Abstract
Sustained disturbances are relevant for environmental biotechnology as they can lead to alternative stable states in a system that may not be reversible. Here, we tested the effect of a sustained organic loading alteration (food-to-biomass ratio, F:M, and carbon-to-nitrogen ratio, C:N) on activated sludge bioreactors, focusing on the stability of nitrification and nitrifiers. Two sets of replicate 5-L sequencing batch reactors were operated at different, low and high, F:M (0.19–0.36 mg COD/mg TSS/d) and C:N (3.5–6.3 mg COD/mg TKN) conditions for a period of 74 days, following 53 days of sludge acclimation. Recovery and resilience were tested during the last 14 days by operating all reactors at low F:M and C:N (henceforth termed F:M–C:N). Stable nitrite accumulation (77%) was achieved through high F:M–C:N loading with a concurrent reduction in the abundance of Nitrospira. Subsequently, only two of the three reactors experiencing a switch back from high to low F:M–C:N recovered the nitrite oxidation function, with an increase in Nitrobacter as the predominant NOB, without a recovery of Nitrospira. The AOB community was more diverse, resistant and resilient than the NOB community. We showed that functional recovery and resilience can vary across replicate reactors, and that nitrification recovery need not coincide with a return to the initial nitrifying community structure.
Similar content being viewed by others
Introduction
Improving stability and optimizing performance of wastewater treatment processes are central tenets of environmental engineering and biotechnology, to help achieve the sustainable development goal of guaranteeing availability and sustainable management of water and sanitation for everyone1. In ecology, disturbances are believed to have direct effects on the stability of ecosystems by altering community structure and function2. Stability is multidimensional3, having several quantifiable aspects like resistance (ability to withstand disturbance), resilience (speed of recovery from disturbance), and recovery (ability to return to prior conditions after disturbance ceases)4. For engineered systems like activated sludge bioreactors, it is important to identify the effect of different disturbances on the microbial community structure so as to relate them to changes in process performance5,6. However, disturbance is also deemed to affect underlying mechanisms of community assembly7 which, if predominantly stochastic, could drive microbial communities to divergent trajectories in terms of composition and function8. Therefore, robust replication9 is valuable to assess the effect of disturbances on the stability of sludge bioreactors.
Sustained disturbances that impose a long-term continuous change of species densities through an alteration of the environment, also called press disturbances10,11, are relevant since they can lead a system to alternative stable states that may or may not be reversible in terms of both community composition and function12. Disturbances could be alterations in the environment that are not directly detrimental for organisms, but still provide opportunities for low abundance members within the community13. In bioreactors, a switch in the substrate feeding scheme employed could then elicit changes in community structure and function. Such disturbances can occur in the form of organic shocks within activated sludge14,15,16 and anaerobic reactor systems17,18. In wastewater treatment, the food-to-biomass ratio (F:M) or sludge loading rate is an important parameter as it determines the growth type and settleability of sludge microorganisms19. Yet, how press disturbances of F:M alterations affect the stability of microbial populations and specific functions in sludge bioreactors remains largely unknown.
Partial nitrification is an important stage in biological nitrogen removal from wastewater via nitritation/anammox20,21, for which the accumulation of nitrite is desired by promoting the growth of ammonia oxidizing bacteria (AOB) while suppressing nitrite oxidizing bacteria (NOB)22. Different strategies have been employed towards this goal, like varying temperature, dissolved oxygen (DO), pH, SRT, and substrate concentrations23. Variations in C:N are known to affect nitrification due to changes in competition for DO and space in biofilms24. However, substrate manipulation in particular has been reported to yield contrasting results in terms of C:N25. Low influent C:N values (2–3 mg COD/mg TN) have been shown to prevent26, but also promote27,28,29 nitrite accumulation in different sludge systems. Other studies have reported nitrite accumulation at high (10 mg COD/mg NH4+-N)30 and also fluctuating influent C:N values (2.5–8 mg COD/mg NH4+-N)31,32. Additionally, although adjustments in C:N also impact F:M values by altering the available carbon for heterotrophic growth, covariations in F:M are rarely considered, and hence there is a knowledge gap about system performance and stability when both factors are controlled simultaneously. To uncover additional nitrite accumulation strategies, more research is needed to understand the effect of variations in influent C:N together with controlled variations of other important operational parameters like F:M.
The aim of this work was to test the effect of a press disturbance of doubling F:M and C:N (henceforth termed F:M–C:N) values in a set of replicated lab-scale sludge bioreactors after an acclimation period (Fig. 1). The focus was on the stability of the nitrite oxidation function and the nitrifying microbial community. Function dynamics were monitored throughout the study by periodic analysis of reactor effluent, as well as detailed temporal studies of reactor cycles at seven different time points. Nitrite accumulation was also tracked due to its relevance for practical applications. Changes in composition of nitrifying organisms and genes involved in nitrification were assessed by metagenomics and 16S rRNA gene amplicon sequencing. The resistance of nitrite oxidation and specific nitrifier abundance was quantified and evaluated by monitoring their transition to a different steady state after the disturbance. We further tested recovery and resilience by shifting the F:M–C:N back to the original pre-disturbance state.
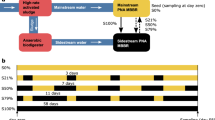
Schematic representation of the experimental design. F:M, food-to-biomass ratio; C:N, carbon-to-nitrogen ratio in the feed. Rectangles represent independent replicate 5-L SBRs. Phases: acclimation (grey), low F:M–C:N (blue, undisturbed), high F:M–C:N (red, press disturbed). Dashed line indicates removal of disturbance through the shift from high to low F:M–C:N.
