Abstract
The false widow spider Steatoda nobilis is associated with bites which develop bacterial infections that are sometimes unresponsive to antibiotics. These could be secondary infections derived from opportunistic bacteria on the skin or infections directly vectored by the spider. In this study, we investigated whether it is plausible for S. nobilis and other synanthropic European spiders to vector bacteria during a bite, by seeking to identify bacteria with pathogenic potential on the spiders. 11 genera of bacteria were identified through 16S rRNA sequencing from the body surfaces and chelicerae of S. nobilis, and two native spiders: Amaurobius similis and Eratigena atrica. Out of 22 bacterial species isolated from S. nobilis, 12 were related to human pathogenicity among which Staphylococcus epidermidis, Kluyvera intermedia, Rothia mucilaginosa and Pseudomonas putida are recognized as class 2 pathogens. The isolates varied in their antibiotic susceptibility: Pseudomonas putida, Staphylococcus capitis and Staphylococcus edaphicus showed the highest extent of resistance, to three antibiotics in total. On the other hand, all bacteria recovered from S. nobilis were susceptible to ciprofloxacin. Our study demonstrates that S. nobilis does carry opportunistic pathogenic bacteria on its body surfaces and chelicerae. Therefore, some post-bite infections could be the result of vector-borne bacterial zoonoses that may be antibiotic resistant.
Similar content being viewed by others
Introduction
Bacterial infections represent a major threat to human health. For example, typhoidal Salmonella causes 27 million annual cases of typhoid fever resulting in 223,000 deaths1, and non-typhoidal Salmonella is responsible for over 93 million cases of gastroenteritis leading to 155,000 annual deaths2. Bacterial infections contribute significantly to sepsis3 and in 2017, 49 million cases of sepsis resulted in 11 million deaths worldwide. Antibiotic resistance further increases the threat to human health with drug-resistant bacteria causing 700,000 annual deaths worldwide4. According to the World Health Organization’s (WHO) global action plan on antimicrobial resistance, it is essential that antibiotic resistance is tackled across every contact zone between humans and the environment5. Contamination of human dwellings, and more specifically food and water storage facilities, is a major issue1. As such, identifying the source of contamination is crucial for reducing the spread of pathogens.
Synanthropic animals (wildlife associated with human habitats) can be major reservoirs and vectors of pathogenic bacteria. Wild, domesticated and captive animals can be colonised by bacteria and act as reservoirs6, transmitting pathogens through physical contact, including bites, stings and scratches7. For example, rats have historically caused epidemics and rat-borne zoonotic pathogens are once again increasing across Europe8. However, some animal groups that can potentially spread pathogenic bacteria in and around human habitats are often overlooked. Recently, venomous snakes were identified as reservoirs for Salmonella with potential to contribute to the health crisis through shedding contaminated faeces around homes and vectoring bacteria during bites1,9. Moreover, a recent study demonstrated that bacteria can survive within the venom and venom glands of snakes and spiders10.
Spiders occupy a varied range of synanthropic niches11. They eat a diverse range of prey, with some capable of catching and consuming large arthropods, fish, lizards, snakes, birds, rodents12,13,14,15,16 and medically important pests, including mosquitos and house flies17,18,19. Some will readily feed on carrion20,21,22. Wild caught specimens of Steatoda nobilis were observed by us feeding on dead prey for up to eight days in laboratory conditions (unpublished data). The innate immune system of arthropods protects against pathogenic microbes23,24,25; however, once dead, the microbes are free to thrive and multiply on the corpse of their host. It is inevitable that spiders will encounter microbes through the environment or through feeding, especially on carrion. The potential therefore exists for spiders to harbour virulent bacteria26,27 and they have been implicated in bite cases that subsequently led to bacterial infections28.
The clinical manifestations arising from spider bites (araneism) are diverse29,30,31,32,33. For example, necrotic araneism (necrosis resulting from spider bite) is most commonly documented from bites by members of the Loxosceles genus, though infrequently other species are involved26,30,34,35,36,37. Bacterial infection following a spider bite could potentiate prolonged and debilitating pathologies26. Indeed, a study showed the presence of Clostridium perfringens in the venom and on the chelicerae of Loxosceles intermedia. When C. perfringens was conjugated with L. intermedia venom and injected into rabbits, their synergism increased the size of the dermonecrotic lesion26. This synergistic activity however has not yet been proven in humans38. The implication of the spider as the source of these bacterial infections remains controversial. Spiders generally avoid humans and bite only as a defensive response to being trapped39. The spiders are then often crushed, escape, or are captured using non-sterile methods, and as a result, comprehensive microbiological analysis is not possible. A previous study that identified bacteria on Tegenaria agrestis (hobo spiders) deemed them to be non-pathogenic40. The authors argue that infections associated with spider bites are typically caused by bacterial species commonly found in the environment and on human skin38,40. Moreover, spider venoms are considered a rich source of antibacterial peptides41 leading to the proposal that these are sterile environments that neutralise bacteria. In this scenario, infections are secondary to the spider bite itself9,10,38,40,42. We therefore face a conundrum in determining if infections are caused by opportunistic bacteria already present on the skin (secondary infections) or are vectored directly from the bite via the chelicerae (vector-borne bacterial zoonoses).
