Abstract
In 2005, the Nigerian Federal Ministry of Health revised the treatment policy for uncomplicated malaria with the introduction of artemisinin-based combination therapies (ACTs). This policy change discouraged the use of Sulphadoxine-pyrimethamine (SP) as the second-line treatment of uncomplicated falciparum malaria. However, SP is used as an intermittent preventive treatment of malaria in pregnancy (IPTp) and seasonal malaria chemoprevention (SMC) in children aged 3–59 months. There have been increasing reports of SP resistance especially in the non-pregnant population in Nigeria, thus, the need to continually monitor the efficacy of SP as IPTp and SMC by estimating polymorphisms in dihydropteroate synthetase (dhps) and dihydrofolate reductase (dhfr) genes associated with SP resistance. The high resolution-melting (HRM) assay was used to investigate polymorphisms in codons 51, 59, 108 and 164 of the dhfr gene and codons 437, 540, 581 and 613 of the dhps gene. DNA was extracted from 271 dried bloodspot filter paper samples obtained from children (< 5 years old) with uncomplicated malaria. The dhfr triple mutant I51R59N108, dhps double mutant G437G581 and quadruple dhfr I51R59N108 + dhps G437 mutant haplotypes were observed in 80.8%, 13.7% and 52.8% parasites, respectively. Although the quintuple dhfr I51R59N108 + dhps G437E540 and sextuple dhfr I51R59N108 + dhps G437E540G581 mutant haplotypes linked with in-vivo and in-vitro SP resistance were not detected, constant surveillance of these haplotypes should be done in the country to detect any change in prevalence.
Similar content being viewed by others
Introduction
In 2005, the Nigerian Federal Ministry of Health (FMoH) revised the treatment policy for uncomplicated malaria with the introduction of artemisinin-based combination therapies (ACTs)1. This treatment policy change discouraged the use of Chloroquine (CQ) and Sulphadoxine-pyrimethamine (SP) as the first-line and second-line treatment of uncomplicated falciparum malaria, respectively. However, SP continues to be used as an intermittent preventive treatment of malaria in pregnancy (IPTp) and seasonal malaria chemoprevention (SMC) in children aged 3–59 months in malaria-endemic countries including Nigeria2,3.
Providing evidence for the continued use of SP as IPTp and SMC in the context of SP resistance in Africa requires constant epidemiological surveillance of parasite resistance levels by monitoring polymorphisms in genes associated with SP resistance. Point mutations such as S436A, A437G, K540E, A581G and A613T/S in dihydropteroate synthetase (dhps) gene and N51I, C59R, S108N and I164L in dihydrofolate reductase (dhfr) gene are observed to play significant roles in SP resistance4,5,6. At a population level, the quintuple mutation in P. falciparum parasites, i.e., triple dhfr mutations of I51, R59, and N108, plus double dhps mutations of G437, and E540 (I51R59N108G437E540) has also been strongly linked with reduced SP efficacy as IPTp, reduced parasite clearance ability in asymptomatic pregnant women and shortened post-treatment prophylactic activity7,8,9.
Since the deployment of SP in Nigeria as IPTp in 200110 and SMC in 201311, there have been various reports of these point mutations in both dhfr and dhps genes associated with SP resistance in various parts of the country11,12. Reports of high prevalence of the triple mutant genotype of dhfr (N51I, C59R and S108N) in addition to the A437G mutation in the dhps gene is common in Nigeria13. However, occurrence of the quintuple dhfr + dhps mutation comprising of N51I, C59R and S108N + A437G and K540E is scarce13.
The goal of this study was, therefore, to: (i) examine the current status of circulating Dhfr and Dhps haplotypes by describing polymorphisms in codons 51, 59, 108 and 164 of Dhfr and codons 437, 540, 581 and 613 of Dhps and (ii) estimate the prevalence of single, double, triple, quadruple and quintuple mutation4 in parasites present in the five (5) Nigerian States.
