Abstract
Early and accurate prediction of the need for intubation may provide more time for preparation and increase safety margins by avoiding high risk late intubation. This study evaluates whether machine learning can predict the need for intubation within 24 h using commonly available bedside and laboratory parameters taken at critical care admission. We extracted data from 2 large critical care databases (MIMIC-III and eICU-CRD). Missing variables were imputed using autoencoder. Machine learning classifiers using logistic regression and random forest were trained using 60% of the data and tested using the remaining 40% of the data. We compared the performance of logistic regression and random forest models to predict intubation in critically ill patients. After excluding patients with limitations of therapy and missing data, we included 17,616 critically ill patients in this retrospective cohort. Within 24 h of admission, 2,292 patients required intubation, whilst 15,324 patients were not intubated. Blood gas parameters (PaO2, PaCO2, HCO3−), Glasgow Coma Score, respiratory variables (respiratory rate, SpO2), temperature, age, and oxygen therapy were used to predict intubation. Random forest had AUC 0.86 (95% CI 0.85–0.87) and logistic regression had AUC 0.77 (95% CI 0.76–0.78) for intubation prediction performance. Random forest model had sensitivity of 0.88 (95% CI 0.86–0.90) and specificity of 0.66 (95% CI 0.63–0.69), with good calibration throughout the range of intubation risks. The results showed that machine learning could predict the need for intubation in critically ill patients using commonly collected bedside clinical parameters and laboratory results. It may be used in real-time to help clinicians predict the need for intubation within 24 h of intensive care unit admission.
Similar content being viewed by others
Introduction
Endotracheal intubation is commonly performed in the critical care setting for airway protection or mechanical ventilation. However, emergent intubation is associated with higher risks than elective intubation1. A clinician needs to balance the risks of emergency intubation against the risks of delaying intubation in a patient who requires it, which is also associated with mortality2.
Except absolute indications such as upper airway obstruction, the decision and timing of intubation are often tailored for individual patients. To facilitate decision making, scoring systems help predict the need for mechanical ventilation during failed non-invasive ventilation or high flow oxygen therapy in patients with acute respiratory failure3,4. However, these scores have not been validated for patients without respiratory failure who require intubation for airway protection. Furthermore, their calculation relies on an accurate assessment of the PaO2/FiO2 ratio, which is not easily attainable in patients on variable performance oxygen devices.
Decision support systems aid clinical decisions by alerting clinicians and proposing treatments based on objective clinical data5. Databases such as Medical Information Mart for Intensive Care (MIMIC) have been used to build models that detect patients ready for discharge, predict the development of acute kidney injury6,7. Predictive model may provide an early warning to clinicians before conventional clinical signs manifest. Previous intubation prediction models have shown good performance up to 3 h prior to intubation, but longer lead time may facilitate time-sensitive intervention to prevent deterioration8. The objective of this study was to develop a tool utilizing bedside clinical and laboratory parameters at an intensive care unit (ICU) admission to predict the need for intubation within the next 24 h.
Methods
Data source
We performed a secondary analysis and built our predictive model on patients included in two databases, the Medical Information Mart for Intensive Care III (MIMIC-III) and the eICU Collaborative Research Database (eICU-CRD)9,10. The MIMIC-III database comprises data from 61,532 ICU stays at the Beth Israel Deaconess Medical Center between 2001 and 2012. The eICU-CRD is populated with > 200,000 admission data from a combination of many critical care units throughout continental United States from 2014 to 2015. These databases contain deidentified data, including high-resolution data of admission and discharge, diagnosis, data from monitors and laboratory results. The databases are released under the Health Insurance Portability and Accountability Act (HIPAA) safe harbor provision.
Study population
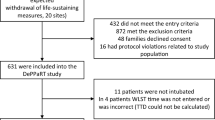
We included all patients aged 18 and above and less than 90 in the eICU-CRD and MIMIC-III database who were not intubated before ICU admission. For patients with multiple ICU and hospital admissions, we only included data from the first ICU admission and first hospital stay. Exclusion criteria included patients with missing airway data or had do-not-resuscitate or do-not-intubate order within 24 h of ICU admission.
