Abstract
Biogeography is the study of where, when, and how modern species evolved and diversified. Acer L. (maple) is one of the most diverse and widespread genera in the Northern Hemisphere. It comprises 124–156 species in the world, approximately 80% species of Acer are native in Asia. The current diversity center of Acer is not congruent with the distribution of the oldest fossils of the genus. Therefore, we herein used 84 species and subspecies to reconstruct the phylogeny and investigate the biogeographic history of Acer using nuclear ITS and three cpDNA fragments (psbA-trnH spacer, rpl16 intron, and trnL-trnF spacer) with maximum likelihood, maximum parsimony, and Bayesian inference methods. The analyses showed that the current diversity center and the origin center of Acer is Asia. Additionally, the North American and Euro-Mediterranean species originated from multiple sources from Asia via the North Atlantic Land Bridge and the Bering Land Bridge, and intercontinental migration has mainly occurred since the Miocene. This study not only provides a novel insight of the origin and dispersal routes of Acer but also exemplifies how past climatic changes affect the diversification-rates of Northern Hemisphere forest trees.
Similar content being viewed by others
Introduction
The biogeographic studies of widely distributed plants can be performed to provide insight into the broader patterns of the evolutionary history and geographic diversification of the flora of the world1. The genus Acer L. forms an important part of the Northern Hemisphere forests, and it comprises ~ 129–156 species2,3. Most extant species of the genus are native to Asia (about 100 species), whereas others occur in North America, Europe, and North Africa2,3,4. Acer fossils dating from the Paleocene to more recent times have been found in the Northern Hemisphere5,6,7,8,9. The oldest described fossil of Acer is of fossilized fruits from the Paleocene collected in Alaska10,11 and is much older than the oldest Asian fossil, which is from the Miocene8. As the current biodiversity center of Acer and the distribution of the oldest fossil are incongruous3,4,7, the two possible explanations of the origin of Acer (from North American or Asia; i.e., the hypotheses ‘Out-of-North-America’ or ‘Out-of-Asia’) are evaluated in the present study as an attempt to explain the historical biogeography of Acer.
Previous studies on the evolutionary relationships and biogeography of Acer have been performed12,13,14,15,16,17,18,19. Wolfe20 suggested that Acer originated in western North America and many lineages spread to eastern Asia during the early Eocene. Hasebe et al.13 attributed the current East-Asian and North American distribution of Acer to multiple migrations. Renner et al.19 also suggested that extant North American species of Acer may have derived from their respective Asian clades at different times. However, the evolutionary trajectory of Acer is still ambiguous. Moreover, the fact that Acer would have experienced climatic oscillations and active orogeny since the Paleocene—for example, the Cenozoic climate cooling21, the establishment of the Asian monsoon22, and the uplift of the Qinghai-Tibetan Plateau23—as well as the Pleistocene climatic oscillations24 must be considered.
In addition, because of the large number of Acer species worldwide, insufficient sampling is often criticized. Incomplete sampling may lead to bias in molecular dating and ancestral area reconstruction25,26. Therefore, an increase in the number of samples of Asian species is necessary to ensure a comprehensive and representative sample size, which in turn would allow the verification of the previous systematic and biogeographic inferences made for the group3,27. Because of the possibility of introgression among closely related species of Acer28, we also used the nuclear ITS to infer the biogeographic history of Acer in addition to three chloroplast DNA (cpDNA) fragments (psbA-trnH spacer, rpl16 intron, and trnL-trnF spacer). Based on the results of phylogenetic analyses, we re-evaluate the taxonomic relationships of main sections and series of Acer, perform molecular dating with fossil corrections, and predict hypothetical distributions, the original center, and probable dispersal routes of Acer species.
Materials and methods
Taxon sampling
Eighty-four representative species or subspecies (including varieties) of Acer represent the vast majority of sections (except for Wardiana) were collected2. Among them, there are 11 subspecies (including varieties) derived from one species (different subspecies of the same species). In addition, sixty-seven species are native to Asia, eight to North America and six to Europe, two species are widely distributed in Europe and Asia, and one species is distributed in North America and Asia (Table S1).
Species identification and classification followed the Maples of the World2. Aesculus L. and Dipteronia Oliv. were chosen as outgroups for phylogenetic analyses29,30. Detailed sampling information is listed in Table S1.