Results
Baseline demographics and clinical data
A total of 271 children from Adamawa (50), Bayelsa (45), Imo (82), Kwara (45) and Sokoto (49) states with uncomplicated falciparum malaria were considered in this analysis. Baseline characteristics of the children are shown in Table 1. Overall, 160 (59.04%) were male. Mean age of all children included in the study was 38.2 ± 16.5 months. Also, mean enrollment body temperature was 37.5 ± 2.5 °C, and 17 of the 271 children (4.4%) had hyperpyrexia (enrollment temperature > 40 °C). Overall geometric asexual parasitemia was 14,673parasite/μL−1 (range: 2003–198,200).
Merozoite surface protein genotyping of Plasmodium falciparum
The family-specific polymorphic length markers of msp-2 and msp-1 were used for genotyping the P. falciparum in children considered for this study. In general, 180 (66.8%) children had polyclonal infections (Table 2). The mean complexity of infection (mCOI) for parasites across all population was 2.3. Allelic family distributions for both msp-2 and msp-1 per State is represented in Table 2. Overall, the 3D7 allelic family was the most amplified (50.7%) in the msp-2 polymorphic marker while the K1 allelic family was the most detected (18.4%) msp-1 polymorphic marker.
Polymorphisms in the dhfr gene
From the 271 children considered, 241 (88.9%), 245 (90.4%) and 219 (80.8%) were infected with parasites that harboured the mutant I51, R59 and N108 alleles respectively. None of the children was infected with parasites habouring the mutant L164 allele. Distribution of the mutant I51, R59 and N108 alleles were significantly higher in polyclonal infections than in monoclonal infections (p < 0.05 for each allele). All isolates obtained from children enrolled in Imo State were infected with parasites that harboured only the mutant I51 and R59 alleles (Fig. 1a,b).
Polymorphisms in the dhps gene
From the 271 children considered, 223 (82.3%) and 45 (16.6%) were infected with parasites that harboured the mutant G437 and G581 alleles respectively. None of the children was infected with parasites that harboured the mutant E540 and T/S 613 alleles. Distribution of the mutant G437 and G581 alleles were significantly higher in polyclonal infections than in monoclonal infections (p < 0.05 for each allele) (Fig. 1c,d).
Dhfr and Dhps haplotype frequencies and distribution
Parasites harbouring the dhfr triple mutant I51R59N108, double mutant I51R59, double mutant R59N108, and single mutant N108 haplotypes were observed in 80.8%, 8.1%, 0.74% and 7.4% respectively of the 271 children considered (Fig. 2). The proportions of children with dhfr triple mutant I51R59N108 and double mutant I51R59 haplotypes in Northern (Adamawa, Sokoto and Kwara) and Southern (Bayelsa and Imo) States was similar (p > 0.05). Conversely, the proportion of children with dhfr single mutant N108 was significantly higher in Northern States versus the Southern States (p > 0.05). Double mutant R59N108 haplotype was only recorded in two children from Kwara and Sokoto States (Northern States) (Fig. 2).
Bar chart showing the distribution of dhfr haplotypes in five Nigerian States. Red colour represents Sokoto State, Yellow represents Kwara State, Purple represents Imo State, Green represents Bayelsa State and Blue represents Adamawa State. The triple mutant haplotype I51R59N108 was the most distributed in all five States.
Parasites harbouring the dhps double mutant G437G581, single mutant G437 and single mutant G581 haplotypes were observed in 13.7%, 66.1% and 2.6% respectively of the 271 children considered (Fig. 3). There was no triple and quadruple dhps mutant haplotype because none of the isolates harboured mutant allele of dhps 540. There was no difference in the distribution of the double mutant G437G581 and single mutant G437 haplotypes in the Northern and Southern States (p > 0.05).
Bar chart showing the distribution of dhps haplotypes in five Nigerian States. Red colour represents Sokoto State, Yellow represents Kwara State, Purple represents Imo State, Green represents Bayelsa State and Blue represents Adamawa State. The single mutant haplotype G437K540A581 was the most distributed in all five States.
