Abstract
The emergence of immune checkpoint inhibitors (ICIs) has revolutionized the treatment of recurrent/metastatic (R/M) head and neck squamous cell carcinoma (HNSCC). Biomarkers of the therapeutic efficacy of ICIs have been extensively investigated. In this study, we aimed to analyze whether molecular phenotypes of circulating tumor cells (CTCs) are associated with treatment responses and clinical outcomes in patients with R/M HNSCC treated with nivolumab. Peripheral blood samples were collected before treatment initiation and after four infusions of nivolumab. CTCs isolated by depletion of CD45-positive cells were analyzed to determine the expression of EPCAM, MET, KRT19, and EGFR using real-time quantitative polymerase chain reaction. CTC-positive samples were analyzed to determine the expression of PIK3CA, CCND1, SNAI1, VIM, ZEB2, CD44, NANOG, ALDH1A1, CD47, CD274, and PDCD1LG2. Of 30 patients treated with nivolumab, 28 (93.3%) were positive for CTCs. In 20 CTC-positive patients, molecular alterations in CTCs before and after nivolumab treatment were investigated. Patients with MET-positive CTCs had significantly shorter overall survival than those with MET-negative CTCs (p = 0.027). The expression level of CCND1 in CTCs of disease-controlled patients was significantly higher than that of disease-progressed patients (p = 0.034). In disease-controlled patients, the expression level of CCND1 in CTCs significantly decreased after nivolumab treatment (p = 0.043). The NANOG expression in CTCs was significantly increased in disease-controlled patients after nivolumab treatment (p = 0.036). Our findings suggest that the molecular profiling of CTCs is a promising tool to predict the treatment efficacy of nivolumab.
Similar content being viewed by others
Introduction
The immune checkpoint inhibitor (ICI), nivolumab, has shown a significant improvement compared with the investigator’s preferred chemotherapy regimen and is currently used as a new therapeutic option in patients with recurrent and metastatic (R/M) head and neck squamous cell carcinoma (HNSCC)1,2,3. Recently, another ICI, pembrolizumab, has also been reported to have clinical benefit in R/M HNSCC4. Thus, the emergence of ICIs has revolutionized in the treatment of R/M HNSCC; however, the objective response rate is still 20%. Therefore, further improvements and identification of biomarkers for therapeutic efficacy are urgently needed.
To date, a variety of candidate biomarkers for R/M HNSCC and other malignancies have been extensively investigated5,6,7,8. Tumor tissue-derived biomarkers such as programmed death-ligand 1 (PD-L1) expression, interferon-γ signature, CD8 T-cell infiltration, and tumor mutational burden are potential sources for the identification of candidate biomarkers. However, the use of tissue specimens in biomarker discovery for ICIs has raised several concerns, such as tissue quality, tissue heterogeneity, and the time of collection. In particular, the tumor microenvironment might fluctuate with disease progression, treatment pressures, and sites of growth.
Recently, accumulating evidence has indicated that liquid biopsy, including circulating tumor cells (CTCs), cell-free DNA, and exosomes, is a reliable method to obtain real-time tumor information9,10,11. Our previous studies on HNSCC have shown that the presence of CTCs was significantly associated with treatment resistance, locoregional recurrence, and shorter progression-free survival12. Notably, the MET and CD274 expression in CTCs showed a poorer and a better prognosis, respectively. Molecular profiling of CTCs may provide new insights to improve patient selection, maximize efficacy, and predict response as one of the more reliable biomarkers. In the present study, we aimed to analyze whether the molecular characteristics of CTCs can be used as biomarkers to predict treatment efficacy in patients with R/M HNSCC treated with nivolumab; we also aimed to investigate the molecular alterations in CTCs during nivolumab treatment.