Molecular methods
Total genomic DNA was extracted from silica-dried leaves using a modified CTAB method31. The primer pairs used for PCR amplification and sequencing are listed in Table S2. PCR amplification was conducted in a LABNET MultiGene 96-well gradient thermal cycler, with a reaction volume of 20 μL containing 10–30 ng of DNA template, 50 mM Tris–HCl, 1.5 mM MgCl2, 1 mM dNTPs, 0.3 μmol/L of each primer pair, and 0.2 U of Taq DNA polymerase (Bernardo Biotech Co., Taiwan). PCR cycles were as follows: 3 min at 94 °C, 30 cycles of 30 s at 94 °C, 30 s at 50–58 °C, and 45–80 s at 72 °C, with a final elongation for 10 min at 72 °C. The PCR products were separated by agarose gel electrophoresis and purified with a Gel Band Purification Kit (TIANGEN Biotech Co., Beijing, China). The purified PCR products were directly sequenced with the PCR primers using the ABI 3730XL DNA analyzer (Applied Biosystems, USA). The sequences are deposited in GenBank under accession numbers KU500429–KU500554 and KU522488–KU522550.
Sequence alignment and phylogenetic analyses
Sequence contigs were assembled and edited using SeqMan implemented in DNAStar version 7.0. Sequence alignments were conducted using BioEdit version 7.1.1132 and manually refined. The 5ʹ and 3ʹ ends of sequences were trimmed to equal alignment lengths for subsequent phylogenetic analyses. The phylogenetic tree with nuclear ITS, three cpDNA fragments and a combination of four DNA fragments were reconstructed separately. A partition homogeneity test33 was performed to test the homogeneous evolutionary rates of four DNA fragments for ensuring the rationality of combining them as one locus. Combining sequences from different sources is a common practice in the phylogenetic analyses26,34,35.
Phylogenetic relationships were reconstructed under maximum parsimony (MP), maximum likelihood (ML), and Bayesian inference (BI) using PAUP*4.0b1036, GARLI v0.95137, and MrBayes 3.238, respectively. For the whole dataset, the best-fitting substitution model for the ML and BI analyses was inferred using Modeltest 3.739. In the MP analysis, all character states were treated as unordered and equally weighted, and a heuristic search was performed with 1000 replicates of random addition of sequences, tree-bisection-reconnection (TBR) branch-swapping, and MULTREES on. Bootstrap analysis was conducted with 1,000 replicates using the same heuristic search settings as described above. The ML analysis was initiated from a BIONJ tree, with support values for the nodes estimated by 1000 bootstrap replicates. In the Bayesian analyses, two independent Markov Chain Monte Carlo (MCMC) runs were initiated, each consisting of one cold and three heated MCMC chains that were run for 5,000,000 generations and sampled every 1000 generations. The first 1000 trees were discarded as burn-in to ensure that the chains had become stationary. The clades division in figures is mainly based on the clustering results of phylogenetic trees and the morphological classification of Acer.
Molecular clock dating
Two fossil calibrations were used to constrain (1) the divergence time of Acer and Dipteronia and (2) the crown age of Acer. Although Acer has a rich fossil record, most fossils are difficult to identify as close relatives of extant species because of the limited morphological differentiation among species. Because of a lack of radiometric dating for fossils, Renner et al.19 was followed to adjust the ages of the fossils to the midpoint of the currently assigned range for each respective geological Epoch or Stage. Therefore, the divergence time of Acer and Dipteronia and the crown age of Acer were calibrated to 62 million years ago (Mya) and 53 Mya with a shape parameter of 4.0 and 1.0, respectively. Divergence times were estimated using BEAST v.1.740, with a relaxed uncorrelated normal clock model of speciation. The MCMC analysis was run for 1,000,000,000 generations. Trees were sampled every 50,000 generations, with the first 4000 trees treated as burn-in.
Ancestral area reconstruction
BioGeoBEARS41 as implemented in R was used to infer the biogeographical history of Acer. This program allows estimation of ancestral ranges under different models such as DEC42,43, a likelihood interpretation of DIVA44 (DIVALIKE) or a likelihood interpretation of BayArea45 (BAYAREALIKE). Additionally, it implements a parameter describing founder-event speciation (+J, post-colonization cladogenesis) likely important in oceanic settings46 and allows the comparison of different models in a statistical framework. The analyses were carried out based on the BEAST Maximum Clade Credibility (MCC) tree with outgroups removed. The study species were mapped in four study areas: (A) Asia, (B) Euro-Mediterranean, (C) western North America, and (D) eastern North America. A random sample of 16,000 post-burn-in trees generated from the BEAST analysis for ancestral area reconstruction was used. The analysis was run on the chronogram obtained under the fossil-calibrated relaxed clock. Biogeographical data were coded according to the distribution of extant taxa included in this study.