The noble false widow spider, Steatoda nobilis, has expanded its range across Europe43, (including Ireland12,44,45 and the UK43), through Western Asia46,47, and the Americas43,48,49,50,51,52,53. This species is increasingly linked to medically significant bites to humans, especially in Ireland and the UK29,48,50,53,54. As range expansion continues, so will the increase in bite cases43. Envenomation symptoms of S. nobilis bites include prolonged moderate to intense pain, swelling and erythema, piloerection, diaphoresis, facial flushing, feverishness, vasodilation of blood capillaries, and minor necrosis29. Two native species of spiders, Amaurobius similis and Eratigena atrica, are commonly found in and around houses and gardens throughout Europe. While both species are capable of biting humans, they are not a common source of complaint by the general public. Irish and British media regularly report on alleged bites by S. nobilis and a BBC report attributes one death to bacterial infection resulting from the bite55. In some media reports, the victims were said to be unresponsive to antibiotics, indicating a potential involvement of antibiotic resistant bacteria. However, media reports typically lack conclusive evidence of spider bites. This led Hambler to call for an urgent evaluation of the potential risk of bacterial transmission from bites by S. nobilis55. In an unpublished case series involving confirmed S. nobilis bites currently being assessed by the authors, three victims were treated for subsequent mild to debilitating bacterial infections, including cellulitis and dermatitis. One victim required hospitalisation and an aggressive course of intravenous antibiotics.
False widow spiders (genus Steatoda), like the closely related black widow spiders (genus Latrodectus), can occasionally subdue small vertebrates13,16,27,56,57 as they possess a fast-acting neurotoxic venom13,58,59,60. It is the presence of α-latrotoxin that can induce neuromuscular paralysis and death in humans following envenomation by Latrodectus species59. The venom protein composition of S. nobilis was recently characterised and revealed that approximately two-thirds of the venom is composed of Latrodectus-like toxins, including the most powerful toxin classes, i.e. α-latrotoxins, α-latroinsectotoxins, and δ-latrocrustotoxins58. Also present are the enzymes (e.g. metalloproteases, serine proteases, and chitinases) that are thought to cause tissue damage and thereby facilitate spread of venom toxins into the prey. In high concentrations, α-latrotoxin can cause localised cell death and, when potentiated by the presence of enzymes, induce necrosis58,thus providing substrate that could facilitate bacterial virulence26.
Previous studies on Latrodectus hesperus demonstrated the potential for spiders to vector bacteria during bite27. Chelicerae excised from 220 specimens recovered five pathogenic antibiotic resistant bacterial species. The microbial colonisers of S. nobilis chelicerae have never been investigated. Such a study would provide data to (1) explain why bacterial infections are increasingly associated with bites by S. nobilis, (2) explain why some patients are unresponsive to frontline antibiotics, and (3) determine if the etiological agent could be vectored directly from the spider’s chelicerae or transferred from the body surface on to the area of the bite site. This could have significant implications for advising first line medical staff who are treating bites by S. nobilis and help in choosing appropriate care and treatment.
The main objectives of this study were to (1) characterise the microbiome of the non-native S. nobilis, along with the native A. similis and E. atrica; (2) identify bacteria species residing on the body surface and chelicerae of the spiders and (3) test the susceptibility of these bacteria to antibiotics.
Results
Isolation and genus identification of bacteria on Amaurobius similis, Eratigena atrica and Steatoda nobilis
In the first stage of this study, we investigated the presence of bacteria on the bodies and chelicerae of 3 spider species. 9 bacteria genera were recovered from A. similis, E. atrica and S. nobilis (Table 1 and data not shown). 5 Salmonella, 1 Bacillus, 2 Staphylococcus, and 1 Escherichia species were recovered from 9 full body samples of A. similis, which included 3 Salmonella and a Staphylococcus sp. identified from bodies of euthanised spiders. Salmonella and Bacillus spp. were also identified on the body of E. atrica and a Staphylococcus sp. on the body of S. nobilis.
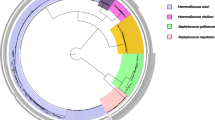
Of particular interest was the identification of 8 different bacteria genera on the chelicerae of these spiders (Table 1, Fig. 1). Bacillus, Raoutella and Staphylococcus spp, were recovered from the chelicerae of both A. similis and E. atrica, among which Staphylococcus spp. were predominant, occurring 7 and 9 times respectively, whereas Paenibacillus spp. were predominant on the chelicerae of S. nobilis being present in 7 out of 8 samples. The second most predominant genus was Bacillus which occurred in 4 samples from A. similis and E. atrica and in 3 samples from S. nobilis. Pseudomonas spp. were recovered from A. similis and S. nobilis, Salmonella and Advenela spp. each occurred once in A. similis and Yersinia occurred once in E. atrica.