Results
Dynamics in bioreactor performance
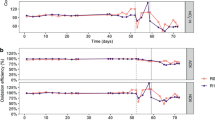
During the acclimation phase, the F:M–C:N values were maintained at 0.21 (mg COD/mg TSS/d) and 3.5 (mg COD/mg TKN), respectively (Table 1). Ammonium concentrations in the effluent decreased gradually while nitrate concentrations increased (Fig. 2). Sludge related parameters like settleability (SVI) and biomass fraction (VSS:TSS) varied during this acclimation period (Fig. S1). Most of these variations decreased after 30 d and trends were stable after 45 d. During the disturbance phase of the study (d54 onwards), sludge was wasted more often to better control the TSS and thus the F:M (Fig. S1), which is the reason why the SRT for the low F:M–C:N reactors is lower than during the acclimation phase. The average F:M and C:N values for the low F:M–C:N reactors were similar to those during the acclimation phase (0.19 and 3.5), while the ones for the high F:M–C:N reactors were controlled to be almost double (0.36 and 6.3). As expected (details in supplementary information), controlling for a higher F:M resulted in a lower solids residence time (SRT) in reactors subjected to this treatment (Table 1). This period showed a clear distinction between high and low F:M–C:N reactors in terms of nitrification (P < 0.014, Table S1), with the high F:M–C:N reactors displaying nitrite accumulation with high NO2–-N effluent concentrations (Fig. 2). To ensure that the partial nitrification was due to different F:M–C:N values and not a lack of available dissolved oxygen, we increased the aeration rate from 1 to 4 L min-1 from d97 onwards without observing significant changes in NO2–-N and NO3–-N effluent compounds. The last two weeks of the study involved shifting operational parameters in the high F:M–C:N reactors to match those of the low F:M–C:N ones (Table 1). During this period a transition towards recovery of the nitrite oxidation function was observed (P > 0.43), with high variability of effluent NO2–-N and NO3–-N across reactors (Fig. 2).
Temporal average effluent concentrations in mg/L of (A) soluble COD, (B) NH4+-N, (C) NO2–-N and (D) NO3–-N. Phases: A, acclimation (n = 4); L, low F:M–C:N (n = 4); H, high F:M–C:N (n = 3). Vertical dashed line indicates the shift from high to low F:M–C:N. Periods of functional resistance development and transient recovery are indicated by curly brackets.
Dynamics in nitrification and nitrifiers
The acclimation phase (d1–d53) displayed negligible nitrite accumulation at 0.3% (± 1.5%). Low F:M–C:N reactors had zero percent nitrite accumulation after d61 and only 1.1% (± 2.0%) during the first week (d54–d60). Conversely, high F:M–C:N reactors showed an initial transient nitrite accumulation of 18% (± 21%) on d54–d60, which subsequently increased and stabilized at 77% (± 6.0%) during the d61-d113 period. Finally, after shifting from high to low F:M–C:N conditions, nitrite accumulation decreased to 55% (± 29%) in the first week (d114–d120), and all the way to zero in the second week (d121-d127) for two of the three reactors (Fig. 2).
Among nitrifiers, the three most abundant bacterial genera detected through both metagenomics and 16S rRNA gene amplicon sequencing were Nitrospira, Nitrosomonas and Nitrobacter. During the disturbance phase, Nitrospira abundance was strongly reduced in high F:M–C:N reactors (to about 0.02%), while Nitrosomonas remained at around half the abundance levels observed for low F:M–C:N reactors (Fig. 3). This coincided with the accumulation of nitrite in high F:M–C:N reactors during the d54–d113 period of the study (Fig. 2). Similar patterns of nitrifier abundance across low and high F:M–C:N replicates were observed for both metagenomics and 16S rRNA gene amplicon sequencing datasets (Fig. 3). Further, relevant genes involved in nitrification (amo, hao and nxr) displayed a reduction in relative abundances measured as transcripts per million (tpm) across high F:M–C:N reactors compared to the low F:M–C:N reactors (Fig. 4). All the aforementioned differences in effluent values as well as nitrifier and nitrification gene abundances were statistically significant from d75 onwards (Table S1), until the disturbance was removed on d114.
Temporal relative abundance of main nitrifier genera in each reactor. Phases: A, acclimation (n = 4); L, low F:M–C:N (n = 4); H, high F:M–C:N (n = 3). Vertical dashed line indicates the shift from high to low F:M–C:N. Closed symbols display 16S rRNA gene amplicon ASV data and open symbols shotgun metagenomics (summarized reads) data. Left, Nitrosomonas; centre, Nitrospira; right, Nitrobacter. There was variable NOB resilience as evident from the increase in Nitrobacter in two (H1 and H2) out of three reactors once the disturbance had been removed.
Temporal dynamics of nitrification genes amo (A), hao (B) and nxr (together with nitrate reductase genes, nar) (C), measured as relative abundances of transcripts per million (tpm) from assembled metagenomics data. Each point represents a different reactor on a given day. Phases: A, acclimation (n = 4); L, low F:M–C:N (n = 4); H, high F:M–C:N (n = 3). Vertical dashed line indicates the shift from high to low F:M–C:N. Lines refer to polynomial regression fitting, while shaded areas represent 95% confidence intervals.
Following the shift from high to low F:M–C:N it was Nitrobacter that rose to be the dominant NOB instead of Nitrospira, but only in two of three reactors (Fig. 3). Variations in performance among replicate reactors were also evident from cycle study profiles before (d110) and after (d124) the shift in feeding regime for the high F:M–C:N reactors (Fig. 5). Two weeks after the change, only two high to low F:M–C:N reactors displayed nitrite oxidation profiles similar to those of the low F:M–C:N reactors. The reactors that recovered functionality were the same as those that registered around 1% of Nitrobacter abundance (Fig. 3). By the end of the study (d124), abundances of Nitrosomonas as well as of amo and hao genes in the high to low reactors recovered to values that were not significantly different from the low F:M–C:N reactors (Figs. 3 and 4, Table S1).
Chemical profiles during a full intermittently-aerated SBR cycle lasting 12 h. Concentrations are shown for all reactors on (A) day 110 and (B) day 124, showing profiles before and after high to low F:M–C:N changes. L, low F:M–C:N reactors (n = 4); H, high F:M–C:N reactors (n = 3). Vertical dashed dotted line indicates the start of the aerobic stage in each cycle. Right panels (B) show that two (H1 and H2) out of three reactors fully recovered the nitrite oxidation function on d124.
Stability metrics for each of the high F:M–C:N replicate reactors (Fig. 6) showed a temporal reduction in resistance for both the nitrite oxidation function and the abundance of Nitrospira. Removal of the disturbance was followed by a temporal recovery of the nitrite oxidation function across all replicates. At the end of the study, this function had been completely restored for the two reactors (H1 and H2) that also displayed the highest nitrite oxidation resilience, as well as an overcompensation-recovery (c > 0) in Nitrobacter abundances. Overall, the ammonia oxidizer Nitrosomonas displayed higher resistance and resilience values than the nitrite oxidizer Nitrospira, which did not recover even after the disturbance had been removed (Fig. 6).
