Clearance of human papillomavirus infection after successful conization in patients with cervical intraepithelial neoplasia. Int. J. Cancer 126, 1903–1909. https://doi.org/10.1002/ijc.24794 (2009).
Ritu, W. et al. Evaluation of the associations between cervical microbiota and HPV infection, clearance, and persistence in cytologically normal women. Cancer Prev. Res. 12, 43–56. https://doi.org/10.1158/1940-6207.capr-18-0233 (2019).
Usyk, M. et al. Cervicovaginal microbiome and natural history of HPV in a longitudinal study. PLoS Pathog. 16, e1008376. https://doi.org/10.1371/journal.ppat.1008376 (2020).
Di Paola, M. et al. Characterization of cervico-vaginal microbiota in women developing persistent high-risk human papillomavirus infection. Sci. Rep. 7, 10200. https://doi.org/10.1038/s41598-017-09842-6 (2017).
Oh, H. Y. et al. The association of uterine cervical microbiota with an increased risk for cervical intraepithelial neoplasia in Korea. Clin. Microbiol. Infect. 21(674), e671-679. https://doi.org/10.1016/j.cmi.2015.02.026 (2015).
Laniewski, P. et al. Features of the cervicovaginal microenvironment drive cancer biomarker signatures in patients across cervical carcinogenesis. Sci. Rep. 9, 7333. https://doi.org/10.1038/s41598-019-43849-5 (2019).
Brotman, R. M. et al. Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J. Infect. Dis. 210, 1723–1733. https://doi.org/10.1093/infdis/jiu330 (2014).
Norenhag, J. et al. The vaginal microbiota, human papillomavirus and cervical dysplasia: A systematic review and network meta-analysis. BJOG 127, 171–180. https://doi.org/10.1111/1471-0528.15854 (2020).
Taylor-Robinson, D. Mollicutes in vaginal microbiology: Mycoplasma hominis, Ureaplasma urealyticum, Ureaplasma parvum and Mycoplasma genitalium. Res. Microbiol. 168, 875–881. https://doi.org/10.1016/j.resmic.2017.02.009 (2017).
Ekiel, A. M., Friedek, D. A., Romanik, M. K., Jozwiak, J. & Martirosian, G. Occurrence of Ureaplasma parvum and Ureaplasma urealyticum in women with cervical dysplasia in Katowice, Poland. J. Korean Med. Sci. 24, 1177–1181. https://doi.org/10.3346/jkms.2009.24.6.1177 (2009).
Vyshenska, D., Lam, K. C., Shulzhenko, N. & Morgun, A. Interplay between viruses and bacterial microbiota in cancer development. Semin. Immunol. 32, 14–24. https://doi.org/10.1016/j.smim.2017.05.003 (2017).
Si, J., You, H. J., Yu, J., Sung, J. & Ko, G. Prevotella as a hub for vaginal microbiota under the influence of host genetics and their association with obesity. Cell Host Microbe 21, 97–105. https://doi.org/10.1016/j.chom.2016.11.010 (2017).
Mitchell, C. et al. Behavioral predictors of colonization with Lactobacillus crispatus or Lactobacillus jensenii after treatment for bacterial vaginosis: A cohort study. Infect. Dis. Obstet. Gynecol. 2012, 706540. https://doi.org/10.1155/2012/706540 (2012).
Anton, L. et al. Common cervicovaginal microbial supernatants alter cervical epithelial function: Mechanisms by which Lactobacillus crispatus contributes to cervical health. Front. Microbiol. 9, 2181. https://doi.org/10.3389/fmicb.2018.02181 (2018).
Doerflinger, S. Y., Throop, A. L. & Herbst-Kralovetz, M. M. Bacteria in the vaginal microbiome alter the innate immune response and barrier properties of the human vaginal epithelia in a species-specific manner. J. Infect. Dis. 209, 1989–1999. https://doi.org/10.1093/infdis/jiu004 (2014).
Mitchell, C. & Marrazzo, J. Bacterial vaginosis and the cervicovaginal immune response. Am. J. Reprod. Immunol. 71, 555–563. https://doi.org/10.1111/aji.12264 (2014).
Behbakht, K. et al. Role of the vaginal microbiological ecosystem and cytokine profile in the promotion of cervical dysplasia: A case–control study. Infect. Dis. Obstet. Gynecol. 10, 181–186. https://doi.org/10.1155/s1064744902000200 (2002).
Bhatia, R. et al. Host chemokine signature as a biomarker for the detection of pre-cancerous cervical lesions. Oncotarget 9, 18548–18558. https://doi.org/10.18632/oncotarget.24946 (2018).
Matamoros, J. A., da Silva, M. I. F., de Moura, P., Leitão, M. & Coimbra, E. C. Reduced expression of IL-1β and IL-18 proinflammatory interleukins increases the risk of developing cervical cancer. Asian Pac. J. Cancer Prev. 20, 2715–2721. https://doi.org/10.31557/apjcp.2019.20.9.2715 (2019).