Comparison of the bacterial community composition and relative abundances from body and chelicerae surfaces of A. similis, E. atrica and S. nobilis. Photographs of the spider species are shown to scale. Data of chelicerae isolates are displayed for A. similis and E. atrica, and combined data of both chelicerae and body isolates are displayed for S. nobilis. The tables display the number of times isolates of each bacterial genus was isolated from a spider species. The pie chart displays the relative abundance of the bacteria genera isolated.
Isolation and species identification of bacteria in the S. nobilis microbiota
To test the hypothesis that spiders can carry pathogens and could play a role in infection following spider bites, bacteria were isolated from the body and chelicerae of S. nobilis and the sequence of the full-length 16S rRNA gene was determined to identify individual isolates to species level. Streptococcus and Staphylococcus were targeted by using selective CNA blood agar and Baird-Parker agar, respectively. Due to the increasing incidence of development in patients of infection associated with S. nobilis bites this species was an ideal candidate for this study. 20 chelicerae, 15 full body (5 of which had been dead for 24–48 h before sampling) and 2 “spider walk” samples of S. nobilis were analysed.
Twenty-five different bacterial isolates were cultured, and identified through 16S rRNA sequence analysis, within the microbiota of S. nobilis: 17 Gram-positive and 8 Gram-negative bacteria. For the majority of sequences, the percentage identity was > 99% with their respective most similar species (Table 2). Among these, 100% identity was found for 3 sequences to Staphylococcus edaphicus, Staphylococcus warneri and Bacillus thuringiensis. Two isolates displayed individual identity of 98% for Bacillus pumilus and Streptococcus anginosus, suggesting the isolates to be closely related to these two species.
Five isolates showed haemolytic activity on blood agar: 4 Bacillus spp. and 1 Micrococcus sp. Twelve isolates were related to human pathogenicity among which were 4 Staphylococcus spp., 3 Bacillus spp., and one each of Rothia, Streptococcus, Dietzia, Pseudomonas and Kluyvera spp. The association with human pathogenicity for each bacterial species was assessed using the bacterial metadatabase BacDive (Table 2).
Bacteria were isolated by each of the 3 sampling methodologies: 2 species were isolated from the agar plate with spider walks, 5 from the chelicerae and 18 from the full bodies (11 from dead spiders and 7 from live spiders). The 2 bacterial species from the spider walks (Kluyvera intermedia and Staphylococcus epidermidis) were different from the species found on other sites. The bacteria detected on the chelicerae were mostly distinct from the bacterial community on the full body, except for Staphylococcus capitis which was present on both sites. Differences in microbiota were also observed between bodies of live and dead spiders, with a wider variety of genera isolated from dead specimens, and primarily Bacillus spp. from live specimens.
Anti-bacterial inactivity of S. nobilis venom
To investigate the hypothesis that bacteria can be transferred from the chelicerae into the host during the bite without being killed by the venom, S. nobilis venom was tested for its antibacterial property through Minimum Inhibitory Concentration (MIC) and agar diffusion assays. MIC assays were performed by testing diluted venom against pathogenic strains of Escherichia coli, Methicillin Resistant Staphylococcus aureus (MRSA) and Listeria monocytogenes. After incubation for 24 h in liquid media the absorbance at OD590 for growth of each pathogen in the presence of the highest concentration of venom (1:100) was 0.61 ± 0.09 (E. coli), 0.68 ± 0.06 (MRSA) and 0.34 ± 0.09 (L. monocytogenes), which was very similar to growth in the absence of venom (0.61 ± 0.01, 0.57 ± 0.06 and 0.35 ± 0.03, respectively). It was necessary for the venom to be diluted in the MIC assay due to limited venom availability. The agar diffusion assay was therefore deployed to assess the antibacterial activity of pure venom, as smaller volumes were sufficient with this method. Furthermore, we investigated the antibacterial ability of the venom against bacteria which are part of the spider microbiota. Two isolates recovered from S. nobilis chelicerae were the target bacteria in the assay: the Gram-negative Pseudomonas azotoformans and the Gram-positive S. capitis. 0.5 µl undiluted venom was applied to solid agar media spread with the bacteria. Alternatively, spiders bit the agar plate of bacteria directly. After 24 h no zone of bacterial clearance was observed on any of the culture plates (data not shown), indicating that the pure undiluted venom did not inhibit the growth of either species. These data demonstrate that the venom did not inhibit growth of either spider commensal bacteria or human pathogens, indicating that bacteria could survive in spider venom during transfer from the chelicerae to the host during a spider bite.