Prevalence of combined dhps and dhfr haplotypes
The prevalence of different alleles on dhfr and dhps genes when combined as single mutant haplotype (dhfr N108), double mutant haplotype (dhfr R59N108), triple mutant haplotype (dhfr I51R59N108), quadruple mutant haplotype (dhfr I51R59N108 + dhps G437), quintuple mutant haplotype (dhfr I51R59N108 + dhps G437E540) and sextuple mutant haplotype (dhfr I51R59N108 + dhps G437E540G581) were considered (Table 3). Although most of the dhfr I51R59N108 triple mutant haplotype was observed in polyclonal infections as only eight were observed in monoclonal infections, there was no significant difference in distribution based on clonality (p > 0.05) or location, i.e., Northern and Southern States (p > 0.05). The quadruple mutant haplotype was observed in 143 (52.8%) of the 271 children (Table 3). Eighty-three (58.0%) of the 143 were observed in polyclonal infections but there was no significant difference in the distribution of this haplotype based on clonality (polyclonal vs. monoclonal) (p > 0.05) or location, i.e., Northern and Southern States (p > 0.05). None of the 271 children was infected with parasites harbouring the quintuple mutation and sextuple mutation as the E540 mutant allele was absent (Table 3).
Discussion
This study assessed the status of circulating dhfr and dhps haplotypes by describing polymorphisms on codons 51, 59, 108 and 164 of dhfr gene and codons 437, 540, 581 and 613 of dhps gene and estimated the prevalence of dhfr + dhps combined mutant haplotypes in 271 parasites obtained from children (< 5 years) children with uncomplicated falciparum malaria in Nigeria 10 years after treatment policy was changed to ACTs.
Polymorphism data from our study showed high prevalence of mutant I51 (88.9%) and N108 (80.8%) dhfr alleles and mutant G437 (82.3%) dhps allele. Similar prevalence of these mutant dhfr and dhps alleles have been recorded in Nigeria11,13 and other West African countries14,15. Although prevalence of these mutant alleles are generally high in West Africa16, lower prevalence (26.5–56.25) have been recorded in other West African countries4,17. The exact reason for the difference in prevalence amongst these West African countries may be as a result of the varied use of SP in these countries18. Also, P. falciparum and other disease etiologies exist as co-infections in patients in these areas. It is equally plausible that the use of other sulpha-related drugs in the treatment of these co-infections may select for these mutations in the P. falciparum genome at varying levels19.
Sulfadoxine-pyrimethamine was previously used as weekly prophylaxis for malaria during which the mutant E540 allele was recorded2,5. However, our study which was conducted when the therapeutic use of SP had changed from weekly prophylaxis to IPTp and SMCs, showed the absence of the mutant E540 allele. Similar trend has been observed in recent studies in Nigeria11,19. The supposed disappearance of this mutant allele is perhaps, as a result of the reduced SP drug pressure in the country due to this treatment policy change.
Pearce et al.20 stated the importance of measuring the frequency of haplotypes as against the prevalence of each point mutation separately as haplotypes are determinants of resistance levels. We observed a high frequency (80.8%) of the dhfr triple mutant haplotype (I51R59N108) which suggests the persistent circulation of similar parasites as those reported in earlier studies post-ACT introduction11,12,18. These parasites are probably selected for as a result of the SP drug pressure, as this drug was not completely withdrawn in the country but rather used as IPTp and SMCs till date. The use of drugs such trimethoprin sulfamethazole targeting dhfr genes in Pneumocystis carnii in an environment where malaria and HIV coinfections is common, could also be responsible for the selection of this haplotype in Plasmodium falciparum populations in Nigeria5. The occurrence of this mutant haplotype at such a high frequency is worrisome as such triple mutations in the dhfr gene has been associated with a 1.5- to threefold higher pyrimethamine resistance in vitro than I51N108 or R59N108 double mutations5. Thus, the efficacy of pyrimethamine as a partner drug in SP’s use as IPTp and SMCs is threatened. Although the double dhps mutant haplotype (G437G581) was observed in our study (12.9%), the absence of the E540 mutation in the dhps gene in combination with either the single mutant G437 or double mutant G437G581 in this study is desired as the double mutant G437E540 haplotype is essential for sulfadoxine resistance5.