Results
Patient characteristics
A total of 30 patients who had histologically confirmed R/M HNSCC were enrolled in this study. Their characteristics are listed in Table 1. The tumor origins included the paranasal sinus (n = 5), oral cavity (n = 3), nasopharynx (n = 2), oropharynx (n = 5), hypopharynx (n = 12), larynx (n = 2), and parotid gland (n = 1). The treatment responses were as follows: complete response (CR) in 0 (0.0%) patients, partial response (PR) in 6 (20.0%), stable disease (SD) in 6 (20.0%), and progressive disease (PD) in 18 (60.0%) patients. The objective response rate and disease control rate were 20.0% and 40.0%, respectively. Eight patients (26.7%) discontinued 4 courses of nivolumab due to rapid disease progression (n = 6) and intolerable immune-related adverse events (n = 2, intestinal lung disease and colitis), respectively. Although we analyzed clinical features between disease-controlled and disease-progressed patients, there was no significant difference in clinical features evaluated.
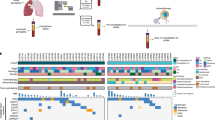
Of the 30 patients with R/M HNSCC treated with nivolumab, 28 (93.3%) were positive for CTCs, 20 of whom underwent peripheral blood sample collection at two time points: before treatment initiation and after 4 infusions of nivolumab (Supplementary Fig. 1).
Molecular phenotype of CTCs and their correlation with treatment responses
To investigate the relationship between the molecular phenotype of CTCs and treatment responses, the patients with CTCs were divided into two groups according to treatment responses: disease-controlled and disease-progressed patients. First, we analyzed whether the expression of epithelial-related markers in CTCs before treatment initiation is correlated with treatment efficacy and clinical outcome. There was no significant correlation between the expression of epithelial-related genes in CTCs and treatment efficacy (data not shown); however, patients with MET-positive CTCs had a significantly shorter overall survival compared with those with MET-negative CTCs (p = 0.027) (Fig. 1 and Table 2). We then investigated which gene expression in CTCs was associated with treatment responses. Notably, CCND1 expression in CTCs from disease-controlled patients was significantly higher than that in disease-progressed patients (p = 0.034, Fig. 2). Furthermore, we investigated whether the levels of gene expression in the CTCs was associated with the prognosis; however, there was no relationship between gene expression of CTCs and prognosis in patients treated with nivolumab (Supplementary Table 1).
Molecular alterations of CTCs following nivolumab treatment
We investigated the change in gene expression in CTCs from individual patients during nivolumab treatment. Although there was no significant change in gene expression including PD-L1 in CTCs in all treated patients by nivolumab treatment (data not shown), the expression level of CCND1 in CTCs in disease-controlled patients, not in disease-progressed patients, was significantly reduced by nivolumab treatment (p = 0.043) (Fig. 3). By contrast, the NANOG expression levels in CTCs in disease-controlled patients were significantly increased by nivolumab treatment (p = 0.036) (Fig. 3).
TCGA analysis
To further elucidate the underlying mechanism of CCND1 that affects the patient’s responses to nivolumab treatment, the relationship between CCND1 expression and genomic alterations in HNSCC was analyzed using the TCGA database. In 515 patients with HNSCC, although no significant difference was observed between CCND1 expression and tumor mutation counts, tumors with high CCND1 expression had significantly higher FGA than those with low CCND1 expression (CCND1 expression and tumor mutation counts; p = 0.638, CCND1 expression and FGA; p = 0.006, Fig. 4a,b). Moreover, there was a weak correlation between the expression of CCND1 and FGA (Fig. 4c, r = 0.238), but not tumor mutation counts (data not shown).
Genomic alterations in head and neck squamous cell carcinoma. Two genomic alterations, tumor mutation counts (a) and fraction genome altered (b), were compared between tumors with low CCND1 expression and tumors with high CCND1 expression. The cutoff value of high and low CCND1 expression was set as the median. The correlation between the expression of CCND1 and fraction genome altered (c) was evaluated.