Temporal-based diversification-rate-shift analyses
The temporal-based analysis that accumulates speciation events (lineages) through time was conducted for inferring the emergence time of species via the lineage-through-time (LTT) analysis. The LTT analysis was performed in R package (APE)47. Effective lineage sizes through time were also estimated using the reversible jump Markov Chain Monte Carlo (rjMCMC)48 by the function mcmc.popsize of APE with one-million-steps Markov chain simulations, and discarding the first 1,000 steps as burn-in, with a setting thinning factor = 1000 for both whole tree topologies.
Result
Sequence characteristics
Sequences with 533, 609, 995, and 1046 base pairs (bp) of ITS, psbA-trnH spacer, rpl16 intron, and trnL-trnF spacer were obtained, respectively. In the 3183 bp of combined sequences, 594 variable sites with 396 parsimonious informative sites were obtained when Aesculus and Dipteronia were used as outgroups.
Phylogenetic relationships
We reconstructed the phylogenetic trees with ITS (Fig. S1), three cpDNA fragments (Fig. S2) and a combination of four DNA fragments. These trees have consistent topologies with only slight differences within some subclades, which is probably due to few informative sites if reconstructing the phylogenetic tree by single genes only. In addition, non-significant incongruence was found in substitution rates among four DNA fragments by the partition homogeneity test (P = 0.08). Therefore, we showed the tree reconstructed by concatenated DNA fragments in Fig. 1. We used three approaches, ML, MP, and BI, to reconstruct the phylogenetic trees of four DNA fragments (Figs. 1, S3, S4). These approaches resulted in roughly congruent phylogenetic topologies except for slight differences in the grouping of certain tips (the best model for the ML and BI analyses was GTR+G+I). The ML tree with more polytomies represented a more conservative tree topology than the other approaches, and it is therefore the main object of this discussion (Fig. 1).
The best maximum likelihood tree combined nuclear ITS and three cpDNA fragments (psbA-trnH, rpl16 and trnL-trnF). The tree is rooted using Aesculus and Dipteronia as outgroups. Bootstrap values of MP, ML above 50% and posterior support value of BI above 0.5 are shown successively. The diagram was generated by Microsoft PowerPoint 2019 (https://www.microsoft.com/zh-cn/microsoft-365/powerpoint).
The result strongly supports the monophyly of Acer (72/92/100) with four major clades, namely clades A, B, C, and D (Fig. 1). In the A2 and A3 branches, the series Lithocarpa of section Lithocarpa formed a clade (80/100/100), whereas the other species belonged to section Macrantha (Fig. 1). Series Cissifolia of section Negundo (65/82/100) and series Arguta of section Glabra (99/100/100) formed monophyletic groups in branches B1 and B2, respectively. Branch B4 was mainly composed of the series Palmata and series Sinensia species of section Palmata. Branch C2 was mostly formed by section Platanoidea species with a relatively high support value (75/99/100). Branch D1 consisted of section Rubra (including several North American species, such as A. rubrum and A. saccharinum) and section Hyptiocarpa (100/100/100). Branch D2 was composed of section Ginnala (100/100/100) except A. carpinifolium. Branch D3 was composed of section Acer (including several European species, such as A. heldreichii, A. monspessulanum, and A. opalus) (88/87/100) except A. pseudoplatanus. Branch D4 was formed by section Trifoliata (58/66/100) except A. pentaphyllum. Branch D5 was mainly composed of the series Trifida of section Pentaphylla and series Penninervia of section Palmata (Fig. 1).
Divergence time estimation
Three main clades (I, II, and III) were inferred by the Bayesian chronogram BEAST (Fig. 2). The results of this analysis were similar to those of the ML, MP, and BI analyses with a slight difference in the placement of some statistically unsupported nodes (Figs. 1, S3, S4). The relaxed molecular clock analysis with two calibration points indicated that the estimated crown ages of clades I, II, and III are 46.03 Mya (95% Highest Posterior Density interval (HPD): 34.71–53.59), 44.50 Mya (95% HPD: 36.40–52.35), and 43.31 Mya (95% HPD: 36.54–50.51), respectively, and roughly in the middle Eocene. Moreover, it indicated that most of the sections and series of Acer originated from the end of the Oligocene to the early Miocene (e.g., sections Palmata and Macrantha) (Fig. 2).
Chronogram for Acer reconstructed based on the combined data matrix (nuclear ribosomal ITS, plastid psbA-trnH spacer, rpl16 intron, and trnL-trnF spacer). Stars indicate the calibration points inferred from fossil records. Numbers on nodes indicate time to the most recent common ancestor (MRCA). Each numbered node bar represents the minimum and maximum ranges of dates for MRCA summarized from both Low and High calibration. The diagram was generated by Microsoft PowerPoint 2019 (https://www.microsoft.com/zh-cn/microsoft-365/powerpoint).