Antibiotic susceptibility testing of strains isolated from S. nobilis
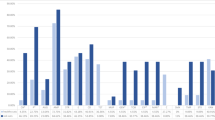
Antibiotic susceptibility testing was performed by disk diffusion assays in accordance with the CLSI standards to determine the range of antibiotic resistance of the bacteria residing on S. nobilis and to determine which antibiotics would be the most effective in treating infection caused by those pathogens following a spider bite. The 25 isolates were tested against 9 antibiotics of 8 different classes, consisting of 8 broad spectrum antibiotics and 1 antibiotic with greater efficacy against Gram-negative bacteria (Colistin B) (Table 3). Of these 25 isolates, 10 are species listed in CLSI guidelines, namely—Staphylococcus (5), Pseudomonas (3), Streptococcus sp. and K. intermedia. For these isolates, resistance and susceptibility were inferred from their EUCAST breakpoints for each antibiotic. For the rest, lack of, or a minimal (≤ 8 mm), zone of clearance around the antibiotic disk was considered as resistant. Resistance to each antibiotic was displayed by at least one isolate, except for ciprofloxacin (Fig. 2, Table 3). Only one isolate was resistant to chloramphenicol (S. edaphicus) or tetracycline [S. capitis (2)], whereas 9 isolates resisted nalidixic acid and 6 were erythromycin resistant (Fig. 2, Table 3). 76% of isolates were resistant to at least one antibiotic and some isolates were multidrug-resistant. Pseudomonas putida, S. capitis (2) and S. edaphicus were notable for resistance to 3 antibiotics. All Staphylococcus isolates showed resistance to gentamicin and nalidixic acid, with the exception of S. capitis (1) for gentamicin and Staphylococcus warneri for nalidixic acid. Dietzia timorensis, Rothia amarae and the Streptococcus sp. isolate had identical resistance profiles with resistance to nalidixic acid and colistin only. These data demonstrate that there is a broad range of antibiotic resistance activity amongst bacteria residing on S. nobilis and the choice of antibiotic treatment for infected bites requires careful consideration.
Discussion
The role of spiders in bacterial transmission has generated much debate27,33,38,40. In recent years, increasing media reports from Ireland and the UK29,33,55 claim that victims of the noble false widow spider Steatoda nobilis frequently suffer debilitating and sometimes fatal bacterial infections55. While these reports are largely unsubstantiated, there have been no studies carried out to validate the true risk of bacterial infections associated with this recently established spider.
In the first part of this study, the microbiomes from S. nobilis (8 chelicerae (C), 1 full body (FB)), A. similis (16 C, 6 FB) and E. atrica (10 C, 2 FB) were partially characterised, revealing diverse bacterial compositions of 9 different genera, most of which were detected on the chelicerae (Table 1). All these bacterial genera contain some species that are associated with human pathogenicity. Since S. nobilis is associated with bites that lead to infections, we focused the next part of this study on the bacteria present on body and chelicerae of S. nobilis (20 C, 15 FB, 2 spider walks (Table 2) and identified the bacteria to species level. In this subsequent investigation, 10 genera were recovered from body and chelicerae of S. nobilis, four in common with those found in the first part of this investigation. Testing the larger sample size of S. nobilis and identifying these isolates to species level allowed us to determine their potential for pathogenicity. The bacteria identified are members of microbiota of animals/humans and/or found in environmental settings. Eleven species are related to human pathogenicity (Table 2) and are recognised as opportunistic bacteria, among which S. epidermidis, K. intermedia, R. mucilaginosa and P. putida are designated as class 2 pathogens. We observed differences between bacterial communities on dead and living spiders. Bacillus spp. were abundant on living spiders, as found in a previous study40. However, for dead specimens only one Bacillus isolate was identified. The diversity of genera was greater on dead spiders and included Dietzia and Serratia spp. This may be explained by the occurrence of saprophytes, such as S. fonticola, P. putida and M. luteus, which thrive on corpses and could outcompete other bacterial species resulting in remodelling of the microbiota diversity and abundance63. Different bacterial communities were observed between sites of the living spiders. In contrast to full body sites, Bacillus spp. were not recovered from chelicerae or spider walks, indicating they probably reside more abundantly on body parts such as the abdomen. From the spider walk only Kluyvera and Staphylococcus spp. were isolated, possibly due to the low sample size. The chelicerae had the most diverse communities, including Pseudomonas, Rothia, Streptococcus, and Staphylococcus spp. This is possibly explained through direct exposure of the chelicerae to, and penetrating into, dead prey, in addition to contact with their legs/feet during grooming.
Staphylococcus spp. were recovered from S. nobilis, A. similis and E. atrica, of which four species recovered from S. nobilis were identified. Among them, S. epidermidis is a known human pathogen and responsible for severe illnesses, including bacteraemia, urinary tract infections, endocarditis, septicaemia and nosocomial sepsis originating from medical devices such as catheters and central lines61. Other Staphylococcus species identified can be opportunistic human pathogens, i.e. can cause severe infection in a host with a weakened immune system, an altered microbiota (such as a disrupted gut microbiota), or breached integumentary barriers, and are considered as typical components of the skin microbiome64,65,66.
Pseudomonas are ubiquitous in the environment and some species are associated with human infections. Pseudomonas spp. were recovered from A. similis and S. nobilis of which three species were identified, and of these one is related to human pathology. P. putida can cause bacteraemia, skin, soft tissue, and urinary tract infections, localised infections, pneumonia, peritonitis, septic arthritis, meningitis, and septicaemia67,68.