Stability metrics of resistance (a), resilience (b) and recovery (c) for the nitrite oxidation function and nitrifier abundances across disturbed replicate reactors, adopted from4. Vertical dashed line indicates removal of disturbance. Dotted and dash-dotted lines represent linear regression fitting per replicate, with the slope (b) being the measure of resilience. Benchmarks: a = 0, maximum resistance; a < 0, low resistance through underperformance; b > 0, recovery; b < 0, further deviation from undisturbed control; c = 0, maximum recovery; c < 0, incomplete recovery; c > 0, overcompensation.
The higher resolution of 16S rRNA gene amplicon sequencing allowed us to taxonomically identify four amplicon sequence variants (ASVs) as Nitrospira, 21 as Nitrosomonas, and one as Nitrobacter (Fig. S2). From these, only two ASVs were identified at the species level. The genus Nitrospira was dominated by the N. defluvii species, with the other three ASVs detected only at low abundances across the low F:M–C:N reactors after d97, the day the aeration rate was increased. The dominant Nitrosomonas ASVs during the disturbance phase were different from the initial ones. Furthermore, N. europaea and Nitrosomonas ASV-70 saw their relative abundances increasing with time across high F:M–C:N reactors (Fig. S2).
Discussion
Disturbance leads to stable nitrite accumulation unveiling system’s resistance
Sludge acclimation served to stabilize important functions like complete nitrification, organic carbon removal, and sludge settling capacity across reactors (Fig. 2, Fig. S1). Ecosystem functionality in terms of COD removal, ammonium removal, and complete nitrification was optimal and stable for the low F:M–C:N reactors, particularly towards the end of the study (Fig. 2). In contrast, press disturbed reactors (high F:M–C:N) experienced a marked reduction in nitrate production. The shift in function was not immediate, as the first seven days showed transient nitrite accumulation, which was variable among replicates, before stabilizing with little within-treatment variability for the remainder of the disturbance phase. Nitrite oxidation was not completely inhibited because 23% of the NOX–-N products still consisted of nitrate during the d61–d113 period. In addition to the calculated resistance metrics (Fig. 6), this percentage also shows the degree to which this particular function was insensitive to disturbance33. The higher variability in function among replicates during the first seven-day transition period could have been due to stochastic effects initially triggered in ecosystems after a disturbance7. Since the disturbance was sustained in this case, selective pressure likely promoted deterministic mechanisms resulting in a less variable function over time (Fig. 2). This is similar to what has been previously reported for studies on sludge bioreactors under sustained 3-chloroaniline disturbance8.
Dynamics of nitrifiers during the disturbance phase
The focus of this work was on the stability of the nitrite oxidation function and the nitrifying microbial community, with overall community dynamics described in detail elsewhere34. Understanding community and activity dynamics of nitrifying bacteria is essential for improving design and operation of wastewater treatment biological processes35. The changes in abundance of different Nitrosomonas ASVs suggested a succession of organisms within this genus (Fig. S2). Additionally, through the use of exact sequence variants36 we observed that Nitrosomonas was the most diverse nitrifying genus with 21 different ASVs detected versus only 4 for Nitrospira (Fig. S2). The diversity of these taxa could have played a role in their observed resistance, resilience and recovery after disturbance, which was higher for the more diverse Nitrosomonas genus than for the less diverse Nitrospira (Fig. 6). The reduction in Nitrospira abundances in the high F:M–C:N reactors (Fig. 3) could have been due to competition for DO with heterotrophs and Nitrosomonas that possess a higher affinity for oxygen37. It is known that AOB have a higher oxygen affinity than NOB such as Nitrobacter38,39. However, some studies showed that Nitrospira can occur in deep layers of biofilms40 and thrive under low DO conditions30,41, suggesting that at least some Nitrospira may have a high affinity for oxygen. Still, the increase in aeration rates from d97 onward did not prevent nitrite accumulation, indicating that a low DO in the system was not the main reason behind our observations. DO concentrations higher than 1 mg/L are enough to achieve optimal nitrification performance42, which was the case for the reactors in this study (Fig. 5). However, nitrifying communities grow in stratified biofilms where AOB are located closer to the water interface and NOB are in the interior zone43,44. The concentration of oxygen deep inside a biofilm or floc is lower than in the mixed liquor. Moreover, stratification in AOB biofilms due to an increase in C:N has been reported37, highlighting that increases in biofilm thickness due to heterotrophic growth further reduce oxygen diffusion inside, which is detrimental to the growth of NOB. In our study, the period during the aerobic phase of a cycle when almost all ammonium had been removed and COD concentrations were either low or remained constant (around 400–500 min, Fig. 5) implies that heterotrophs and AOB were not competing with NOB for oxygen anymore. Although this should have provided sufficient oxygen to NOB to be active during the remainder of the aerobic phase, an increase in nitrate production was not observed. A possible reason for this could be nitrite accumulation, which was reported to be toxic to NOB at high concentrations45. Hence, the observed NOB reduction at high F:M–C:N could have been due to a combination of competition for DO with heterotrophs and Nitrosomonas, reduced oxygen diffusion into the nitrifier biofilm due to heterotrophic growth, and nitrite accumulation due to AOB activity. Further research is needed to understand the underlying mechanism of why a high F:M–C:N leads to a reduction in NOB abundance and a concomitant accumulation of nitrite in bioreactors.
AOB growth rates are normally higher than those of NOB at 30 °C, which implies that the SRT can be reduced to achieve partial nitrification46. In our study, increasing F:M while keeping TSS constant implied an SRT reduction of 35% in the high F:M–C:N reactors compared to the low F:M–C:N reactors. It is conceivable that part of the observed reduction in nitrifiers was due to washout given their low growth rates. It was suggested based on mathematical modelling that a reduction in SRT has a stronger effect on NOB than on AOB47. However, the aerobic SRT of 5.1 d used to operate the high F:M reactors is common in activated sludge processes performing complete nitrification19,48, and around the operating SRT of 5–6 d at the full-scale plant that provided the sludge inoculum for this study. SRT values of 4–8 d have been suggested as optimum for nitrification in practice48,49, while complete nitrification has been reported for SRT values as low as 2 d50. Thus, our observed changes in nitrifiers and nitrification function were driven by controlled changes in F:M and C:N values and not by washout of NOB due to a low SRT.