Data
We collected demographics data (sex, age, specialty), physiological parameters (heart rate, blood pressure, respiratory rate, SpO2, GCS), laboratory variables (glucose, lactate, pH, PaCO2, PaO2), sequential organ function assessment (SOFA) score, airway device, ventilator data, oxygen therapy, and vasopressor use. Oxygen therapy was supplementary oxygen using any method other than endotracheal devices. These variables were selected because our aim was to develop a model based on data, observations and interventions which were consistently available at the time of ICU admission. The data points closest to the time of ICU admission were used. Patients without a full set of core parameters of heart rate, systolic blood pressure, diastolic blood pressure, mean arterial pressure, respiratory rate, and temperature within 1 h of ICU admission were excluded. Patients who had > 2 missing data of SpO2, Glasgow Coma Score (GCS), shock index, pulse pressure, glucose, PaO2, PaCO2, or HCO3- within 2 h of admission were also excluded.
Missing data is a major limitation in database studies because it reduces sample size and introduces bias by patient selection and imputation11,12,13,14,15. For example, patients who are critically ill may have more blood tests, but this does not mean that more blood tests itself cause a higher severity of illness. Prediction models also perform better when missing data is addressed12. In addition, assumptions become increasingly accurate with more covariates. In this case, multiple imputations can help to overcome these biases14,15. Previously, algorithmic variants based on computationally intensive techniques such as Singular Value Decomposition, K-nearest neighbors (KNN), and relatively less complex methods such as mean and median imputation were used. More recently, imputation with deep learning models such as the Autoencoder (AE) has improved the performance of predictive models13. Autoencoder is a type of neural network that learns an appropriate representation of input with minimized reconstruction errors16. In this study, we used AE to impute missing data for SpO2, GCS, shock index, pulse pressure, glucose, PaO2, PaCO2, or HCO3−. These missing values were imputed by using data on gender, age, physiological parameters and laboratory variables recorded within 2 h of ICU admission. AE was constructed with a modified mean square error between the reconstructed layer and the input data based only on present features17,18,19,20. To do this, first we removed data points that were present to make them “missing” completely at random. Then we trained the AE to impute the missing data based on minimizing the mean square error between the value of the imputed data against the actual value of the removed features. The imputation processes of the training set and the test set were performed separately to avoid information leak into each dataset with Keras 2.2.4 and Tensorflow 1.15.0 in Python21,22. After partitioning of data, AE was conducted for each training and test set which converged around 0.05 error rate, and the datasets were used for machine learning classifiers. For comparison, we experimented with other forms of imputation but found that AE outperformed KNN imputation in outcomes of machine learning classification in our dataset (Supplementary Table S4 online). We also performed modelling on a subset of patients who did not have any missing data to assess efficacy of imputation.
Model
Time of intubation was defined as the first record for airway of any tracheal device (endotracheal tube, tracheostomy, naso-endotracheal tube) or mechanical ventilation data. Patients who had time of intubation within 24 h from ICU admission were classified as intubated and the remaining patients were classified as non-intubated. Since the aim was to provide decision support for clinicians to assess the risk of the need for intubation upon ICU admission, we limited our prediction time window to within the first 24 h of ICU stay. Our rationale was an extension of the prediction window beyond 24 h whilst using only data at a single time point (ICU admission) would likely weaken the utility of the model since increases in lead time decrease model performance8.
We used random forest (RF) for our prediction task as it allows for conventional clinical interpretations of feature importance, along with comparisons using logistic regression (LR) with L2 penalty. Only data recorded before intubation time was used for predictive models. After unity-based data normalization, the entire intubated cohort of 2,292 patients was split into a training set and test set with a 6:4 ratio. The same number of non-intubated patients were used for the test set and all remaining patients were used for the training set. Due to the class imbalance, both models were trained with adjusting weights inversely proportional to class frequencies in the data. The training epochs and parameters were chosen based on error rate convergence and the best performance with shuffled and randomly selected data. To confirm the stability of overall process in random data partitioning, missing data imputation and machine learning classifiers were repeated 12 times. Since our aim was to develop a model that alerts physicians to patients with increased risk of needing intubation at ICU admission, optimal model performance was defined as the highest sensitivity without compromising specificity and accuracy. Sensitivity analysis was performed to find the best RF model threshold that achieved this goal. We used Scikit-learn 0.20.3 library for data pre-processing and models23. The primary objective was to predict the need for intubation within 24 h of ICU admission.