We also observed high levels of polyclonal infections in this study as most of the States considered had > 60% proportion of children with polyclonal infections. Further analysis of data revealed that the mutant dhfr (I51, R59 and N108) and mutant dhps (G437 and G581) alleles were significantly higher in polyclonal infections than in monoclonal infections (p < 0.05 for each mutant allele). Also, the dhfr triple mutant I51R59N108 haplotype was significantly higher in polyclonal infections (p < 0.05). These observations may be problematic as high levels of polyclonality is linked to increased parasite transmission and diversity. This may result in the increase in spread of these mutant alleles and haplotype within the country. This can jeopardise the use of SPs as IPTp and SMCs in Nigeria.
Unpublished P. falciparum microsatellite data21 confirmed the high intra-population diversity observed using the msp-1 and msp-2 polymorphic genes but also revealed low population differentiation in these parasites from the five parasite populations (Nigeria States). This suggests that, despite the high parasite diversity observed, parasites were genetically similar across the country. This may be responsible for the observed similarities in the distribution of the combined dhfr + dhps triple mutant haplotype (dhfr I51R59N108: p > 0.05) and quadruple mutant haplotype (dhfr I51R59N108 + dhps G437: p > 0.05) in both Northern and Southern States of the country considered in this study.
The quintuple mutant haplotype (dhfr I51R59N108 + dhps G437E540) that was earlier reported (2005) in Nigeria5 was absent in this study. This is possibly due to the reduced SP drug pressure as a result of policy change from SP as second-line treatment of malaria to ACTs. Both the quintuple and sextuple mutant haplotypes have been strongly linked with in vivo and in vitro SP resistance22 in Southern and East Africa23,24 and their absence in this current study, is not only beneficial, but consistent with reports from other West African countries4,25. Nevertheless, continuous monitoring for re-emergence of dhfr I51R59N108 + dhps G437E540 and emergence of dhfr I51R59N108 + dhps G437E540G581 haplotypes should be maintained in the country to detect any change in the recorded prevalence. This would ensure that alternative control measures are rapidly put in place to prevent the spread of these haplotypes within the country, which if not checked, will lead to reduced efficacy of SP as IPTp and SMCs.
Methods
Study site
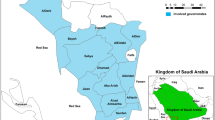
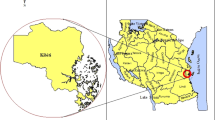
This study is part of a national drug therapeutic efficacy testing (DTET) study for monitoring antimalarial efficacies of artemether-lumefantrine (AL), artesunate-amodiaquine (AA) and dihydroartemisinin-piperaquine (DHP) in the treatment of acute uncomplicated falciparum infections in children under the age of five years. Samples considered for analysis in this study were obtained from five Nigerian States which were sub-classified into Northern region (Adamawa, Sokoto and Kwara States) and Southern region (Bayelsa and Imo States) (Fig. 4). These States are parts of sentinel locations for the National Malaria Elimination Program (NMEP) of the Federal Ministry of Health in Nigeria for the year 2014/2015 Drug Therapeutic Efficacy Testing (DTET) study26.
This map shows the States in Nigeria were analysed samples were obtained. These five States represents five of the six geopolitical zones in the country, i.e., North-West: Sokoto, North-East: Adamawa, North-Central: Kwara, South-South: Bayelsa and South-East: Imo. (Map generated using Datawrapper: https://www.datawrapper.de).
Patients enrolment criteria and sample collection
Description of patient enrolment at sentinel locations was initially discussed in an earlier study26. Two to three drops of finger-prick blood were blotted on 3 mm Whatman filter paper (Whatman International Limited, Maidstone, United Kingdom) before treatment (Day 0) and during follow-up on days 1–3, 7, 14, 21, 28, 35 and 42 post-treatment. Blood samples impregnated on filter papers were allowed to air-dry appropriately at room temperature and stored in airtight envelopes with silica gels.