Importance of parameter covariations for nitrification studies
We showed that a combined high F:M–C:N approach led to stable and reproducible nitrite accumulation (77%) after seven days of transition. A comparison with earlier studies where either F:M or C:N was the parameter of interest reveals inconsistent outcomes. For example, similar to our results, conditions at high F:M resulted in higher nitrite accumulation compared to low F:M in studies on full-scale sludge systems that focused on the effect of varying F:M directly51 or indirectly through changes in SRT52,53. Likewise, nitrite accumulation was found at high influent C:N (10 mg COD/mg NH4+-N) in a pilot-scale study using a completely stirred tank reactor30. However, contrary to our results, low influent C:N values (1–3 mg COD/mg NH4+-N) in high-strength industrial wastewaters were reported to yield partial nitrification in a review of full-scale anammox processes46. Also, nitrite accumulation was reported at low C:N values in studies using a 35-L SBR (~ 2 mg COD/mg TN)27, a lab-scale 6.3-L SBR (3.33 mg COD/mg NH4+-N)29, and a pilot-scale continuous-flow A/O/A reactor (3.19 mg COD/mg NH4+-N)28. On the other hand, there was complete nitrification without nitrite accumulation at low influent C:N (3 ± 1 mg COD/mg TN) in pilot-scale membrane batch reactors26, similar to our results. Increasing C:N molar ratios from 2 to 5 was shown to significantly reduce nitrification rates in a laboratory denitrification-nitrification system54. Moreover, nitrite accumulation was even reported for fluctuating low and high influent C:N values (2.5–8.0 mg COD/mg NH4+-N) in two different pilot-scale studies using SBRs31,32. These multiple conflicting findings in the literature suggest that other factors in addition to C:N variations affect nitrification. As changes in C:N also affect F:M values by altering the available carbon for heterotrophic growth, we recommend to evaluate these interconnected parameters simultaneously. Further studies employing different bioreactor configurations and influent types (including raw sewage) are required to assess the repeatability of the use of a high F:M–C:N strategy to achieve stable nitrite accumulation in sludge bioreactors.
Further, control of TSS is critical to operational control strategies in practice, based on either F:M or SRT48,49. In our study design, doubling the amount of COD in the influent almost doubled both F:M and C:N values, as operational TSS and influent TKN were controlled to remain close to constant. Doubling the influent COD also doubled the OLR, and higher COD concentrations also increased the biomass produced per unit time; thus more sludge had to be wasted to keep the TSS constant, reducing the SRT as a consequence (details in supplementary information). This point serves to illustrate that important operational parameters like SRT, C:N and OLR also co-vary with F:M48; thus it is important to account for such covariations during experimentation with sludge bioreactor systems.
Recovery and resilience of nitrite oxidation and nitrifiers
Since disturbances often occur in biotechnological systems, it is desirable to understand the mechanisms of recovery after disturbance55, for which experimental replication is important to ensure reproducibility9 and capture fluctuations in process instability56. Hence, we tested whether returning the press-disturbed reactors to their previous low F:M–C:N level would lead to recovery and resilience. The switch back had significant effects on the nitrite oxidation function and the abundances of nitrifier communities and nitrification genes. Relative abundances of Nitrosomonas genera recovered quickly; yet high F:M–C:N reactors exhibited variable functional resilience because after ten days of returning to pre-disturbance conditions, only two of three reactors completely recovered the nitrification function. This inconsistency among independent replicate reactors could be due to the stochastic growth after disturbance typically associated with r-strategists57 and ruderal organisms58. According to the r/K ecological framework, early niche colonization stages should favour r-strategists, whereas K-strategies should prevail at a later stage when many organisms attempt to colonize59. In our study, the variable nitrification recovery after removing the high F:M–C:N disturbance seemed to be due to stochastic colonization by r-strategist NOB (Nitrobacter), replacing the function performed by K-strategist NOB (Nitrospira) before the disturbance. Besides having a fast growth rate60 and prevailing at alternating conditions61, Nitrobacter has been shown to thrive at higher DO62 and nitrite51 concentrations, which explains why its abundance increased in the previously high F:M–C:N reactors after the shift to low F:M–C:N conditions. However, as part of a secondary succession process after disturbance, it is possible that Nitrospira would have recovered as a dominant NOB if more time had been allowed in the study. Further, these reactors constituted a closed system, which implies that the Nitrobacter colonizers came from low-abundance seed-bank populations. Disturbance was shown to open niches for bacterial colonization in open sludge systems63. Here, we showed that recruitment of organisms from the existing seed-bank is also possible after disturbance. As immigration of nitrifiers into sludge systems has been shown to alter the local community composition and function64,65, studies of the effect of disturbance in closed sludge systems can help disentangle the effect of immigrating populations.
In this study, functional resilience differed from nitrifying community resilience. Reactors with recovered function after returning to low F:M–C:N conditions (Fig. 5) still remained distinct from the control reactors in terms of NOB composition (Fig. 3). The fact that an altered community can perform the same functions as the original one supports the idea of functional redundancy66. This finding is similar to what was found in a full-scale sludge system after a press disturbance53, but contrary to what was reported for lake microbial systems after a pulse disturbance67. These contrasting reports highlight the complexity of assessing disturbance-diversity-function relationships68, as they depend not only on the system assessed but also on the disturbance frequency (i.e., pulse or press). Finally, the variability in the recovery of the nitrification function and fluctuations in nitrifier populations could only be captured thanks to the replication employed in the design of our study. As we move towards stable operation for microbial resource management55, future disturbance studies on sludge bioreactors should be robustly designed to ensure process reproducibility and highlight operational ranges where functional variability can be encountered. Overall, this study exemplifies how controlled disturbance studies on sludge communities using parameters within the range of plant operation can lead to insights that are both ecologically and practically meaningful.