Discussion
The primary objective of this study was to explore whether the molecular profiling of CTCs could predict treatment efficacy in patients with R/M HNSCC treated with nivolumab. We succeeded in achieving this objective to some extent, and the following points are of particular importance: (1) patients with MET-positive CTCs have a poorer prognosis compared with those with MET-negative CTCs, (2) CTCs before treatment initiation in the disease-controlled patients showed significantly higher CCND1 expression and significantly reduced by nivolumab treatment compared with those in disease-progressed patients, and (3) the expression of NANOG in CTCs significantly increased in the disease-controlled patients.
The first point probably indicated that MET-positive CTCs are resistant not only to chemotherapy and/or radiotherapy but also to immunotherapy. In our previous study on patients with treatment-naïve HNSCC, patients with MET-positive CTCs had a shorter progression-free survival than those with MET-negative CTCs12. c-MET overexpression and amplification in HNSCC have been shown to drive tumorigenesis, proliferation, invasion, and metastasis, and is commonly associated with resistance to therapy13,14,15,16. With regard to the immunological role of c-Met expression in tumor cells, Saigi et al. demonstrated that c-Met activation induces PD-L1 expression via a pathway independent of JAK/STAT activation17. On the contrary, Wang et al. suggested that c-MET expression in tumor cells suppresses the function of NK cells and T cells via the production of indoleamine-2,3-dioxygenase18. In addition, the activation of HGF/Met signaling in HNSCC increases glycolysis, which would result in the suppression of T-cell functions19. Thus, c-Met expression in tumor cells contributes to immune suppression through a variety of mechanisms. Although the clinical application of ICIs is currently being expanded to a variety of malignancies, a combination of c-Met targeting therapies with ICIs should be considered in patients with MET-positive CTCs to improve the therapeutic efficacy of cancer immunotherapy.
The second point suggests that CCND1 expression in tumor cells might be a good target for T cells reactivated by ICIs. Among several gene expressions related to cell growth, epithelial-mesenchymal transition, cancer stemness, and immune regulation, CCND1 expression in CTCs is significantly associated with disease control. CCND1 is known to play a critical role in cell cycle regulation, and high expression of CCND1 is associated with treatment resistance and poor prognosis in HNSCC, as with MET20,21,22. Therefore, the overexpression of cyclin D1 is associated with genome instability, and consequently results in the increase of somatic mutations and neoantigens. In the TCGA database analysis of HNSCC, no significant difference was observed in the in mutation counts between the low CCDN1 group and high CCDN1 group; however, the FGA was significantly higher in the high CCND1 group than in the low CCND1 group. The FGA is the percentage of genome affected by gains or losses in the copy number, which may increase the neoantigen loads and induce tumor (neoantigen)-specific T cells. Another possibility is that CCND1 protein itself may function as a tumor antigen recognized by T cells. In fact, several epitopes presented by tumor cells and recognized by both CD4 + and CD8 + T cells have been identified23,24,25,26. Thus, a preexisting immune response against tumor cells expressing CCND1 may be boosted by nivolumab treatment and thus eliminate such tumor cells. More interestingly, CCND1 expression in CTCs was significantly decreased in patients with disease control after four infusions of nivolumab, suggesting that tumor-specific T cells may lyse tumor cells expressing CCND1.
The last point may indicate that the cancer stemness phenotype represents an immune-resistant phenotype of CTCs. Of patients treated with nivolumab, disease-controlled patients showed significantly increased expression of cancer stemness markers, NANOG. This finding suggests that the induction of an effective antitumor immune response by nivolumab treatment presumably allows the concentration of CTCs to display the cancer stem cell (CSC) phenotype due to lysis of non-CSC tumor cells. Although CSCs are known to be subpopulations that are capable of self-renewal and differentiation27,28, in terms of immunological characterization of CSCs, CSCs can evade immune surveillance by reducing immunogenicity and immune suppressive activity compared with their non-CSC counterparts29,30,31,32. Indeed, our previous study revealed that CD44-positive cancer stem-like cells in HNSCC produced various immune suppressive cytokines and enhanced the regulatory T-cell response compared with CD44-negative cells33. Hence, development of new treatment strategies targeting CSC-related molecules is needed to further modulate the antitumor immune responses reactivated by ICIs.