Two species of Rothia were recovered from S. nobilis, of which one is related to human pathology. R. mucilaginosa is a common constituent of the oral and upper respiratory microbiota. It is commonly associated with teeth and gum disease, but is now considered an emerging opportunistic pathogen, especially in immunocompromised patients associated with endocarditis, pneumonia, arthritis, meningitis, skin and soft-tissue infections, prosthetic joint infections, and endophthalmitis. For example, it was isolated from five cancer patients who developed bacteraemia69,70.
The native spiders A. similis and E. atrica, are common synanthropic spiders throughout Europe and neither species is considered to pose a threat to the general public. Bites are not thought to be common and therefore the risk of transmission resulting in infection is likely to be low40. Some bacteria genera from T. agrestis that were reported previously, were also identified in our study, e.g. Bacillus, Paenibacillus, Pseudomonas and Staphylococcus, and of those isolates identified to species level, Bacillus thuringiensis is present in both datasets. This species is now recognised as pathogenic, and our study shows it can display antibiotic resistance. In our study after allowing S. nobilis to walk on petri dishes with BHI agar, we recovered K. intermedia and S. epidermidis, indicating that spiders have potential to shed bacteria on the surfaces they touch. Following the rapid expansion of T. agrestis in North America, local media reported that the species was responsible for envenomation-led necrotic lesions. This claim has since been debunked, as with other spiders such as yellow sac spiders from the genus Cheiracanthium30. However, the claims that S. nobilis may be involved in severe envenomation events in Ireland and the UK appear to have some merit. As the geographical range of S. nobilis expands and their overall density increases in heavily urbanised areas, envenomations are becoming a more common occurrence. As a result, the transmission of pathogenic microbes during a bite event is now a cause for concern. The role of bacterial araneism is controversial; however, it is accepted that it is experimentally plausible for spiders to vector bacteria, and a confirmed infection vectored directly from a spider bite is discussed in the literature28,38. We demonstrated here that (1) a wide range of bacteria ubiquitous in the environment are carried on spider chelicerae and exoskeleton (Fig. 1), and (2) some are potentially pathogenic and involved in a wide range of clinical manifestations. In total, 11 species of potentially pathogenic bacteria were isolated from bodies or chelicerae of S. nobilis. We believe this clearly demonstrates the potential for bacteria to be vectored during bites and that it is just as likely that infections arise zoonotically as from commensal bacteria present on the skin (as is the current consensus)9,10,38.
In the case of S. nobilis, vectored infection may be facilitated by the venom’s ability to kill localised skin cells58, potentially disrupt normal immune response30, and provide substrate for bacteria to thrive. Moreover, S. nobilis typically bite humans when accidently trapped or squashed between the skin and clothing/bed sheets29,54. Therefore, the site around the bite could be contaminated by bacteria present on either the chelicerae or the body of the spider. Previous studies reveal spider venoms as rich sources of antibacterial peptides41 that could neutralise bacteria in paralyzed prey38,71. However, recent advances in venomics studies confirms that spider venoms are not sterile and should be viewed as microenvironments9. The results here demonstrate that S. nobilis venom has no inhibitory effect on bacterial growth, suggesting that the venom is unlikely to eliminate bacteria from the chelicerae.
Since the development of penicillin and subsequent antibiotics in the 1940s, there has been a rise in antibiotic resistant bacteria72 which currently kill over 700,000 people annually 4. Therefore, it is important to determine how antibiotic resistant bacteria move through the environment and establish contact zones between humans and the environment5. Pathogenic bacteria recovered from the chelicerae of black widow spiders27 included multiple antibiotic resistant strains, with fluoroquinolones and aminoglycosides recommended as the most efficient antibiotics for treating infections arising from black widow bites. Out of three confirmed bite cases by S. nobilis that resulted in dermatitis (data unpublished), one of the victims was unresponsive to antibiotic treatment. We tested the susceptibility of 25 bacteria recovered from S. nobilis against nine antibiotics used by front line medical staff and 19 antibiotic-resistant strains were identified (Table 3). The most resistant isolates were P. putida, which showed resistance to three broad range antibiotics (amoxicillin, erythromycin and cefoxitin), S. capitis (2) which also showed resistance to three completely different class of antibiotics (gentamicin, tetracycline and nalidixic acid) and S. edaphicus which showed resistance to gentamicin, chloramphenicol and nalidixic acid. S. capitis and S. edaphicus are the only isolates in this study to show resistance against tetracycline and chloramphenicol, respectively. In terms of resistance shown by the recovered isolates to each antibiotic (Fig. 2A), 9 of the isolates showed resistance to nalidixic acid followed by erythromycin (6), cefoxitin (5), gentamicin and amoxycillin (4), colistin (3), tetracycline (1) and chloramphenicol (1). All bacteria recovered from S. nobilis were susceptible to ciprofloxacin. An abundance of multidrug-resistant isolates were identified with 3 isolates resistant to 3 different antibiotics and 7 isolates resistant to 2 antibiotics (Fig. 2B). These data support the fundamental need to identify bacteria from spider bite victims. Additionally, there is a need for catalogues of the microbiota of spiders and cross-reference databanks with pathogenicity and antibiotic-resistance to better inform appropriate treatment for infections associated with spider bites.