Methods
Experimental design
The study was conducted using seven 5-L bioreactors inoculated with activated sludge from a water reclamation plant in Singapore and operated as sequencing batch reactors (SBR) on continuous 12-h cycles with intermittent aeration (Fig. 1). Initially, four reactors were acclimated to lab conditions and fed with complex synthetic wastewater for 53 days. The complex synthetic feed was adapted from Hesselmann et al.69. At the start of the experiment (d54), the sludge of the acclimation reactors was thoroughly mixed and redistributed across eight reactors. From these, four were randomly selected and designated as high F:M and C:N reactors, receiving double the carbon substrate in terms of chemical oxygen demand (COD) amount in its feed as a press disturbance for 60 days. One of these reactors broke prematurely, reducing the count to three replicates. The remaining four reactors were operated as before at low F:M and C:N. During the last two weeks of the study (d114–d127), the feed for the high F:M and C:N reactors was adjusted to equal that of low F:M and C:N reactors (Table 1). For the sake of simplicity, we refer to both F:M and C:N as F:M–C:N. Details about sludge inoculum, acclimation phase and complex synthetic wastewater preparation are available as supplementary information.
Operational parameters
The reactor temperature was maintained at 30 °C and sludge was continuously mixed with a magnetic stirrer. In each cycle, SBR phases were: 5 min feed, 200 min anoxic/anaerobic react, 445 min aerobic react, 50 min sludge settle, and 20 min supernatant drain. The DO concentration was controlled at 2–6 mg/L during the aerobic phase. The pH ranged from 6 to 9, owing to alkalinity provided in the feed. After sludge settling the supernatant effluent was discharged (about half the working volume of each reactor, or 2.5 L), followed by the replacement of the same volume with synthetic wastewater during the feeding phase at the beginning of the next 12-h cycle. Two cycles per day corresponded to a hydraulic retention time (HRT) of 24 h. Effluent and influent compositions were measured 2–3 times per week in accordance with Standard Methods70. The targets were soluble COD, total alkalinity, and nitrogen species (ammonium, nitrite and nitrate) in the liquid phase using colorimetric tests and ion chromatography. Nitrite accumulation percentage in the effluent was calculated as the ratio of nitrite concentration and the sum of nitrate and nitrite concentrations. Total organic carbon and total Kjeldahl nitrogen (TKN) were also measured in the influent. To control the F:M, sludge biomass was measured as total (TSS) and volatile suspended solids (VSS) twice a week, after which sludge wastage was done to target 1500 mg/L of TSS. Sludge volume index (SVI) was calculated from the liquid and sludge volumes measured in the reactors after 30 min settling and the TSS values obtained in the same cycle. Microbial community function was also investigated in the form of intensive sampling (every 30–60 min) over seven 12-h cycle studies conducted throughout the experiment. Detailed equations and explanations for F:M, C:N and SRT calculations, as well as analytical methods, are available as supplementary information.
Bioreactor arrangement
Each of the SBRs employed in this study was equipped with: a magnetic stir plate to ensure mixed liquor homogeneity, a pair of EasySense pH and DO probes with their corresponding transmitters (Mettler Toledo), a dedicated air pump, a dedicated feed pump, a solenoid valve for supernatant discharge, and a surrounding water jacket connected to a re-circulating water heater. The different portions of the cycle were controlled by a computer software specifically designed for these reactors (VentureMerger, Singapore).
16S rRNA amplicon sequencing and reads processing
Bacterial 16S rRNA amplicon sequencing was done in two steps (for details see Supplementary Information). Primer set 341f/785r targeted the V3–V4 variable regions of the 16S rRNA gene71. The libraries were sequenced on an Illumina MiSeq platform (v.3) with 20% PhiX spike-in and at a read-length of 300 bp paired-end. Sequenced sample libraries were processed following the DADA2 bioinformatics pipeline72 using the version 1.3.3 of the dada2 R-package. DADA2 allows inference of exact amplicon sequence variants (ASVs) providing several benefits over traditional OTU clustering methods36. Illumina sequencing adaptors and PCR primers were trimmed prior to quality filtering. Sequences were truncated after 280 and 255 nucleotides for forward and reverse reads, respectively, length at which average quality dropped below a Phred score of 20. After truncation, reads with expected error rates higher than 3 and 5 for forward and reverse reads were removed. After filtering, error rate learning, ASV inference and denoising, reads were merged with a minimum overlap of 20 bp. Chimeric sequences (0.18% on average) were identified and removed. For a total of 104 samples, 19,679 reads were kept on average per sample after processing, representing 49.2% of the average input reads. Taxonomy was assigned using the SILVA database (v.132)73. Adequacy of sequencing depth after reads processing was corroborated with rarefaction curves at the ASV level (Fig. S3).
Metagenomics sequencing and reads processing
Libraries were sequenced in one lane on an Illumina HiSeq2500 sequencer in rapid mode at a final concentration of 11 pM and a read-length of 250 bp paired-end. In total, around 325 million paired-end reads were generated, with 3.4 ± 0.4 million paired-end reads on average per sample (total 48 samples). Illumina adaptors, short reads, low quality reads or reads containing any ambiguous bases were removed using cutadapt74. High quality reads (91.0 ± 1.4% of the raw reads) were randomly subsampled to an even depth of 4,678,535 for each sample prior to further analysis. Taxonomic assignment of metagenomics reads was done following the method described by Ilott et al.75. High quality reads were aligned against the NCBI non-redundant (NR) protein database (March 2016) using DIAMOND76 with default parameters. The lowest common ancestor approach implemented in MEGAN Community Edition v.6.5.577 was used to assign taxonomy to the NCBI-NR aligned reads with the following parameters (maxMatches = 25, minScore = 50, minSupport = 20, paired = true). On average, 36.8% of the high-quality reads were assigned to cellular organisms, of which 98.4% were assigned to the bacterial domain. Adequacy of sequencing depth was corroborated with rarefaction curves at the genus taxonomic level (Fig. S3). Identification and quantification of genes involved in nitrogen metabolism was performed using SqueezeMeta pipeline78. Read pairs were co-assembled using Megahit79. Open reading frames (ORFs) were then predicted from contigs using Prodigal80. Functional annotation was performed using DIAMOND76 against the KEGG database81. Read mapping against contigs was then performed using Bowtie282 in order to quantify the abundance of genes among the different samples, and transformed in transcripts per million (tpm) values to be consistent with prior works83. Nitrification genes were then quantified as follows: amo as the tpm sum from entries K10944 (pmoA-amoA), K10945 (pmoB-amoB) and K10946 (pmoC-amoC); hao as the tpm from entry K10535; and nxr/nar as tpm sum from entries K00370 (narG, narZ, nxrA) and K00371 (narH, narY, nxrB).