Taken collectively, ICIs can boost the immune responses against tumor cells in certain patients and exert a strong selective pressure on tumor cells in peripheral blood as well as within the tumor microenvironment. Despite the small sample size, molecular alterations in CTCs may in fact be a result of the selective pressure of antitumor immune responses induced by nivolumab. Thus, our findings suggest that the molecular profiling of CTCs may be a promising tool to predict the treatment efficacy of nivolumab and provide new insights into more precise immunotherapies.
Methods
This study was approved by the Gunma University Ethical Review Board for Medical Research Involving Human Subjects (HS2017-152), and all experiments were performed in accordance with the approved guidelines. Written informed consent was obtained from each patient.
Patients and blood collection
Patients who had histologically confirmed R/M HNSCC and treated with nivolumab were eligible for this study. We evaluated several clinical variables, including age, sex, primary site, local recurrence, lymph node metastasis, distant metastasis, performance status, previous radiotherapy, PD-L1 expression, and completion of 4 course of nivolumab. Treatment response was evaluated according to RECIST, version 1.1, during weeks 8–10. If the patients had an unacceptable level of drug-related toxic effects or rapid disease progression before evaluation, the therapeutic effects were evaluated at that point. Blood samples were collected using the K2EDTA Vacutainer (BD Bioscience) before the first nivolumab infusion and if possible, after fourth nivolumab infusion. The samples were analyzed within 4 h of collection.
Detection and gene expression analysis of CTCs
CTC detection and gene expression analysis were performed as described previously34,35. In brief, the peripheral blood mononuclear cells were isolated from blood samples (7.5 mL), and the contaminating erythrocytes were lysed with a red blood cell lysis buffer (Roche). The cell suspension was incubated with a human CD45 depletion cocktail for 15 min and then with magnetic particles for 10 min (EasySep Human CD45 Depletion Kit II, STEMCELL Technologies). Tubes containing cells were placed on a magnet for 10 min twice, and the unbound cells (CTCs) were transferred to new tubes.
Total RNA from the CTCs was extracted using an RNeasy Micro kit (Qiagen) according to the manufacturer’s instructions. cDNA synthesis was performed using the QuantiTect Reverse Transcription kit (Qiagen) with preamplification using the TaqMan PreAmp Master Mix kit (Applied Biosystems). The preamplified products were then analyzed using real-time quantitative polymerase chain reaction (Applied Biosystems) to quantify the 16 target genes. Sixteen primers for the fifteen targets (epithelial cell adhesion molecule [EPCAM], MET, keratin 19 [KRT19], epidermal growth factor receptor [EGFR], phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha [PIK3CA], cyclin D1 [CCND1], snail family transcriptional repressor 1 [SNAI1], vimentin [VIM], zinc finger E-box binding homeobox 2 [ZEB2], CD44, nanog homeobox [NANOG], aldehyde dehydrogenase 1 family member A1 [ALDH1A1], CD47, CD274, and programmed cell death 1 ligand 2 [PDCD1LG2]) and ACTB (β-actin) as control were purchased from Applied Biosystems (TaqMan Gene Expression Assays). All samples were analyzed in triplicates. Detection of at least one of the four epithelial-related genes (EPCAM, MET, KRT19, and EGFR) was defined as CTC positivity. The Ct values of the target genes were normalized to a reference gene (ACTB). The expression levels and changes of the target genes in CTCs were indicated as 40-delta Ct value and − ΔΔCt value, respectively.
The Cancer Genome Atlas (TCGA) analysis
We analyzed the relationship between CCND1 expression and mutation counts and fraction genome altered (FGA) in HNSCC using the TCGA database. The CCND1 expression Z-scores obtained based on the batch-normalized expression values from Illumina HiSeqV2, mutation count, and FGA values were downloaded from cBioPortal (https://www.cbioportal.org/study/clinicalData?id=hnsc_tcga_pan_can_atlas_2018). The cutoff value of high and low CCND1 expression was set as the median.