Conclusion
Our study demonstrates that the non-native S. nobilis and two native spider species, A. similis and E. atrica, carry opportunistic pathogenic bacteria on their body surfaces and chelicerae. Bacteria may be vectored directly from the spider, and as a result, post-bite infections may be the result of vector-borne bacterial zoonoses. Some of the bacteria carried by spiders are multidrug-resistant. Furthermore, our results showed that the venom of S. nobilis has no inhibitory effects against bacterial growth, indicating that it is most likely not a barrier to bacterial infection resulting from a spider bite. We believe this study provides a baseline for future research targeting synanthropic spider species to determine bacterial compositions and develop a database of bacterial species isolated from spiders, and to determine links to human disease.
Methods
Spider and venom collection
Specimens of Amaurobius similis, Eratigena atrica, and Steatoda nobilis were collected in Ireland, from garden walls and park railings in Lucan, Co. Dublin, Edgeworthstown, Co. Longford, Galway city, Co. Galway and Ferrybank, Co Waterford. Specimens were collected using sterile forceps, placed immediately into sterile tubes, and transported to the lab. Species identities were confirmed using identification guides specific to S. nobilis12 and Collins Field Guide for all other spiders62.
Using aseptic techniques, the specimens were dispatched, and the chelicerae were either clipped or swabbed. For whole body cultures, spiders were either submerged in media or swabbed. For surface colonisation analysis, spiders walked directly on Brain Heart Infusion (BHI) agar. The most common method for euthanising arthropods is dispatchment. A select number of spiders were euthanised using CO2, and immediately processed to determine if bacteria was recoverable by this alternate method.
For venom extractions, S. nobilis specimens were anesthetized using CO2 for 2 min and venom was extracted by electrostimulation with repeated pulses delivered at 15–20 V. Venom droplets were collected from the venom pores located on the outer subterminal part of the chelicerae using 5 µl microcapillary tubes modified with a tapered end for maximum efficiency. Venom from approximately 100 specimens was pooled and then flash-frozen in liquid nitrogen and stored at − 80 °C.
Preliminary testing for microbiomes from A. similis, E. atrica, and S. nobilis and 16S rRNA gene amplification, sequencing, and analysis
Whole bodies or chelicerae from three species of spiders: A. similis, E. atrica, and S. nobilis were transferred into 750 μl (10% dilution) of Luria Bertani (LB) broth, Nutrient broth (NB), Tryptic Soy broth (TSB), MRS broth and BHI broth, and incubated at both 37 °C and 10 °C. Whole culture from each spider or chelicerae were pelleted, DNA was extracted collectively from each sample using the QIAGEN Dneasy Blood & Tissue Kit and V3-V4 region of 16S rRNA was amplified using 341F 5′-CCTACGGGAGGCAGCAG-3′73, and 806R 5′-GGACTACHVGGGTWTCTAAT-3′74. The amplified product was then sent to GATC Biotech for sanger sequencing. A BLAST search was carried out with the obtained sequence using the NCBI rRNA/ITS database (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Bacterial isolation from S. nobilis and 16S rRNA gene amplification, sequencing and analysis
For isolating surface bacteria, S. nobilis spiders were washed individually with 5 ml BHI broth for 5 min. Some spiders were washed immediately after dispatchment, while others were processed 24–48 h after death. The wash media was then incubated at 37 °C overnight. For isolating bacteria from chelicerae, clipped chelicerae from each individual spider were inoculated into BHI broth and incubated at 37 °C. After 24 h incubation, the cultures were diluted and plated on BHI agar and incubated 48 h to 72 h at 37 °C. Selective media, Baird-Parker agar and TS-blood agar supplemented with colistin and nalidixic acid, were also inoculated with overnight cultures and incubated 48 h to 72 h at 37 °C. Colonies with different morphologies were selected for further analysis.
The 16S rRNA gene was amplified using Taq polymerase (Bioline) and universal primers, 27F (5′-AGAGTTTGATCATGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) using Colony PCR75. The PCR product was purified using the Wizard SV Gel and PCR Clean-Up System (Promega) and sequenced using primers, 27F 1492R (Eurofins Genomics, Germany).
A BLAST search was carried out with the obtained sequence using the NCBI rRNA/ITS database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Closest bacterial species were identified using Blast tree view produced by Blast pairwise alignment.
Inhibitory effects of S. nobilis venom against pathogens
Antibacterial activity of S. nobilis venom was assessed by agar diffusion assay and Minimum Inhibitory Concentration (MIC) assay. Agar diffusion assay was carried out against S. capitis (2) and P. azotoformans (isolated from S. nobilis chelicerae). Spiders were stimulated to aseptically bite Mueller–Hinton agar spread with 100 μl of adjusted overnight bacterial culture (0.8 OD590). We could observe the fangs penetrating the agar in a biting motion and also observe venom being expelled from the fangs. The restraining of the spider was enough to stimulate the bite and therefore no other manual stimulation was required. In addition, 0.5 μl neat venom was spotted onto the agar plate containing bacteria. Plates were incubated for 24 h at 37 °C and then assessed for zones of bacterial clearance.