DNA extraction
DNA was extracted from dry blood spot (DBS), i.e., blood impregnated filter papers of day 0 samples (before treatment) for the detection of polymorphisms of the drug resistance markers and to determine parasite genetic diversity as previously described27. A Qiagen DNA extraction kit (Qiagen, Hilden, Germany) was used to extract DNA from DBS, following the manufacturer's protocol. Briefly, a quarter of the DBS was used for extraction, and DNA content was eluted in a final volume of 60 μl with buffer AE.
Genotyping Plasmodium falciparum using the msp-1 and msp-2 gene
The polymorphic length markers msp-2 and msp-1 were amplified by nested PCR as previously described28. The Glurp polymorphic marker was not considered in this study due to the low PCR amplification reported in Nigeria29. PCR amplification was performed on a thermocycler (Eppendorf Vapo. Protect Mastercycler pro, Germany) in a final volume of 25 μl. Two per cent (2%) agarose gel was used for the resolution of PCR amplicons. The amplified products were sized against a 100-base pair (bp) DNA molecular weight marker (New England Biolabs, Beverly, MA) and visualised using a gel visualisation box (Syngene, UK). Interpretations were made based on the number of parasite clones present in a sample. Briefly, infections were defined as polyclonal if parasites from a single patient showed more than one allelic family or more than one amplicon fragment in a single allelic family of the gene. Infections were defined as monoclonal if an isolate had a single amplicon fragment in one allelic family and the other allelic family(ies) was (were) not amplified6.
High resolution melting drug resistance assay
High resolution melting (HRM) assay was performed as previously described30. Briefly, the 10X primer–probe mix and reaction mix was prepared. One microliter of the quantified DNA sample was dispensed in PCR well containing 9.0 μl reaction mix. PCR cycling and melting conditions used were those described earlier30. Standard software included with the instruments was used for unlabeled probe analysis to visualise melting peaks based on different melting temperatures, indicative of different base pairs, and compared with wild type and mutant controls to call alleles for both dhfr (Supplementary Figs. S1 and S2) and dhps (Supplementary Figs. S3 and S4) assays. Parasite genomic controls used in this study, i.e., 3D7, HB3, DD2 and Tm90C6B were graciously donated by BEI resources (MR4, BEI resources, USA).
Statistical analysis
Data were double-entered and analysed using version 6 of Epi-Info software and the statistical program SPSS for Windows version 20.0. Proportions were compared by calculating χ2 using Yates’ correction, Fisher’s exact or Mantel Haenszel tests. Normally distributed, continuous data were analysed by Student’s t-test and analysis of variance (ANOVA). Mann–Whitney U tests and the Kruskal Wallis tests (or by Wilcoxon ranked sum test) were used to compare data that did not conform to normal distribution. P values of < 0.05 were taken to indicate significant differences.
Ethical approval
The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the National Health Research Ethics Committee, Federal Ministry of Health (FMOH), Abuja, Nigeria. Informed consent was obtained from parents and legal guardians of participants prior to enrollment in study.
References
Federal Ministry of Health. National Antimalarial Treatment Guidelines (Federal Ministry of Health, Abuja, 2005).
Meremikwu, M., Donegan, S., Sinclair, D., Esu, E. & Oringanje, C. Intermittent preventive treatment for malaria in children living in areas with seasonal transmission. Cochrane Database Syst. Rev. 2012(2), 003756–003756 (2012).
WHO. World Malaria Report, 2013 (WHO, Geneva, 2013).
Ndiaye, D. et al. High-resolution melting: A useful field-deployable method to measure dhfr and dhps drug resistance in both highly and lowly endemic plasmodium populations. Malar. J. 16, 153 (2017).
Happi, C. et al. Polymorphisms in Plasmodium falciparum dhfr and dhps genes and age related in vivo sulfadoxine–pyrimethamine resistance in malaria-infected patients from Nigeria. Acta Trop. 95(3), 183–193 (2005).