The LTT plot showed a trend of exponential lineage growth in the width of the phylogenetic tree on a logarithmic scale suggesting that the diversity rate of all Acer species remained stable (Fig. 3A,C). Figure 3B,D shows the diversity rate diverging approximately 30 Mya among different distributions, with a notable decline in North America and Europe.
Temporal analysis of diversification rates. The lineage-through-time (LTT) of Acer species and reversible jump Markov chain Monte Carlo (rjMCMC) simulation of Acer (gray curves: bold line indicates mode value, and thin lines indicate 95% confidence intervals; left y-axis). (A) All species (dark curve, left y-axis). (B) Species from different regions (color curve, left y-axis). (C) All species except 11 subspecies (including varieties) which derived from one species (different subspecies of the same species) (dark curve, left y-axis). (D) Species (except 11 subspecies) from different regions (color curve, left y-axis). The diagrams of (A–D) were generated by Microsoft PowerPoint 2019 (https://www.microsoft.com/zh-cn/microsoft-365/powerpoint).
Biogeographic analyses
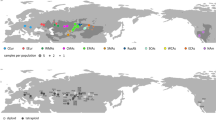
The best biogeographic model for the genus Acer as evaluated by BioGeoBears was DEC+J (AIC = 147.67, lnL = − 70.83) (Table 1). This analysis showed that the most probable ancestral area of Acer is Asia (region A), whereas the extant Euro-Mediterranean and North American species were distributed in different branches (Fig. 4). Moreover, the analysis showed that at least five independent dispersal events from Asia to North America in clades I–III are needed to explain the present differences in the Asia-North America distribution pattern (Fig. 4). The timing of most dispersal events was mainly in Miocene and a few in the Eocene (Fig. 4). In clades II, the effective number of lineages after 40 Mya in European and American species has declined, indicating that their number of species has not increased.
Ancestral area reconstruction for the genus Acer based on the combined data matrix (nuclear ribosomal ITS, plastid psbA-trnH spacer, rpl16 intron, and trnL-trnF spacer). The one- and two-letter codes in the inset represent different geographic ranges. The map shows the geographical extents of the four areas ((A) Asia, (B) Euro-Mediterranean, (C) western North America, (D) eastern North America) considered in the analysis. The pie charts on the nodes show the most likely ancestral areas (correspond to the colors in the inset) as reconstructed by BioGeoBEARS. Current distributions are indicated before the species names. Branches 1–5 represent five migration events of Acer. The diagram was generated by Microsoft PowerPoint 2019 (https://www.microsoft.com/zh-cn/microsoft-365/powerpoint).
Discussion
Phylogenetic relationships
Similar to the results of previous studies, the present results support the monophyly of Acer16,17,18,19 (Fig. 1). Based on leaf and fruit characteristics, Ogata49 defined three series in section Macrantha. However, the phylogenetic inference herein obtained in section Macrantha is divided into two parts in clade A (Fig. 1) does not support such taxonomic treatment; it is however congruent with the taxonomic proposal of Zhang et al.50. Series Palmata, Sinensia, and Penninervia of section Palmata in the B4 and D5 branches do not form a monophyletic group. Reciprocal paraphyly may reflect the frequent interbreeding among Acer species51 or the previous misclassification. Renner et al.19 found a phylogenetic admixture between species of sections Rubra and Hyptiocarpa based on cpDNA only. However, both ITS and cpDNA used in the present study suggest a monophyly of section Rubra separate from section Hyptiocarpa (Fig. 1). These incongruent phylogenetic inferences may be a result of incomplete lineage sorting caused by ancestral interbreeding. The Euro-Mediterranean and North America species did not form the monophyletic group (Fig. 1), which is consistent with the results of Renner et al.19. This may indicate the multiple origins of the Euro-Mediterranean and North America species.
Diversification rate of Acer
The diversification rates of Acer in the Euro-Mediterranean and North American regions began to decline approximately 30 Mya (mid-Oligocene) (Fig. 3B,D). This may have been caused by the limitation of migration in the Euro-Mediterranean and North American regions because of global climate cooling with the onset of permanent Antarctic ice sheets52. Compared to Asia, the Euro-Mediterranean and North America regions have a low number of species, which may also contribute to the decline of species diversification. In addition, most areas of Euro-Mediterranean and North America were covered by ice sheets during glacial periods; this may have led to a higher extinction rate among species in these areas than in those in Asia, which did not experience large areas of glacier cover52.