To assess the feature importance in the RF model, we used the local model-specific feature importance from the RF and the local model-agnostic SHAP (SHapley Additive exPlanation) values23,24. These complementary approaches facilitate the interpretation of feature evaluation. Feature importance was calculated from how much each feature (variable) contributed to decreasing impurity over the trees and datasets23,25. In contrast, SHAP values attribute to each feature the change in the expected model prediction when conditioning on that feature.
Statistics
Median with interquartile range (IQR) were used to describe continuous variables. The Kolmogorov–Smirnov test was used to test for normality. Mann–Whitney U test was used for non-parametric comparisons between continuous variables. We used the chi-square test to compare discrete variables. Sensitivity, specificity, positive predictive value, negative predictive value, positive likelihood ratio, negative likelihood ratio and Area under the curve (AUC) of the receiver operating curve (ROC) were used to assess the performance of LR and RF. Model performance was also assessed separately with or without specifying non-surgical and surgical patients. Statistical analysis was performed with SciPy 1.2.2 library in Python26.
Results
Baseline demographics
The combined database from eICU-CRD and MIMIC-III contained 185,887 patients. Of these, 17,616 patients fulfilled inclusion and exclusion criteria (Fig. 1). Up to 13% (2,292/17,616) of patients were intubated within 24 h of ICU admission. Of those who did not require intubation within 24 h, 5.1% (777/15,324) were intubated after 24 h. Baseline characteristics of our cohort are shown in Table 1. Baseline characteristics after imputation by AE are shown in Supplementary Table S1A and S1B online. The median SpO2 of patients given oxygen therapy was 97% (95 to 99).
Model performance
Our final model with RF had AUC 0.86 (95% CI 0.85–0.87) to predict intubation within 24 h of ICU admission. Comparatively, LR model only had AUC 0.77 (95% CI 0.76–0.78). Sensitivity analysis of our RF model using different thresholds is shown in (Supplementary Table S5 online). Overall, a threshold of 0.4 resulted in the best compromise in sensitivity of 0.88 (95% CI 0.86–0.90), specificity of 0.66 (95% CI 0.63–0.69), accuracy of 0.77 (95% CI 0.76–0.78), and AUC of 0.86 (95% CI 0.85–0.87). Other performance indicators of the RF model is shown in Table 2. The ROC curves for each fold and mean are shown in Fig. 2. RF model showed good calibration over the whole range of intubation risk prediction (Fig. 3). Analysis of feature importance is shown (Fig. 4 and Supplementary Table S2 and S3 online). The SHAP values and feature importance from random forest showed similar and consistent patterns. Gender, pulse pressure and use of vasopressor were relatively less important features. The performance of LR and RF in a smaller cohort of 2,345 patients without missing data is consistently lower than models trained on imputed data (Supplementary Table S4 online).
Discussion
Using data derived from 17,616 patients, we developed a model which could predict the need for intubation in critically ill patients within 24 h of ICU admission with sensitivity 0.88, specificity 0.66 and AUC 0.86. The model only uses bedside parameters that are routinely available at the time of critical care admission. Our predictive model may be used clinically to alert physicians on patients at increased risks of needing intubation within 24 h of ICU admission without additional workload for medical or nursing staff.
Risk factors associated with the need for intubation in specific populations such as patients with inhalation injury or acute poisoning have been reported27,28. However, intubation risk prediction models have generally focused on patients with respiratory failure3,4,8,29. Our model had better performance for both non-surgical and surgical critically ill patients when compared to single center studies on patients with respiratory failure admitted to surgical and trauma ICUs29,30. In another study also using MIMIC-III data, Ren et al.’s gradient boosting model had AUC 0.89 (95% CI 0.87 to 0.91) to predict intubation with a lead time of 3 h8. Although it had better performance than our model, it required at least twice as many predictive parameters. More than half were of these parameters were based on laboratory tests, which may not be readily available at admission. Detailed handling of this missing data and effects of imputation was not reported in their study. Furthermore, parameter values from two time points were used in their model. Instead of a 3 h prediction window, our model risk predicts intubation within the first day of ICU stay using commonly available physiological data and point of care test results at a single time point (ICU admission). Another advantage of our model is external validity based on model training of a 17,616 patient cohort combined from MIMIC-III and eICU-CRD databases consisting of different medical, surgical and mixed ICUs. Our model was also internally validated by random selections of patients into 12 different training and test cohorts. We showed that the model generated is stable across these training cohorts, which reduced the chance of noise or overfitting. It had consistent performance across the entire range of intubation risk prediction (Fig. 3).