Wang, P. et al. Resistance to antifolates in Plasmodium falciparum monitored by sequence analysis of dihydropteroate synthetase and dihydrofolate reductase alleles in a large number of field samples of diverse origins. Mol. Biochem. Parasitol. 89, 161–177 (1997).
Desai, M. et al. Impact of sulfadoxine-pyrimethamine resistance on effectiveness of intermittent preventive therapy for malaria in pregnancy at clearing infections and preventing low birth weight. Clin. Infect. Dis. 62(3), 323–333 (2016).
Nankabirwa, J. et al. Efficacy, safety, and tolerability of three regimens for prevention of malaria: A randomized, placebo-controlled trial in Ugandan Schoolchildren. PLoS ONE 5(10), e13438 (2010).
Gesase, S. et al. High resistance of Plasmodium falciparum to sulphadoxine/pyrimethamine in Northern Tanzania and the emergence of dhps resistance mutation at codon 581. PLoS ONE 4(2), e4569 (2009).
Onoka, C. A., Hanson, K. & Onwujekwe, O. E. Low coverage of intermittent preventive treatment for malaria in pregnancy in Nigeria: Demand-side influences. Malar. J. 11(1), 82. https://doi.org/10.1186/1475-2875-11-82 (2012).
Oguike, M. et al. Molecular determinants of sulfadoxine-pyrimethamine resistance in Plasmodium falciparum in Nigeria and the regional emergence of dhps 431V. Int. J. Parasitol. Drugs Drug Resist. 6, 220–229 (2016).
Esu, E. et al. Intermittent screening and treatment with artemether–lumefantrine versus intermittent preventive treatment with sulfadoxine–pyrimethamine for malaria in pregnancy: A facility-based, open-label, non-inferiority trial in Nigeria. Malar. J. 17(1), 251 (2018).
Quan, H. High multiple mutations of Plasmodium falciparum-resistant genotypes to sulphadoxine-pyrimethamine in Lagos, Nigeria. Infect. Dis. Poverty 9(1), 91 (2020).
Chauvin, P. et al. Prevalence of Plasmodium falciparum parasites resistant to sulfadoxine/pyrimethamine in pregnant women in Yaoundé, Cameroon: Emergence of highly resistant dhfr/dhfr alleles. Antimicrob. Chemother. 70(9), 2566–2571 (2015).
Abugri, J. et al. Prevalence of chloroquine and antifolate drug resistance alleles in Plasmodium falciparum clinical isolates from three areas in Ghana. AAS Open Res. 1, 1 (2018).
Lucchi, N. et al. Increasing prevalence of a novel triple-mutant dihydropteroate synthase genotype in Plasmodium falciparum in western Kenya. Antimicrob. Agents Chemother. 59(7), 3995–4002 (2015).
Tahita, M. et al. Prevalence of the dhfr and dhps mutations among pregnant women in rural burkina faso five years after the introduction of intermittent preventive treatment with sulfadoxine-pyrimethamine. PLoS ONE 10(9), e0137440–e0137440 (2015).
Iwalokun, B., Iwalokun, S., Adebodun, V. & Balogun, M. Carriage of mutant dihydrofolate reductase and dihydropteroate synthase genes among Plasmodium falciparum isolates recovered from pregnant women with asymptomatic infection in Lagos, Nigeria. Med. Princ. Pract. 24(5), 436–443 (2015).
Ikpa, F., Shaa, K. & Auta, K. Molecular markers of sulfadoxine-pyrimethamine resistant malaria prior to intermittent preventive treatment among pregnancies in Makurdi, Nigeria. Int. J. Bio. Chem. Sci. 8(5), 1961–1968 (2015).
Pearce, R., Drakeley, C., Chandramohan, D., Mosha, F. & Roper, C. Molecular determination of point mutation haplotypes in the dihydrofolate reductase and dihydropteroate synthase of Plasmodium falciparum in three districts of Northern Tanzania. Antimicrob. Chemother 1, 1347–1354 (2003).
Ajogbasile, F. et al. Microsatellite loci analysis reveals high intra-population diversity and low population differentiation in parasite from nine Nigeria States 10 years post adoption of ACTs. (2020).