Biogeographic patterns of Acer
The phylogenetic analyses showed that the most probable ancestral area of Acer was Asia (Fig. 4), even though the oldest fossil of Acer dating to the Tertiary period was discovered in North America10,11. Asia was found to be the modern diversity center of Acer with many primitive species53. The entire area of the north–south trending river valleys in southwest China acted as an important migration corridor, offering protection during cycles of climate oscillation54. Therefore, the origin center of Acer is speculated to be in Asia, from where the genus was later dispersed to Europe and North America (Fig. 4). In clade II, the North American species is most close to the root of clade II, makes it possible that the North American species was the origin of species of clade II. However, Asian origin still has the highest probability in the assessment of the ancestral area of clade II because the geographic distributions of branches other than clade II are also inferred to be in Asia. Although we cannot completely rule out the hypothesis of the Extinction of North American ancestors, from the geographic distribution and phylogenetic positions of extant species, the Out-of-Asia is the most parsimonious hypothesis of the biogeographic pattern of Acer. In addition, the decline in the effective number of lineages of European and American species after 40 Mya may be due to the fact that the extinction rate was similar to the speciation rate, and the immigration rate (from Asia) of the European and American species has declined after 40 Mya.
Modern genera with global distributions have been hypothesized to have originated in various areas and attained their present distribution via multiple pathways. At least three possible dispersal routes have been proposed to explain the intercontinental distribution of Acer: migration across the NALB55, migration across the BLB56, and long-distance dispersal1,56,57.
Our results allow us to hypothesize that, in the middle to late Eocene, Acer species migrated from Asia to North America forming A. glabrum, A. negundo, and A. spicatum in branch 1 (Fig. 4). The BLB connected East Asia and North America from the late Eocene to the middle Oligocene, providing a corridor for species migration1. In branches 2–5, migration events are hypothesized to have occurred mainly in the Miocene (Fig. 4). A. circinatum (branch 2), A. rubrum (branch 4), A. saccharinum (branch 4), and A. macrophyllum (branch 5), currently distributed in North America, probably migrated from Asia via the BLB. The BLB provided a route for temperate migration between Asia and North America until ca. 3.5 Mya1,58. Many species have been suggested to have migrated using the BLB, such as Penthorum L. (Penthoraceae)59, Circaea L. (Onagraceae)60, and Saxifraga rivularis L. (Saxifragaceae)61. Therefore, migration events using the BLB seem to be the most reasonable explanation for the intercontinental distribution of taxa in Asia and North America. However, the Aleutian land bridge, south of the BLB in the Tertiary, has also been proposed as an explanation for these migration routed1.
The NALB between Eurasia and eastern North America is another possible migration route (Fig. 4). Acer species from branch 3 are hypothesized to have migrated to Europe and formed four endemic species (A. pseudoplatanus, A. monspessulanum, A. opalus, and A. heldreichii). Then, some species may have migrated from Europe to North America and formed A. saccharum subsp. nigrum and A. saccharum subsp. grandidentatum (Fig. 4). The NALB was the shortest route connecting Europe and North America. Tiffney55,56 suggested that plant migration via the NALB was possible during the Paleocene and Eocene. Even in the Miocene, the islands could have served as “stepping stones” for the migration of deciduous taxa from Europe to North America1. In summary, we propose that Acer originated in Asia, dispersed to Europe, and spread to North America through different routes (namely, NALB and BLB).
During the Oligocene–Miocene transition, the global climate warming trend reduced the extent of the Antarctic ice sheets62; this warm phase peaked in the mid-Miocene Climatic Optimum (17–15 Mya)63,64. Donoghue and Smith65 summarized the discontinuous distribution pattern of 66 plant groups and concluded that most temperate forest plants originated and diversified in Asia. These plants spread out from Asia several times since 30 Mya, and Miocene was an active periods of species diffusion. In addition, the considerable increase of insect diversity in the Miocene may accelerate the diversification rate of plant species66. Therefore, it is reasonable to conclude that climate change during the Miocene may have been a major driving force for the migration of Acer.
Conclusion
In this study, we not only reduced the taxonomic problem surrounding Acer by reconstructing a phylogeny but also raised the hypothesis of Asian origin and multiple long-distance dispersal routes for the species in this genus. Regarding large-scale biogeographic patterns, the case of this genus shows the impact of climate change on dispersal and speciation, which in turn affects the diversification rate among continents. Asia, the origin center and the region less covered by ice sheets during glacial periods, became a hotbed for the growth of Acer and its speciation. The diverse landforms, such as Hengduan Mountain, are also beneficial to species diversification as they provide rich niches. In conclusion, this study provides an outstanding example for the discontinuous distribution in the north temperate zone.
Data availability
All data sets are provided in the Supplementary Information and deposited in GenBank.