Stability measures
Stability metrics of resistance, resilience and recovery were calculated for each replicate reactor at the disturbed high F:M–C:N level (H1, H2 and H3) as described by Hillebrand et al.4. Metrics were applied to the nitrite oxidation function, measured as NO3–N production, and to the relative abundances of the main nitrifier genera Nitrosomonas, Nitrospira and Nitrobacter, detected via 16S rRNA gene amplicon sequencing. Resistance (a) was measured as the initial log response ratio of a parameter (Fi) from each disturbed replicate versus the average value of the undisturbed reactors: a = ln(Fi,H1-3/Fi,L). Recovery (b) was measured in the same way but as the final log response ratio, after disturbance was removed. Resilience (c) was measured as the slope of regression of the aforementioned log response ratio over the recovery time, following disturbance removal.
Statistical analyses
Welch’s ANOVA was employed for univariate testing. All reported p-values were corrected for multiple comparisons using a False Discovery Rate (FDR) of 5%84. Regression analyses were performed in R v3.5.1 using the ggplot2 package.
Data availability
DNA sequencing data are available at NCBI BioProjects PRJNA559245. See supplementary information for details about sludge inoculum and acclimation phase, complex synthetic wastewater preparation, chemical analysis, calculation of parameters (F:M, C:N, SRT), DNA extraction and purification, and 16S rRNA gene and metagenomics library preparation and sequencing.
References
Osborn, D., Cutter, A. & Ullah, F. in Stakeholder Forum, Commissioned by the UN Development Program. Geneva, Switzerland.
Cain, M., Bowman, W. & Hacker, S. Ecology 3rd edn. (Sinauer Associates Inc., Sunderland, 2014).
Donohue, I. et al. On the dimensionality of ecological stability. Ecol. Lett. 16, 421–429 (2013).
Hillebrand, H. et al. Decomposing multiple dimensions of stability in global change experiments. Ecol. Lett. 21, 21–30 (2018).
Briones, A. & Raskin, L. Diversity and dynamics of microbial communities in engineered environments and their implications for process stability. Curr. Opin. Biotechnol. 14, 270–276 (2003).
Wang, Q., Ding, C., Tao, G. & He, J. Analysis of enhanced nitrogen removal mechanisms in a validation wastewater treatment plant containing anammox bacteria. Appl. Microbiol. Biotechnol. 103, 1255–1265 (2019).
Zhou, J. & Ning, D. Stochastic community assembly: does it matter in microbial ecology?. Microbiol. Mol. Biol. Rev. 81, 1–32 (2017).
Santillan, E., Seshan, H., Constancias, F., Drautz-Moses, D. I. & Wuertz, S. Frequency of disturbance alters diversity, function, and underlying assembly mechanisms of complex bacterial communities. NPJ Biofilms Microbiomes 5, 1–8 (2019).
Prosser, J. I. Replicate or lie. Environ. Microbiol. 12, 1806–1810 (2010).
Bender, E. A., Case, T. J. & Gilpin, M. E. Perturbation experiments in community ecology: theory and practice. Ecology 65, 1–13 (1984).
Shade, A. et al. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 3, 1–19 (2012).
Botton, S., van Heusden, M., Parsons, J. R., Smidt, H. & van Straalen, N. Resilience of microbial systems towards disturbances. Crit. Rev. Microbiol. 32, 101–112 (2006).
Rykiel, E. J. Towards a definition of ecological disturbance. Aust. J. Ecol. 10, 361–365 (1985).
Hu, B., Wheatley, A., Ishtchenko, V. & Huddersman, K. The effect of shock loads on SAF bioreactors for sewage treatment works. Chem. Eng. J. 166, 73–80 (2011).
Bassin, J. P. et al. Effect of increasing organic loading rates on the performance of moving-bed biofilm reactors filled with different support media: assessing the activity of suspended and attached biomass fractions. Process Saf. Environ. Prot. 100, 131–141 (2016).
Seetha, N., Bhargava, R. & Kumar, P. Effect of organic shock loads on a two-stage activated sludge-biofilm reactor. Bioresour. Technol. 101, 3060–3066 (2010).
Ketheesan, B. & Stuckey, D. C. Effects of hydraulic/organic shock/transient loads in anaerobic wastewater treatment: a review. Crit. Rev. Environ. Sci. Technol. 45, 2693–2727 (2015).
Senturk, E., Ince, M. & Onkal Engin, G. The effect of shock loading on the performance of a thermophilic anaerobic contact reactor at constant organic loading rate. J. Environ. Health Sci. Eng. 12, 1–6 (2014).
Gray, N. F. Biology of Wastewater Treatment 2nd edn, Vol. 4 (Imperial College Press, London, 2004).
Laureni, M. et al. Mainstream partial nitritation and anammox: long-term process stability and effluent quality at low temperatures. Water Res. 101, 628–639 (2016).
Wang, Q. & He, J. Newly designed high-coverage degenerate primers for nitrogen removal mechanism analysis in a partial nitrification-anammox (PN/A) process. FEMS Microbiol. Ecol. 96, fiz202 (2019).
Ma, B. et al. Suppressing nitrite-oxidizing bacteria growth to achieve nitrogen removal from domestic wastewater via anammox using intermittent aeration with low dissolved oxygen. Sci. Rep. 5, 1–9 (2015).
Sinha, B. & Annachhatre, A. P. Partial nitrification—operational parameters and microorganisms involved. Rev. Environ. Sci. Bio. Technol. 6, 285–313 (2007).
Okabe, S., Oozawa, Y., Hirata, K. & Watanabe, Y. Relationship between population dynamics of nitrifiers in biofilms and reactor performance at various C:N ratios. Water Res. 30, 1563–1572 (1996).