Statistical analysis
GraphPad Prism version 8.0 for Windows (GraphPad Software, San Diego, CA, USA) was used to perform all statistical analyses. Chi-squared test for independence and Fisher’s exact test were used to examine differences in categorical variables. The Welch’s t-test was used to examine the differences in the means between the two groups. The Kaplan–Meier curves were plotted and compared using the log-rank test. The optimal cut-off values of gene expression in CTCs for overall survival were determined based on receiver operating characteristics curve analysis. In TCGA analysis, the Welch’s t-test was used to determine the significant difference between the high CCND1 group and low CCND1 group. The Pearson correlation coefficient was used to measure the strength of a linear association between two variables, CCND1 expression level and mutation counts or FGA. Two-sided p-values < 0.05 were considered significant.
Abbreviations
- ICI:
-
Immune checkpoint inhibitor
- HNSCC:
-
Head and neck squamous cell carcinoma
- R/M:
-
Recurrent/metastatic
- PD-L1:
-
Programmed death-ligand 1
- CTC:
-
Circulating tumor cell
- TCGA:
-
The Cancer Genome Atlas
- FGA:
-
Fraction genome altered
- CSC:
-
Cancer stem cell
References
Ferris, R. L. et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 375, 1856–1867. https://doi.org/10.1056/NEJMoa1602252 (2016).
Cramer, J. D., Burtness, B. & Ferris, R. L. Immunotherapy for head and neck cancer: recent advances and future directions. Oral Oncol. 99, 104460. https://doi.org/10.1016/j.oraloncology.2019.104460 (2019).
Kao, H. F. & Lou, P. J. Immune checkpoint inhibitors for head and neck squamous cell carcinoma: current landscape and future directions. Head Neck 41, 4–18. https://doi.org/10.1002/hed.25930 (2019).
Burtness, B. et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet 394, 1915–1928. https://doi.org/10.1016/S0140-6736(19)32591-7 (2019).
Arora, S. et al. Existing and emerging biomarkers for immune checkpoint immunotherapy in solid tumors. Adv. Ther. 36, 2638–2678. https://doi.org/10.1007/s12325-019-01051-z (2019).
Oliva, M. et al. Immune biomarkers of response to immune-checkpoint inhibitors in head and neck squamous cell carcinoma. Ann. Oncol. 30, 57–67. https://doi.org/10.1093/annonc/mdy507 (2019).
Xiao, Q. et al. Genetic and epigenetic biomarkers of immune checkpoint blockade response. J. Clin. Med. 9, 286. https://doi.org/10.3390/jcm9010286 (2020).
Gavrielatou, N., Doumas, S., Economopoulou, P., Foukas, P. G. & Psyrri, A. Biomarkers for immunotherapy response in head and neck cancer. Cancer Treat. Rev. 84, 101977. https://doi.org/10.1016/j.ctrv.2020.101977 (2020).
Campos, C. D. M., Jackson, J. M., Witek, M. A. & Soper, S. A. Molecular profiling of liquid biopsy samples for precision medicine. Cancer J. 24, 93–103. https://doi.org/10.1097/PPO.0000000000000311 (2018).
Soda, N., Rehm, B. H. A., Sonar, P., Nguyen, N. T. & Shiddiky, M. J. A. Advanced liquid biopsy technologies for circulating biomarker detection. J. Mater. Chem. B. 7, 6670–6704. https://doi.org/10.1039/C9TB01490J (2019).
Wang, J., Chang, S., Li, G. & Sun, Y. Application of liquid biopsy in precision medicine: opportunities and challenges. Front. Med. 11, 522–527. https://doi.org/10.1007/s11684-017-0526-7 (2017).