References
Wen, J. Evolution of Eastern Asian and Eastern North American disjunct distributions in flowering plants. Annu. Rev. Ecol. Syst. 30, 421–455 (1999).
van Gelderen, D. M., Jong, P. C., Oterdoom, H. J. & Dudley, T. R. Maples of the World (Timber Press, Portland, 1994).
Xu, T. Z., Chen, Y. S., de Jong, P. C., Oterdoom, H. J. & Chang, C. S. Aceraceae. In Flora of China (eds Wu, Z. Y. et al.) 515–553 (Science Press, Beijing, 2008).
Xu, T. Z. The systematic evolution and distribution of the genus Acer. Acta Bot. Yunnanica 20, 383–393 (1998).
Akhmet, M. A. Tertiary maples of Eastern Asia. Paleontol. J. 5, 362–371 (1971).
Tanai, T. & Ozaki, K. The genus Acer from the upper Miocene in Tottori prefecture, Western Japan. J. Fac. Sci. 17, 575–606 (1977).
Wolfe, J. A. & Tanai, T. Systematics, phylogeny, and distribution of Acer (maples) in the Cenozoic of Western North America. J. Fac. Sci. 22, 1–246 (1987).
Li, X. X. et al. Fossil Floras of China Through the Geological Ages (Guangdong Science and Technology Press, Guangdong, 1995).
Chen, Y. F., Wong, W. O., Hu, Q., Liufu, Y. Q. & Xie, Z. M. A new fossil-species of Acer (Sapindaceae) from the Ningming Basin in Guangxi, South China. Phytotaxa 298, 158 (2017).
Taylor, D. W. Paleobiogeographic relationships of angiosperms from the Cretaceous and early Tertiary of the North American area. Bot. Rev. 56, 279–417 (1990).
Manchester, S. R. Biogeographical relationships of North American Tertiary floras. Ann. Mo. Bot. Gard. 86, 472–522 (1999).
Ackerly, D. D. & Donoghue, M. J. Leaf size, sapling allometry, and Corner’s rules: phylogeny and correlated evolution in maples (Acer). Am. Nat. 152, 767–791 (1998).
Hasebe, M., Ando, T. & Iwatsuki, K. Intrageneric relationships of maple trees based on the chloroplast DNA restriction fragment length polymorphisms. J. Plant. Res. 111, 441–451 (1998).
Suh, Y., Heol, K. & Park, C. W. Phylogenetic relationships of maples (Acer L.; Aceraceae) implied by nuclear ribosomal ITS sequences. J. Plant Res. 113, 193–202 (2000).
Pfosser, M. F. et al. The origin of species of Acer (Sapindaceae) endemic to Ullung island, Korea. Syst. Bot. 27, 351–367 (2002).
Grimm, G. W., Renner, S. S., Stamatakis, A. & Hemleben, V. A. Nuclear ribosomal DNA phylogeny of Acer inferred with maximum likelihood, splits graphs, and motif analysis of 606 sequences. Evol. Bioinform. 2, 7–22 (2006).
Li, J. H., Yue, J. & Shoup, S. Phylogenetics of Acer (Aceroideae, Sapindaceae) based on nucleotide sequences of two chloroplast non-coding regions. Harvard Pap. Bot. 11, 101–115 (2006).
Renner, S. S., Beenken, L., Grimm, G. W., Kocyan, A. & Ricklefs, R. E. The evolution of dioecy, heterodichogamy, and labile sex expression in Acer. Evolution 61, 2701–2719 (2007).
Renner, S. S., Grimm, G. W., Schneeweiss, G. M., Stuessy, T. F. & Ricklefs, R. E. Rooting and dating maples (Acer) with an uncorrelated-rates molecular clock: implications for North American/Asian disjunctions. Syst. Biol. 57, 795–808 (2008).
Wolfe, J. A. Vicariance Biogeography of Angiosperms in Relation to Paleobotanical Data (Columbia University Press, New York, 1981).
Raymo, M. E. & Ruddiman, W. F. Tectonic forcing of late Cenozoic climate. Nature 359, 117–122 (1992).
An, Z., Kutzbach, J. E., Prell, W. L. & Porter, S. C. Evolution of Asian monsoons and phased uplift of the Himalaya—Tibetan plateau since Late Miocene times. Nature 411, 62–66 (2001).
Favre, A. et al. The role of the uplift of the Qinghai—Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev. 90, 236–253 (2015).
Yu, G., Zhang, M., Rao, D. & Yang, J. Effect of Pleistocene climatic oscillations on the phylogeography and demography of red knobby newt (Tylototriton shanjing) from Southwestern China. PLoS ONE 8, e56066 (2013).