Ge, S. et al. Detection of nitrifiers and evaluation of partial nitrification for wastewater treatment: a review. Chemosphere 140, 85–98 (2015).
Ma, J. et al. Analysis of nitrification efficiency and microbial community in a membrane bioreactor fed with low COD/N-ratio wastewater. PLoS ONE 8, 1–10 (2013).
Tan, C., Ma, F. & Qiu, S. Impact of carbon to nitrogen ratio on nitrogen removal at a low oxygen concentration in a sequencing batch biofilm reactor. Water Sci. Technol. 67, 612–618 (2012).
Zhang, T. et al. Achieving partial nitrification in a continuous post-denitrification reactor treating low C/N sewage. Chem. Eng. J. 335, 330–337 (2018).
She, Z. et al. Partial nitrification and denitrification in a sequencing batch reactor treating high-salinity wastewater. Chem. Eng. J. 288, 207–215 (2016).
Regmi, P. et al. Control of aeration, aerobic SRT and COD input for mainstream nitritation/denitritation. Water Res. 57, 162–171 (2014).
Ge, S., Peng, Y., Qiu, S., Zhu, A. & Ren, N. Complete nitrogen removal from municipal wastewater via partial nitrification by appropriately alternating anoxic/aerobic conditions in a continuous plug-flow step feed process. Water Res. 55, 95–105 (2014).
Jiang, H. et al. A pilot-scale study on start-up and stable operation of mainstream partial nitrification-anammox biofilter process based on online pH-DO linkage control. Chem. Eng. J. 350, 1035–1042 (2018).
Pimm, S. L. The complexity and stability of ecosystems. Nature 307, 321 (1984).
Santillan, E., Constancias, F. & Wuertz, S. Press disturbance alters community structure and assembly mechanisms of bacterial taxa and functional genes in mesocosm-scale bioreactors. mSystems 5, e00471–e00420 (2020).
Nowka, B., Daims, H. & Spieck, E. Comparison of oxidation kinetics of nitrite-oxidizing bacteria: nitrite availability as a key factor in niche differentiation. Appl. Environ. Microbiol. 81, 745–753 (2015).
Callahan, B. J., McMurdie, P. J. & Holmes, S. P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643 (2017).
Okabe, S., Aoi, Y., Satoh, H. & Suwa, Y. Nitrification. In Nitrification in Wastewater Treatment (eds Ward, B. B. et al.) 405–418 (ASM Press, Washington, DC, 2011).
Blackburne, R., Yuan, Z. & Keller, J. Partial nitrification to nitrite using low dissolved oxygen concentration as the main selection factor. Biodegradation 19, 303–312 (2008).
Garrido, J. M., van Benthum, W. A. J., van Loosdrecht, M. C. M. & Heijnen, J. J. Influence of dissolved oxygen concentration on nitrite accumulation in a biofilm airlift suspension reactor. Biotechnol. Bioeng. 53, 168–178 (1997).
Almstrand, R., Daims, H., Persson, F., Sörensson, F. & Hermansson, M. New methods for analysis of spatial distribution and coaggregation of microbial populations in complex biofilms. Appl. Environ. Microbiol. 79, 5978–5987 (2013).
Law, Y. et al. High dissolved oxygen selection against nitrospira sublineage I in full-scale activated sludge. Environ. Sci. Technol. 53, 8157–8166 (2019).
Gonzalez, C., Garcia, P. A. & Munoz, R. Effect of feed characteristics on the organic matter, nitrogen and phosphorus removal in an activated sludge system treating piggery slurry. Water Sci. Technol. 60, 2145–2152 (2009).
Lydmark, P., Lind, M., Sörensson, F. & Hermansson, M. Vertical distribution of nitrifying populations in bacterial biofilms from a full-scale nitrifying trickling filter. Environ. Microbiol. 8, 2036–2049 (2006).
Okabe, S., Satoh, H. & Watanabe, Y. In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 65, 3182–3191 (1999).
Anthonisen, A., Loehr, R., Prakasam, T. & Srinath, E. Inhibition of nitrification by ammonia and nitrous acid. Journal (Water Pollut. Control Fed.), 835–852 (1976).
Lackner, S. et al. Full-scale partial nitritation/anammox experiences: an application survey. Water Res. 55, 292–303 (2014).
Wu, J., He, C., van Loosdrecht, M. C. M. & Pérez, J. Selection of ammonium oxidizing bacteria (AOB) over nitrite oxidizing bacteria (NOB) based on conversion rates. Chem. Eng. J. 304, 953–961 (2016).
Tchobanoglous, G. B., Franklin, L. & Stensel, H. D. Wastewater engineering: treatment and reuse 4th edn. (McGraw Hill, New York, 2003).
Smith, R. C., Elger, S. O. & Mleziva, S. Implementation of solids retention time (SRT) control in wastewater treatment. Xylem Anal. 20, 1–6 (2015).
Simsek, H., Kasi, M., Ohm, J.-B., Murthy, S. & Khan, E. Impact of solids retention time on dissolved organic nitrogen and its biodegradability in treated wastewater. Water Res. 92, 44–51 (2016).
Wu, Y.-J. et al. Impact of food to microorganism (F/M) ratio and colloidal chemical oxygen demand on nitrification performance of a full-scale membrane bioreactor treating thin film transistor liquid crystal display wastewater. Bioresour. Technol. 141, 35–40 (2013).
Meerburg, F. A. et al. High-rate activated sludge communities have a distinctly different structure compared to low-rate sludge communities, and are less sensitive towards environmental and operational variables. Water Res. 100, 137–145 (2016).
Vuono, D. C. et al. Disturbance and temporal partitioning of the activated sludge metacommunity. ISME J. 9, 425–435 (2015).
Ballinger, S. J., Head, I. M., Curtis, T. P. & Godley, A. R. The effect of C/N ratio on ammonia oxidising bacteria community structure in a laboratory nitrification-denitrification reactor. Water Sci. Technol. 46, 543–550 (2002).
Cabrol, L. et al. Management of microbial communities through transient disturbances enhances the functional resilience of nitrifying gas-biofilters to future disturbances. Environ. Sci. Technol. 50, 338–348 (2016).
Wells, G. F. et al. Comparing the resistance, resilience, and stability of replicate moving bed biofilm and suspended growth combined nitritation–anammox reactors. Environ. Sci. Technol. 51, 5108–5117 (2017).