Tada, H., Takahashi, H., Kuwabara-Yokobori, Y., Shino, M. & Chikamatsu, K. Molecular profiling of circulating tumor cells predicts clinical outcome in head and neck squamous cell carcinoma. Oral Oncol. 102, 104558. https://doi.org/10.1016/j.oraloncology.2019.104558 (2020).
Hartmann, S., Bhola, N. E. & Grandis, J. R. HGF/Met signaling in head and neck cancer: impact on the tumor microenvironment. Clin. Cancer Res. 22, 4005–4013. https://doi.org/10.1158/1078-0432.CCR-16-0951 (2016).
Lim, Y. C., Kang, H. J. & Moon, J. H. C-Met pathway promotes self-renewal and tumorigenecity of head and neck squamous cell carcinoma stem-like cell. Oral Oncol. 50, 633–639. https://doi.org/10.1016/j.oraloncology.2014.04.004 (2014).
Lo Muzio, L. et al. Effect of c-Met expression on survival in head and neck squamous cell carcinoma. Tumour Biol. 27, 115–121. https://doi.org/10.1159/000092716 (2006).
Szturz, P. et al. Understanding c-MET signalling in squamous cell carcinoma of the head & neck. Crit. Rev. Oncol. Hematol. 111, 39–51. https://doi.org/10.1016/j.critrevonc.2017.01.004 (2017).
Saigi, M. et al. MET-oncogenic and JAK2-inactivating alterations are independent factors that affect regulation of PD-L1 expression in lung cancer. Clin. Cancer Res. 24, 4579–4587. https://doi.org/10.1158/1078-0432.CCR-18-0267 (2018).
Wang, D. et al. The hepatocyte growth factor antagonist NK4 inhibits indoleamine-2,3-dioxygenase expression via the c-Met-phosphatidylinositol 3-kinase-AKT signaling pathway. Int. J. Oncol. 48, 2303–2309. https://doi.org/10.3892/ijo.2016.3486 (2016).
Boschert, V. et al. The influence of met receptor level on HGF-induced glycolytic reprogramming in head and neck squamous cell carcinoma. Int. J. Mol. Sci. 21, 471. https://doi.org/10.3390/ijms21020471 (2020).
Ramos-García, P. et al. Prognostic and clinicopathological significance of cyclin D1 expression in oral squamous cell carcinoma: a systematic review and meta-analysis. Oral Oncol. 83, 96–106. https://doi.org/10.1016/j.oraloncology.2018.06.007 (2018).
Ramos-García, P. et al. Predictive value of CCND1/cyclin D1 alterations in the malignant transformation of potentially malignant head and neck disorders: systematic review and meta-analysis. Head Neck 41, 3395–3407. https://doi.org/10.1002/hed.25834 (2019).
Feng, Z. et al. CCND1 as a predictive biomarker of neoadjuvant chemotherapy in patients with locally advanced head and neck squamous cell carcinoma. PLoS ONE 6, e26399. https://doi.org/10.1371/journal.pone.0026399 (2011).
Dengjel, J. et al. Unexpected abundance of HLA class II presented peptides in primary renal cell carcinomas. Clin. Cancer Res. 12, 4163–4170. https://doi.org/10.1158/1078-0432.CCR-05-2470 (2006).
Dao, T. et al. Identification of a human cyclin D1-derived peptide that induces human cytotoxic CD4 T cells. PLoS ONE 4, e6730. https://doi.org/10.1371/journal.pone.0006730 (2009).
Dengjel, J. et al. Identification of a naturally processed cyclin D1 T-helper epitope by a novel combination of HLA class II targeting and differential mass spectrometry. Eur. J. Immunol. 34, 3644–3651. https://doi.org/10.1002/eji.200425510 (2004).
Kondo, E. et al. Cyclin D1-specific cytotoxic T lymphocytes are present in the repertoire of cancer patients: implications for cancer immunotherapy. Clin. Cancer Res. 14, 6574–6579. https://doi.org/10.1158/1078-0432.CCR-08-0825 (2008).