Chin, S. W., Shaw, J., Haberle, R., Wen, J. & Potter, D. Diversification of almonds, peaches, plums and cherries—molecular systematics and biogeographic history of Prunus (Rosaceae). Mol. Phylogenet. Evol. 76, 34–48 (2014).
Reck-Kortmann, M. et al. Multilocus phylogeny reconstruction: new insights into the evolutionary history of the genus Petunia. Mol. Phylogenet. Evol. 81, 19–28 (2014).
Tanai, T. Revisions of tertiary Acer from East Asia. J. Fac. Sci. 20, 291–390 (1983).
Saeki, I., Dick, C. W., Barnes, B. V. & Murakami, N. Comparative phylogeography of red maple (Acer rubrum L.) and silver maple (Acer saccharinum L.): impacts of habitat specialization, hybridization and glacial history. J. Biogeogr 38, 992–1005 (2011).
Judd, W. S., Sanders, R. W. & Donoghue, M. J. Angiosperm family pairs: preliminary phylogenetic analyses. Harvard Pap. Bot. 1, 1–51 (1994).
Gadek, P. A. et al. Sapindales: molecular delimitation and infraordinal groups. Am. J. Bot. 83, 802–811 (1996).
Doyle, J. J. & Doyle, J. A. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15 (1987).
Hall, T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acid. Sci. 41, 95–98 (1999).
Farris, J. S., Källersjö, M., Kluge, A. G. & Bult, C. Testing significance of incongruence. Cladistics 10, 315–319 (1994).
Von Mering, S. & Kadereit, J. W. Phylogeny, biogeography and evolution of TriglochinL. (Juncaginaceae)—morphological diversification is linked to habitat shifts rather than to genetic diversification. Mol. Phylogenet. Evol. 83, 200–212 (2015).
Chen, X. H. et al. Biogeographic diversification of Mahonia (Berberidaceae): Implications for the origin and evolution of East Asian subtropical evergreen broadleaved forests. Mol. Phylogenet. Evol. 151, 106910 (2020).
Swofford, D. L. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b10 (Sinauer Associates, Sunderland, 2002).
Zwickl, D. J. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. (Ph. D. dissertation, University of Texas, Austin, Texas, USA, 2006).
Ronquist, F. et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012).
Posada, D. & Buckley, T. R. Model selection and model averaging in phylogenetics: advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst. Biol. 53, 793–808 (2004).
Drummond, A. J., Suchard, M. A., Xie, D. & Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012).
Matzke, N. J. BioGeoBEARS: Biogeography with Bayesian (and likelihood) Evolutionary Analysis in R Scripts (CRAN The Comprehensive R Archive Network, Berkeley, 2013).
Ree, R. H., Moore, B. R., Webb, C. O. & Donoghue, M. J. A likelihood framework for inferring the evolution of geographic range on phylogenetic trees. Evolution 59, 2299–2311 (2005).
Ree, R. H. & Smith, S. A. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 57, 4–14 (2008).
Ronquist, F. Dispersal-vicariance analysis: a new approach to the quantification of historical biogeography. Syst. Biol. 46, 195–203 (1997).
Landis, M. J., Matzke, N. J., Moore, B. R. & Huelsenbeck, J. P. Bayesian analysis of biogeography when the number of areas is large. Syst. Biol. 62, 1–16 (2013).
Matzke, N. J. Probabilistic historical biogeography: new models for founder-event speciation, imperfect detection, and fossils allow improved accuracy and model-testing. Front. Biogeogr. 5, 242–248 (2013).
Paradis, E., Claude, J. & Strimmer, K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004).
Opgen-Rhein, R., Fahrmeir, L. & Strimmer, K. Inference of demographic history from genealogical trees using reversible jump Markov chain Monte Carlo. BMC Evol. Biol. 5, 6 (2005).
Ogata, K. A systematic study of the genus Acer. Jpn. J. Bot. 19, 277–323 (1967).
Zhang, Z., Li, C. & Li, J. Conflicting phylogenies of section Macrantha (Acer, Aceroideae, Sapindaceae) based on chloroplast and nuclear DNA. Syst. Bot. 35, 801–810 (2010).
Li, J. H. Phylogenetic evaluation of series delimitations in section Palmata (Acer, Aceroideae, Sapindaceae) based on sequences of nuclear and chloroplast genes. Aliso 29, 43–49 (2011).
Liu, Z. et al. Global cooling during the Eocene-Oligocene climate transition. Science 323, 1187–1190 (2009).
Xu, T. Z. Phytogeography of the family Aceraceae. Acta Bot. Yunnanica 18, 43–50 (1996).