Naidoo, I. & Roper, C. Mapping partially resistant, fully resistant, and super resistant malaria. Trends Parasitol. 29(10), 505–515 (2013).
Bwijo, B. et al. High prevalence of quintuple mutant dhps/dhfr genes in Plasmodium falciparum infections seven years after introduction of sulfadoxine and pyrimethamine as first line treatment in Malawi. Acta Trop. 85, 363–373 (2003).
Roper, C., Pearce, R., Nair, S., Nosten, F. & Anderson, T. Intercontinental spread of pyrimethamine-resistant malaria. Science 305, 1124 (2004).
Ndiaye, D. et al. Polymorphism in dhfr/dhps genes, parasite density and ex vivo response to pyrimethamine in Plasmodium falciparum malaria parasites in Thies, Senegal. Int. J. Parasitol Drugs Drug resist. 3, 135–142 (2013).
Ebenebe, J. et al. Efficacy of artemisinin-based combination treatments of uncomplicated falciparum malaria in under-five-year-old nigerian children ten years following adoption as first-line antimalarials. Am. J. Trop. Med. Hyg. 99(3), 649–664 (2018).
Bankole, B. et al. Characterization of Plasmodium falciparum structure in Nigeria with malaria SNPs barcode. Malar. J. 17(1), 472 (2018).
Viriyakosol, S. et al. Genotyping of Plasmodium falciparum isolates by the polymerase chain reaction and potential uses in epidemiological studies. Bull. World Health Organ. 73(1), 85–95 (1995).
Kolawole, O., Mokuolu, O., Olukosi, A. & Oloyede, T. Population genomics diversity of Plasmodium falciparum in malaria patients attending Okelele Health Centre, Okelele, Ilorin, Kwara State, Nigeria. Afr. Health Sci. 16(3), 704–711 (2016).
Daniels, R. et al. Rapid, field-deployable method for genotyping and discovery of single-nucleotide polymorphisms associated with drug resistance in Plasmodium falciparum. Antimicrob. Agents Chemother. 56(6), 2976–2986 (2012).
Acknowledgement
The authors thank all the patients, their parents or guardians for volunteering to participate in the study. We also acknowledge the principal investigators (PI) in each of the five sentinel locations considered in this study. The Clinical and molecular aspects of this study were funded by grants from the U.S President’s Malaria Initiative (USPMI), African Center of Excellence for Genomics of Infectious Diseases (ACEGID), World Bank (ACE019) and The National Institute of Health Grants (5U01HG007480-03 and U54HG007480). We thank the Department of Immunology and Infectious Diseases, Harvard TH Chan School of Public Health, Boston, MA, USA for the training on Molecular SNP Barcoding and HRM analysis.
Author information
Authors and Affiliations
Contributions
A.T.K., F.V.A., J.N.U., P.J.E.: Performed the experiment. A.T.K., K.A., P.E.O.: Analysed data. A.S.: Performed the clinical studies. A.T.K., K.A., O.A.F.: Wrote the manuscript. C.T.H., O.A.F., D.F.W.: Conceived and designed experiment. All authors contributed and agreed on the content of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kayode, A.T., Ajogbasile, F.V., Akano, K. et al. Polymorphisms in Plasmodium falciparum dihydropteroate synthetase and dihydrofolate reductase genes in Nigerian children with uncomplicated malaria using high-resolution melting technique. Sci Rep 11, 471 (2021). https://doi.org/10.1038/s41598-020-80017-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-80017-6
This article is cited by
-
A snapshot of the prevalence of dihydropteroate synthase-431V mutation and other sulfadoxine-pyrimethamine resistance markers in Plasmodium falciparum isolates in Nigeria
Malaria Journal (2023)
-
Seasonal malaria chemoprevention in Africa and China’s upgraded role as a contributor: a scoping review
Infectious Diseases of Poverty (2023)
-
Assessment of genetic polymorphisms associated with malaria antifolate resistance among the population of Libreville, Gabon
Malaria Journal (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.