Nassar, D. & Blanpain, C. Cancer stem cells: basic concepts and therapeutic implications. Annu. Rev. Pathol. 11, 47–76. https://doi.org/10.1146/annurev-pathol-012615-044438 (2016).
Nguyen, L. V., Vanner, R., Dirks, P. & Eaves, C. J. Cancer stem cells: an evolving concept. Nat. Rev. Cancer 12, 133–143. https://doi.org/10.1038/nrc3184 (2012).
Clara, J. A., Monge, C., Yang, Y. & Takebe, N. Targeting signalling pathways and the immune microenvironment of cancer stem cells - a clinical update. Nat. Rev. Clin. Oncol. 17, 204–232. https://doi.org/10.1038/s41571-019-0293-2 (2020).
Codony-Servat, J. & Rosell, R. Cancer stem cells and immunoresistance: clinical implications and solutions. Transl. Lung Cancer Res. 4, 689–703. https://doi.org/10.3978/j.issn.2218-6751.2015.12.11 (2015).
Qian, X. et al. Biology and immunology of cancer stem(-like) cells in head and neck cancer. Crit. Rev. Oncol. Hematol. 95, 337–345. https://doi.org/10.1016/j.critrevonc.2015.03.009 (2015).
Maccalli, C., Volontè, A., Cimminiello, C. & Parmiani, G. Immunology of cancer stem cells in solid tumours: a review. Eur. J. Cancer 50, 649–655. https://doi.org/10.1016/j.ejca.2013.11.014 (2014).
Chikamatsu, K., Takahashi, G., Sakakura, K., Ferronse, S. & Masuyama, K. Immunoregulatory properties of CD44+ cancer stem-like cells in squamous cell carcinoma of the head and neck. Head Neck 33, 208–215. https://doi.org/10.1002/hed.21420 (2011).
Chikamatsu, K. et al. Expression of immune-regulatory molecules in circulating tumor cells derived from patients with head and neck squamous cell carcinoma. Oral Oncol. 89, 34–39. https://doi.org/10.1016/j.oraloncology.2018.12.002 (2019).
Tada, H., Takahashi, H., Ida, S., Nagata, Y. & Chikamatsu, K. Epithelial- mesenchymal transition status of circulating tumor cells is associated with tumor relapse in head and neck squamous cell carcinoma. Anticancer Res. 40, 3559–3564. https://doi.org/10.21873/anticanres.14345 (2020).
Acknowledgements
This work was supported in part by a Grant-in-Aid for Scientific Research (Grant No. 20K18243 to Hideyuki T., no. 20K09747 to M.S., no. 19K18794 to S.I., no 19K18758 to I.M., and no. 20H03834 to K.C.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Author information
Authors and Affiliations
Contributions
Conception and design: Hideyuki T. and K.C., Acquisition of data: Hiroe T., Y.N., M.U., M.S., S.I., I.M., and T.M., Analysis and interpretation of data: Hiroe T., Hideyuki T., and R. K-I., Writing the manuscript: Hiroe T. and K.C. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tada, H., Takahashi, H., Kawabata-Iwakawa, R. et al. Molecular phenotypes of circulating tumor cells and efficacy of nivolumab treatment in patients with head and neck squamous cell carcinoma. Sci Rep 10, 21573 (2020). https://doi.org/10.1038/s41598-020-78741-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-78741-0
This article is cited by
-
Innovations in Molecular Biomarkers and Biomaterial-Based Immunotherapies for Head & Neck Cancer
Current Surgery Reports (2024)
-
A new marker constructed from immune-related lncRNA pairs can be used to predict clinical treatment effects and prognosis: in-depth exploration of underlying mechanisms in HNSCC
World Journal of Surgical Oncology (2023)
-
The organoid as reliable cancer modeling in personalized medicine, does applicable in precision medicine of head and neck squamous cell carcinoma?
The Pharmacogenomics Journal (2023)
-
Application of liquid biopsy as multi-functional biomarkers in head and neck cancer
British Journal of Cancer (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.