Otani, S. et al. Cytokine expression profiles in cervical mucus from patients with cervical cancer and its precursor lesions. Cytokine 120, 210–219. https://doi.org/10.1016/j.cyto.2019.05.011 (2019).
Marks, M. A. et al. Differences in the concentration and correlation of cervical immune markers among HPV positive and negative perimenopausal women. Cytokine 56, 798–803. https://doi.org/10.1016/j.cyto.2011.09.012 (2011).
Yin, X. et al. Time intervals between prior cervical conization and posterior hysterectomy influence postoperative infection in patients with cervical intraepithelial neoplasia or cancer. Med. Sci. Monit. 24, 9063–9072. https://doi.org/10.12659/msm.911892 (2018).
Brotman, R. M. et al. Association between the vaginal microbiota, menopause status, and signs of vulvovaginal atrophy. Menopause 21, 450–458. https://doi.org/10.1097/GME.0b013e3182a4690b (2014).
Ravel, J. et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 108(Suppl 1), 4680–4687. https://doi.org/10.1073/pnas.1002611107 (2011).
Virtanen, S., Kalliala, I., Nieminen, P. & Salonen, A. Comparative analysis of vaginal microbiota sampling using 16S rRNA gene analysis. PLoS ONE 12, e0181477. https://doi.org/10.1371/journal.pone.0181477 (2017).
Mitra, A. et al. Comparison of vaginal microbiota sampling techniques: Cytobrush versus swab. Sci. Rep. 7, 9802. https://doi.org/10.1038/s41598-017-09844-4 (2017).
Smith, B. C. et al. The cervical microbiome over 7 years and a comparison of methodologies for its characterization. PLoS ONE 7, e40425. https://doi.org/10.1371/journal.pone.0040425 (2012).
Kyrgiou, M., Mitra, A. & Moscicki, A. B. Does the vaginal microbiota play a role in the development of cervical cancer?. Transl. Res. 179, 168–182. https://doi.org/10.1016/j.trsl.2016.07.004 (2017).
Bradshaw, C. S. et al. The influence of behaviors and relationships on the vaginal microbiota of women and their female partners: The WOW Health Study. J. Infect. Dis. 209, 1562–1572. https://doi.org/10.1093/infdis/jit664 (2014).
Brotman, R. M. et al. Association between cigarette smoking and the vaginal microbiota: A pilot study. BMC Infect. Dis. 14, 471. https://doi.org/10.1186/1471-2334-14-471 (2014).
Ravel, J. et al. Daily temporal dynamics of vaginal microbiota before, during and after episodes of bacterial vaginosis. Microbiome 1, 29. https://doi.org/10.1186/2049-2618-1-29 (2013).
Azuma, Y. et al. Human papillomavirus genotype distribution in cervical intraepithelial neoplasia grade 2/3 and invasive cervical cancer in Japanese women. Jpn. J. Clin. Oncol. 44, 910–917. https://doi.org/10.1093/jjco/hyu112 (2014).
Herlemann, D. P. et al. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571–1579. https://doi.org/10.1038/ismej.2011.41 (2011).
Joshi, N. & Fass, J. Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files (Version 1.33) [Software]. https://github.com/najoshi/sickle (2011).
Magoc, T. & Salzberg, S. L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. https://doi.org/10.1093/bioinformatics/btr507 (2011).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. https://doi.org/10.1038/s41587-019-0209-9 (2019).
Callahan, B. J. et al. DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. https://doi.org/10.1038/nmeth.3869 (2016).
Marks, M. A., Eby, Y., Howard, R. & Gravitt, P. E. Comparison of normalization methods for measuring immune markers in cervical secretion specimens. J. Immunol. Methods 382, 211–215. https://doi.org/10.1016/j.jim.2012.05.012 (2012).
Acknowledgements
We thank Ms. Yumiko Usui and Ms. Yuko Nakagawa, Department of Obstetrics and Gynecology, Fujita Health University, for editing the manuscript. We thank Mr. Nakano and Mr. Ijichi, Bioengineering Lab. Co., Ltd, Kanagawa, Japan for helping write the manuscript. We thank NAI for editing a draft of this manuscript. This work was partly supported by KAKENHI from the Ministry of Education, Culture, Sports, Science and Technology, Japan (Grant No. 17K11300) and a Fujita Health University Research Grant-in-Aid.
Author information
Authors and Affiliations
Contributions
Designed the experiments: T.F., Performed the experiments: A.I., I.K., H.N., Analyzed the data: T.F., R.K., A.I., R.I., T.T., Contributed reagents/materials/analysis tools: R.K., R.K., E.N., T.F., Contributed to the writing of the manuscript: T.F., A.I., I.K., H.N.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kawahara, R., Fujii, T., Kukimoto, I. et al. Changes to the cervicovaginal microbiota and cervical cytokine profile following surgery for cervical intraepithelial neoplasia. Sci Rep 11, 2156 (2021). https://doi.org/10.1038/s41598-020-80176-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-80176-6
This article is cited by
-
Diagnostic and prognostic potential of the microbiome in ovarian cancer treatment response
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.