Pianka, E. R. R-selection and K-selection. Am. Nat. 104, 592–579 (1970).
Santillan, E., Seshan, H., Constancias, F. & Wuertz, S. Trait-based life-history strategies explain succession scenario for complex bacterial communities under varying disturbance. Environ. Microbiol. 21, 3751–3764 (2019).
Macarthur, R. H. & Wilson, E. O. The Theory of Island Biogeography 224 (Princeton, Princeton University Press, 1967).
Blackburne, R., Vadivelu, V. M., Yuan, Z. & Keller, J. Kinetic characterisation of an enriched Nitrospira culture with comparison to Nitrobacter. Water Res. 41, 3033–3042 (2007).
Dytczak, M. A., Londry, K. L. & Oleszkiewicz, J. A. Activated sludge operational regime has significant impact on the type of nitrifying community and its nitrification rates. Water Res. 42, 2320–2328 (2008).
Huang, Z., Gedalanga, P. B., Asvapathanagul, P. & Olson, B. H. Influence of physicochemical and operational parameters on Nitrobacter and Nitrospira communities in an aerobic activated sludge bioreactor. Water Res. 44, 4351–4358 (2010).
Vuono, D. C., Munakata-Marr, J., Spear, J. R. & Drewes, J. E. Disturbance opens recruitment sites for bacterial colonization in activated sludge. Environ. Microbiol. 18, 87–99 (2016).
Jauffur, S., Isazadeh, S. & Frigon, D. Should activated sludge models consider influent seeding of nitrifiers? Field characterization of nitrifying bacteria. Water Sci. Technol. 70, 1526–1532 (2014).
Yu, L. et al. Natural continuous influent nitrifier immigration effects on nitrification and the microbial community of activated sludge systems. J. Environ. Sci. 74, 159–167 (2018).
Allison, S. D. & Martiny, J. B. H. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA 105, 11512–11519 (2008).
Shade, A. et al. Lake microbial communities are resilient after a whole-ecosystem disturbance. ISME J. 6, 2153–2167 (2012).
Santillan, E. Disturbance-Performance-Diversity Relationships and Microbial Ecology in Bioreactors for Wastewater Treatment. Ph.D. thesis, University of California, Davis (2018).
Hesselmann, R. P. X., Werlen, C., Hahn, D., van der Meer, J. R. & Zehnder, A. J. B. Enrichment, phylogenetic analysis and detection of a bacterium that performs enhanced biological phosphate removal in activated sludge. Syst. Appl. Microbiol. 22, 454–465 (1999).
APHA-AWWA-WEF. Standard Methods for the Examination of Water and Wastewater 22nd edn. (AWWA, Mumbai, 2005).
Thijs, S. et al. Comparative evaluation of four bacteria-specific primer pairs for 16S rRNA gene surveys. Front. Microbiol. 8, 1–15 (2017).
Callahan, B. J. et al. DADA2: High resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Glöckner, F. O. et al. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 261, 169–176 (2017).
Chen, C., Khaleel, S. S., Huang, H. & Wu, C. H. Software for pre-processing Illumina next-generation sequencing short read sequences. Sour. Code Biol. Med. 9, 8–8 (2014).
Ilott, N. E. et al. Defining the microbial transcriptional response to colitis through integrated host and microbiome profiling. ISME J. 10, 2389–2404 (2016).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Huson, D. H. et al. MEGAN community edition: interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comp. Biol. 12, 1–12 (2016).
Tamames, J. & Puente-Sánchez, F. SqueezeMeta, a highly portable, fully automatic metagenomic analysis pipeline. Front. Microbiol. 9 (2019).
Li, D., Liu, C.-M., Luo, R., Sadakane, K. & Lam, T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676 (2015).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11, 119 (2010).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Puente-Sánchez, F., García-García, N. & Tamames, J. SQMtools: automated processing and visual analysis of ’omics data with R and anvi’o. BMC Bioinform. 21, 358 (2020).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Ser. B. (Method.) 57, 289–300 (1995).
Acknowledgements
This research was supported by the Singapore National Research Foundation and Ministry of Education under the Research Centre of Excellence Program. We thank D.I. Drautz-Moses for her support with the 16S rRNA gene amplicon and metagenomics library preparation and sequencing pipelines employed, and T.J. Qiang and N.A.B.A. Latiff for their assistance with molecular work. S.S. Thi and A.F.B.M. Batcha are acknowledged for their support with analytical chemistry equipment. We thank J. Coppens for insightful discussions on nitrification. E.S. was partially supported by a Fulbright Fellowship.
Author information
Authors and Affiliations
Contributions
E.S. and S.W. conceived the study. E.S. designed the experiment. S.W. obtained the funding for the study. E.S. and W.X.P. performed the experiments and conducted laboratory and molecular analyses (except library preparation and sequencing). E.S. performed the 16S rRNA gene bioinformatics analyses. F.C. performed the metagenomics bioinformatics analyses. E.S. interpreted the data, generated the results, and elaborated the main arguments in the manuscript. E.S. and S.W. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Santillan, E., Phua, W.X., Constancias, F. et al. Sustained organic loading disturbance favors nitrite accumulation in bioreactors with variable resistance, recovery and resilience of nitrification and nitrifiers. Sci Rep 10, 21388 (2020). https://doi.org/10.1038/s41598-020-78314-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-78314-1
This article is cited by
-
Microbial community-based protein from soybean-processing wastewater as a sustainable alternative fish feed ingredient
Scientific Reports (2024)
-
Microbiome assembly predictably shapes diversity across a range of disturbance frequencies in experimental microcosms
npj Biofilms and Microbiomes (2022)
-
Disturbance-based management of ecosystem services and disservices in partial nitritation-anammox biofilms
npj Biofilms and Microbiomes (2022)
-
PCR based fingerprinting techniques to transpire the microbial populations diversity of biofilm based nitrifying packed bed bioreactor in the bioremediation of aquaculture wastewater
Green Technology, Resilience, and Sustainability (2022)
-
Relevance of Candidatus Nitrotoga for nitrite oxidation in technical nitrogen removal systems
Applied Microbiology and Biotechnology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.