Proposed scoring systems such as HACOR and ROX predict the need for intubation in patients with respiratory failure treated with non-invasive ventilation (NIV) and high flow nasal cannula (HFNC), with AUC 0.88 (95% CI 0.85–0.90) and AUC 0.74 (95% CI 0.64–0.84), respectively3,4. Although the performance of our RF model’s AUC 0.86 (95% CI 0.85–0.87) is within this range, there are important differences. First, HACOR and ROX models are only applicable to patients with severe respiratory failure who are already on NIV or HFNC. In contrast, patients with respiratory failure only accounted for a small proportion in our cohort, as reflected by the low prevalence of those who were given oxygen therapy (16.3%, 2,872/17,616). Nevertheless, we found that our model did not lose predictive performance even when expanding the cohort from non-surgical to include all ICU patients (Supplementary Table S6). There was consensus between the model using all patients and the model for only non-surgical patients, which balances local system performance and generalization31. Furthermore, our model had better sensitivity 0.88 (95% CI 0.86–0.90) but comparative specificity 0.66 (95% CI 0.63–0.69) to HACOR (sensitivity 0.62 and specificity 0.93) and ROX (sensitivity 0.70 and specificity 0.72)3,4. Second, HACOR and ROX utilize PaO2/FiO2 ratio in risk prediction as accurate FiO2 can be obtained in patients NIV and HFNC. In contrast, FiO2 component cannot be accurately estimated in patients on variable performance oxygen devices. Thus we built our model on the absence or presence of oxygen therapy rather than dependence on reliable PaO2/FiO2. Third, the ROX index was only shown to be useful 12 h after initiation of HFNC.
The most important features in our model included blood gas parameters, GCS and RR, which are similar to previous intubation risk prediction models3,4,8. We used two independent feature assessments, feature importance from random forest and SHAP showed consistent important feature patterns. Since GCS, RR and blood gas results are important clinical features of the neurological and respiratory assessment, it is not a surprise that they are the most contributing features of an intubation prediction model. Yet counterintuitively, we found that intubated patients had a higher median PaO2 prior to intubation compared to those who did not require intubation. We postulate this may be because patients who appeared more unwell were perhaps more likely to be given supplementary oxygen. Indeed, our finding of elevated PaO2 in patients who required intubation is consistent with Ren et al.’s intubation prediction model for patients with respiratory failure8. In contrast, the importance of SpO2 as a predictor of intubation risk was relatively low. This is different to Politano et al.’s model on surgical and trauma patients with respiratory failure which utilizes SpO229. In a neonatal model, intubation for respiratory decompensation was also modelled by reduced SpO232. Reduced importance of SpO2 in our model is likely because the proportion of patients with respiratory failure in our cohort is relatively small. Furthermore, since the goal of oxygen therapy is to maintain oxygenation, it’s possible that only a minority of patients with severe respiratory failure who were not intubated before ICU admission would manifest abnormal SpO2 on arrival to ICU. Indeed, patients who were given oxygen in our cohort had median SpO2 of 97%. Again, the surprisingly low importance of SpO2 is not unique to our model, as SpO2 was found to be of lower importance than urine output or age in Ren et al.’s model8.
Often machine learning models are studied using a single-centered specific time frame and potentially biased retrospective data, and have been proposed as tools that can be implemented in practice without careful consideration in the medical field33. In this project, we demonstrated the scalability, generalisability and clinical interpretability of this model using multicenter databases and easily collectable bedside parameters at ICU admission, and taking into account the effects of interpolation of missing values and comparisons of performance with multiple evaluation indicators.