The average volume of venom that each spider produces is approximately 0.22 µl (with a maximum of approximately 0.6 µl). Due to the limited amount of venom available, MICs were carried out by diluting the samples to achieve usable volumes. The MICs were performed against clinical isolates E. coli DSM10973, MRSA BH1CC and L. monocytogenes EGD-e. An overnight culture was adjusted with Muller-Hinton broth to an inoculum density of 1 × 106 cfu ml−1. Starting with a 1:100 dilution of the venom, twofold serial dilutions of the venom, was tested against all the pathogens in a final inoculum of 5 × 105 cfu ml−1. After 24 h incubation at 37 °C, absorbance at 590 nm was measured using a microplate reader (Tecan) with Magellan software.
Antibiotic susceptibility testing
Disk diffusion assays were carried out to determine antibiotic susceptibility. Experiments were conducted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines76. 6 mm disks preloaded with each antibiotic (Oxoid) were placed onto Mueller–Hinton agar plates that had been spread with 100 µl overnight bacterial culture (1 × 108 cfu ml−1). Plates were incubated at 37 °C for 18 h and the clear zone around each disk was measured using a ruler and interpreted according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints77 Bacterial spread plates without antibiotic disks were used as negative control and the bacteria grew as a lawn each time. Three independent experiments were performed in duplicate.
Ethical statement
No ethical approval was required to work with spiders. The three bite victims have provided the authors with written consent to use their case history and other relevant details to produce manuscripts intended for publication in scientific journals. They are aware that such publications may be available to the public both in print and on the Internet.
Data availability
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.
References
Pulford, C. V. et al. The diversity, evolution and ecology of Salmonella in venomous snakes. PLoS Negl. Trop. Dis. 13, e0007169 (2019).
Gal-Mor, O., Boyle, E. C. & Grassl, G. A. Same species, different diseases: how and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front. Microbiol. 5, 0391. https://doi.org/10.3389/fmicb.2014.00391 (2014).
Brandenburg, K., Heinbockel, L., Correa, W. & Lohner, K. Peptides with dual mode of action: Killing bacteria and preventing endotoxin-induced sepsis. Biochim. Biophys. Acta 1858, 971–979 (2016).
Rappuoli, R., Bloom, D. E. & Black, S. Deploy vaccines to fight superbugs. Nature 552, 165–167 (2017).
Pärnänen, K. M. et al. Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 5, eaau9124 (2019).
Dunbar, J. P. et al. Trunk vertebrae osteomyelitis in a spectacled caiman (Caiman crocodilus). Herpetol. Bull. 134, 15–18 (2015).
Cantas, L. & Suer, K. The important bacterial zoonoses in “one health” concept. Front. Public Health 2, 144 (2014).
Strand, T. M. & Lundkvist, Å. Rat-borne diseases at the horizon. A systematic review on infectious agents carried by rats in Europe 1995–2016. Infect. Ecol. Epidemiol. 9, 1553461 (2019).
Ul-Hasan, S. et al. The emerging field of venom-microbiomics for exploring venom as a microenvironment, and the corresponding Initiative for Venom Associated Microbes and Parasites (iVAMP). Toxicon 4, 100016 (2019).
Esmaeilishirazifard, E. et al. Microbial adaptation to venom is common in snakes and spiders. bioRxiv 2018, 348433 (2018).
Nentwig, W. Introduction, establishment rate, pathways and impact of spiders alien to Europe. Biol. Invasions. 17, 2757–2778 (2015).
Dugon, M. M. et al. Occurrence, reproductive rate and identification of the non-native noble false widow spider Steatoda nobilis (Thorell, 1875) in Ireland. Biol. Environ. 117, 77–89 (2017).
Dunbar, J., Ennis, C., Gandola, R. & Dugon, M. Biting off more than one can chew: First record of the non-native noble false widow spider Steatoda nobilis (Thorell, 1875) feeding on the native viviparous lizard Zootoca vivipara. Biol. Environ. 118B, 45–48 (2018).
Hódar, J. A. & Sánchez-Piñero, F. Feeding habits of the blackwidow spider Latrodectus lilianae (Araneae: Theridiidae) in an arid zone of south-east Spain. J. Zool. 257, 101–109 (2002).
Kuhn-Nentwig, L., Stöcklin, R. & Nentwig, W. Venom composition and strategies in spiders: is everything possible?. Adv. Insect Physiol. 40, 1–86 (2011).
O’Shea, M. & Kelly, K. Predation on a weasel skink (Saproscincus mustelinus)(Squamata: Scincidae: Lygosominae) by a redback spider (Latrodectus hasselti)(Araneae: Araneomorpha: Theridiidae), with a review of other Latrodectus predation events involving squamates. Herpetofauna 44, 49–55 (2017).