Qiu, Y. X., Fu, C. X. & Comes, H. P. Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol. Phylogenet. Evol. 59, e225–e244 (2011).
Tiffney, B. H. The Eocene North Atlantic land bridge: its importance in tertiary and modern phytogeography of the Northern hemisphere. J. Arnold Arboretum 66, 243–273 (1985).
Tiffney, B. H. Perspectives on the origin of the floristic similarity between eastern Asia and eastern North America. J. Arnold Arboretum 66, 73–94 (1985).
Wen, J., Ickert-Bond, S., Nie, Z. L. & Li, R. Timing and modes of evolution of Eastern Asian-North American biogeographic disjunctions in seed plants. In Darwin's Heritage Today—Proceedings of the Darwin 200 Beijing Intern, 252–269 (2010).
Hopkins, D. M. The Bering Land Bridge (Stanford University Press, Palo Alto, 1967).
Xiang, Q. Y., Soltis, D. E., Soltis, P. S., Manchester, S. R. & Crawford, D. J. Timing the Eastern Asian–Eastern North American floristic disjunction: molecular clock corroborates paleontological estimates. Mol. Phylogenet. Evol. 15, 462–472 (2000).
Xie, L., Wagner, W. L., Ree, R. H., Berry, P. E. & Wen, J. Molecular phylogeny, divergence time estimates, and historical biogeography of Circaea (Onagraceae) in the Northern Hemisphere. Mol. Phylogenet. Evol. 53, 995–1009 (2009).
Westergaard, K. B., Jørgensen, M. H., Tove, M. G., Alsos, I. G. & Brochmann, C. The extreme Beringian/Atlantic disjunction in Saxifraga rivularis (Saxifragaceae) has formed at least twice. J. Biogeogr. 37, 1262–1276 (2010).
Dupont-Nivet, G. et al. Tibetan plateau aridification linked to global cooling at the Eocene-Oligocene transition. Nature 445, 635–638 (2007).
Vincent, E., Killingley, J. S. & Berger, W. H. Miocene oxygen and carbon isotope stratigraphy of the tropical Indian Ocean. Geol. Soc. Am. Mem. 163, 103–129 (1985).
Flower, B. P. & Kennett, J. P. Middle Miocene deepwater paleoceanography in the southwest Pacific: relations with East Antarctic ice sheet development. Paleoceanography 10, 1095–1112 (1995).
Donoghue, M. J. & Smith, S. A. Patterns in the assembly of temperate forests around the Northern Hemisphere. Philos. Trans. R. Soc. Lond. B 359, 1633–1644 (2004).
Wappler, T. Insect herbivory close to the oligocene-miocene transition—a quantitative analysis. Palaeogeogr. Palaeocl. 292, 540–550 (2010).
Acknowledgements
We thank Shi-Yao Chai from Baotou Teachers’ College for her insightful comments on this paper. We also gratefully thank Editage (www.editage.cn) for English language editing. This research was financially supported by the program “The biogeographical feature and competitive hybridization of Maple (Acer L.) in East Asia” of National Natural Science Foundation of China (41901063) and Research Program of Science and Technology at Universities of Inner Mongolia Autonomous Region (NJZZ19185) to Jian Gao, “Reintroduction Technologies and Demonstration of Extremely Rare Wild Plant Population” of National Key Research and Development Program (2016YFC0503106) to Jun-Qing Li, and Ministry of Science and Technology in Taiwan (MOST 108-2628-B-003-001) to Pei-Chun Liao.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: P.-C.L., J.-Q.L. and J.G. Performed the experiments: J.G. and B.-H.H. Contributed reagents/materials/analysis tools: J.G., B.-H.H., T.Y., Y.-Y.Z. and J.-Q.L. Analyzed the data: J.G., B.-H.H. and P.-C.L. Wrote the paper: J.G. and P.-C.L. All authors participated in the discussion, read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gao, J., Liao, PC., Huang, BH. et al. Historical biogeography of Acer L. (Sapindaceae): genetic evidence for Out-of-Asia hypothesis with multiple dispersals to North America and Europe. Sci Rep 10, 21178 (2020). https://doi.org/10.1038/s41598-020-78145-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-78145-0
This article is cited by
-
Fossil samaras of Acer L. (Sapindaceae) from the Upper Pliocene of western Yunnan, southwestern China
Palaeobiodiversity and Palaeoenvironments (2023)
-
Terminal velocities and falling patterns correlate with morphology of diaspores in wind-dispersed forestry species
Trees (2022)
-
Can we foresee future maple invasions? A comparative study of performance-related traits and invasiveness of eight Acer species
Plant Ecology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.