This study has several key limitations. Firstly, data extracted from ICUs in the United States may not reflect international practice. Nevertheless our model was derived from a large multicenter derivation cohort of nonspecific critically ill patients. Secondly, the complexity of the RF makes an analysis of the construct counterintuitive to the clinician. But most of the important features were clinically relevant. Thirdly, we imputed missing data, which could affect the outcomes of our models. However, we showed that baseline characteristics remain largely unchanged after missing data imputation (Supplementary Table S1A and S1B online). Therefore, even if patients had missing data, imputation may be used to fill missing data and still provide risk prediction using our model. Fourth, we were unable to utilize diagnosis into the models as diagnostic code were performed later in the ICU stay. Nevertheless, future intubation risk models may be enriched by addition of provisional diagnosis recorded at ICU admission. Fifth, we limited our prediction time to within 24 h. It is possible that some patients who actually required intubation were only intubated after 24 h due to delay. However this bias effect is likely minimal in our cohort since only 5.1% of patients classified as non-intubated required intubation after the initial 24 h. Finally, certain clinical parameters, such as the paradoxical movement of abdominal muscles, are associated with respiratory failure34. Unfortunately it was not possible to consistently extract physical examination findings from the databases. Further studies may perform analysis of clinical progress notes to increase the performance of prediction models.
Conclusion
We developed a tool to predict the need for intubation in critically ill patients within first 24 h of admission to ICU. Since it only uses simple routinely captured bedside parameters, it may be used in real-time to the predict need for intubation upon ICU admission.
Data availability
The datasets analysed during the current study are available in the PhysioNet repository, MIMIC-III: https://physionet.org/content/mimiciii/1.4/ and eICU-CRD: https://physionet.org/content/eicu-crd/2.0/. The datasets generated during the current study along with scripts to create the analyses and processed datasets are available in the Github repository, https://github.com/ucabhkw/INTML20.
References
Schwartz, D. E., Matthay, M. A. & Cohen, N. H. Death and other complications of emergency airway management in critically ill adults a prospective investigation of 297 tracheal intubations. Anesthesiol. J. Am. Soc. Anesthesiol. 82, 367–376 (1995).
Bauer, P. R. et al. Association between timing of intubation and outcome in critically ill patients: A secondary analysis of the ICON audit. J. Crit. Care 42, 1–5 (2017).
Duan, J., Han, X., Bai, L., Zhou, L. & Huang, S. Assessment of heart rate, acidosis, consciousness, oxygenation, and respiratory rate to predict noninvasive ventilation failure in hypoxemic patients. Intensive Care Med. 43, 192–199 (2017).
Roca, O. et al. Predicting success of high-flow nasal cannula in pneumonia patients with hypoxemic respiratory failure: The utility of the ROX index. J. Crit. Care 35, 200–205 (2016).
Haug, P. J. et al. Decision support in medicine: Examples from the HELP system. Comput. Biomed. Res. 27, 396–418 (1994).
McWilliams, C. J. et al. Towards a decision support tool for intensive care discharge: Machine learning algorithm development using electronic healthcare data from MIMIC-III and Bristol, UK. BMJ Open 9, e025925 (2019).
Zimmerman, L. P. et al. Early prediction of acute kidney injury following ICU admission using a multivariate panel of physiological measurements. BMC Med. Inform. Decis. Mak. 19, 16 (2019).
Ren, O. et al. Predicting and understanding unexpected respiratory decompensation in critical care using sparse and heterogeneous clinical data. in 2018 IEEE International Conference on Healthcare Informatics (ICHI) 144–151 (IEEE, 2018).
Johnson, A. E. W. et al. MIMIC-III, a freely accessible critical care database. Sci. Data 3, 160035 (2016).
Pollard, T. J. et al. The eICU collaborative research database, a freely available multi-center database for critical care research. Sci. Data 5, 1–13 (2018).
Sterne, J. A. C. et al. Multiple imputation for missing data in epidemiological and clinical research: Potential and pitfalls. BMJ 339, 157–160 (2009).
Wells, B. J., Nowacki, A. S., Chagin, K. & Kattan, M. W. Strategies for handling missing data in electronic health record derived data. eGEMs (Gen. Evid. Methods Improv. Patient Outcomes) 1, 7 (2013).
Beaulieu-jones, B. K. & Moore, J. H. Missing data imputation in the electronic health record using deeply learned autoencoders * The pooled resource Open-access ALS clinical trials Consortium †. Pacific Symp. Biocomput.
Gondara, L. & Wang, K. MIDA: Multiple imputation using denoising autoencoders. Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics) 10939 LNAI, 260–272 (2018).
Janssen, K. J. M. et al. Missing covariate data in medical research: To impute is better than to ignore. J. Clin. Epidemiol. 63, 721–727 (2010).
Vincent, P., Larochelle, H., Lajoie, I., Bengio, Y. & Manzagol, P.-A. Stacked denoising autoencoders: Learning useful representations in a deep network with a local denoising criterion. J. Mach. Learn. Res. 11, 3371–3408 (2010).
Bengio, Y., Lamblin, P., Popovici, D. & Larochelle, H. Greedy layer-wise training of deep networks. in Advances in Neural Information Processing Systems 153–160 (2007).
Ranzato, M., Poultney, C., Chopra, S. & Cun, Y. L. Efficient learning of sparse representations with an energy-based model. in Advances in Neural Information Processing Systems 1137–1144 (2007).
Vincent, P., Larochelle, H., Bengio, Y. & Manzagol, P.-A. Extracting and composing robust features with denoising autoencoders. in Proceedings of the 25th international conference on Machine learning 1096–1103 (2008).
Beaulieu-Jones, B. K. & Greene, C. S. Semi-supervised learning of the electronic health record for phenotype stratification. J. Biomed. Inform. 64, 168–178 (2016).
Chollet, F. and others. Keras. (2015).
Abadi, M. et al. TensorFlow: A System for Large-Scale Machine Learning. in 12th {USENIX} Symposium on Operating Systems Design and Implementation ({OSDI} 16) 265–283 ({USENIX} Association, 2016).
Varoquaux, G. et al. Scikit-learn. GetMobile Mob Comput. Commun. 19, 29–33 (2015).
Lundberg, S. M. & Lee, S. I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 2017, 4766–4775 (2017).
Qi, Y. Random forest for bioinformatics. In Ensemble Machine Learning 307–323 (Springer, Berlin, 2012).
Virtanen, P. et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
Onishi, S., Osuka, A., Kuroki, Y. & Ueyama, M. Indications of early intubation for patients with inhalation injury. Acute Med. Surg. 4, 278–285 (2017).
Buswell, L., Hayes, A. & Baombe, J. BET 2: Predicting the need for endotracheal intubation in poisoned patients. Emerg. Med. J. 36, 573–575 (2019).
Politano, A. D. et al. Predicting the need for urgent intubation in a surgical/trauma intensive care unit. Surgery 154, 1110–1116 (2013).
Blackburn, H. N. et al. External validation in an intermediate unit of a respiratory decompensation model trained in an intensive care unit. Surgery 161, 760–770 (2017).
Futoma, J., Simons, M., Panch, T., Doshi-Velez, F. & Celi, L. A. The myth of generalisability in clinical research and machine learning in health care. Lancet Digit. Heal. 2, e489–e492 (2020).
Clark, M. T. et al. Predictive monitoring for respiratory decompensation leading to urgent unplanned intubation in the neonatal intensive care unit. Pediatr. Res. 73, 104–110 (2013).
Kelly, C. J., Karthikesalingam, A., Suleyman, M., Corrado, G. & King, D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 17, 1–9 (2019).
Boles, J.-M. et al. Weaning from mechanical ventilation. Eur. Respir. J. 29, 1033–1056 (2007).
Author information
Authors and Affiliations
Contributions
B.M.K.S., G.H.K. and L.L. conceived and designed the study, performed data acquisition/interpretation, and provided substantial input to the manuscript. P.H. approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Siu, B.M.K., Kwak, G.H., Ling, L. et al. Predicting the need for intubation in the first 24 h after critical care admission using machine learning approaches. Sci Rep 10, 20931 (2020). https://doi.org/10.1038/s41598-020-77893-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-77893-3
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.