Barin, A., Arabkhazaeli, F., Rahbari, S. & Madani, S. The housefly, Musca domestica, as a possible mechanical vector of Newcastle disease virus in the laboratory and field. Med. Vet. Entomol. 24, 88–90 (2010).
Ndava, J., Llera, S. D. & Manyanga, P. The future of mosquito control: The role of spiders as biological control agents: A review. Int. J. Mosq. 5, 6–11 (2018).
Wang, Y.-C. et al. Transmission of Salmonella between swine farms by the housefly (Musca domestica). J. Food. Prot. 74, 1012–1016 (2011).
Barrantes, G. & Weng, J.-L. Carrion feeding by spiderlings of the cob-web spider Theridion evexum (Araneae, Theridiidae). J. Arachnol. 35, 557–561 (2007).
Sandidge, J. S. Arachnology: scavenging by brown recluse spiders. Nature 426, 30 (2003).
Vetter, R. S. Scavenging by spiders (Araneae) and its relationship to pest management of the brown recluse spider. J. Econ. Entomol. 104, 986–989 (2011).
Baxter, R. H., Contet, A. & Krueger, K. Arthropod innate immune systems and vector-borne diseases. Biochemistry 56, 907–918 (2017).
Kavanagh, K. & Reeves, E. P. Insect and mammalian innate immune responses are much alike. Microbe 2, 596 (2007).
Savitzky, A. H. et al. Sequestered defensive toxins in tetrapod vertebrates: principles, patterns, and prospects for future studies. Chemoecology 22, 141–158 (2012).
Monteiro, C. L. B. et al. Isolation and identification of Clostridium perfringens in the venom and fangs of Loxosceles intermedia (brown spider): enhancement of the dermonecrotic lesion in loxoscelism. Toxicon 40, 409–418 (2002).
Ahrens, B. & Crocker, C. Bacterial etiology of necrotic arachnidism in black widow spider bites. J. Clin. Toxicol 1, 106 (2011).
Peel, M. M. et al. Isolation, identification, and molecular characterization of strains of Photorhabdus luminescens from infected humans in Australia. J. Clin. Microbiol. 37, 3647–3653 (1999).
Dunbar, J. P., Afoullouss, S., Sulpice, R. & Dugon, M. M. Envenomation by the noble false widow spider Steatoda nobilis (Thorell, 1875)–five new cases of steatodism from Ireland and Great Britain. Clin. Toxicol. 56, 433–435 (2018).
Dunbar, J. P., Sulpice, R. & Dugon, M. M. The kiss of (cell) death: can venom-induced immune response contribute to dermal necrosis following arthropod envenomations?. Clin. Toxicol. 57, 677–685 (2019).
Hauke, T. J. & Herzig, V. Dangerous arachnids—Fake news or reality?. Toxicon 138, 173–183 (2017).
Swansen, D., Vetter, R. S. & White, J. Clinical manifestations and diagnosis of widow spider bites. UpToDate (2018).
Vetter, R. S. Clinical consequences of toxic envenomation by spiders. Toxicon 152, 65–70 (2018).
Hogan, C. J., Barbaro, K. C. & Winkel, K. Loxoscelism: old obstacles, new directions. Ann. Emerg. Med. 44, 608–624 (2004).
Patel, K. D., Modur, V., Zimmerman, G. A., Prescott, S. M. & McIntyre, T. M. The necrotic venom of the brown recluse spider induces dysregulated endothelial cell-dependent neutrophil activation. Differential induction of GM-CSF, IL-8, and E-selectin expression. J. Clin. Investig. 94, 631–642 (1994).
Ribeiro, M. et al. Pattern of inflammatory response to Loxosceles intermedia venom in distinct mouse strains: a key element to understand skin lesions and dermonecrosis by poisoning. Toxicon 96, 10–23 (2015).
Rivera, I.-G. et al. Sphingomyelinase D/ceramide 1-phosphate in cell survival and inflammation. Toxins 7, 1457–1466 (2015).
Vetter, R., Swanson, D., Weinstein, S. & White, J. Do spiders vector bacteria during bites? The evidence indicates otherwise. Toxicon 93, 171–174 (2015).
Benoit, R. & Suchard, J. R. Necrotic skin lesions: spider bite-or something else?. Consultant 46, 1386–1386 (2006).
Gaver-Wainwright, M. M., Zack, R. S., Foradori, M. J. & Lavine, L. C. Misdiagnosis of spider bites: bacterial associates, mechanical pathogen transfer, and hemolytic potential of venom from the hobo spider, Tegenaria agrestis (Araneae: Agelenidae). J. Med. Entomol. 48, 382–388 (2011).
Saez, N. J. et al. Spider-venom peptides as therapeutics. Toxins 2, 2851–2871 (2010).
Agnarsson, I. Morphological phylogeny of cobweb spiders and their relatives (Araneae, Araneoidea, Theridiidae). Zool. J. Linnean. Soc. 141, 447–626 (2004).
Bauer, T. et al. Steatoda nobilis, a false widow on the rise: a synthesis of past and current distribution trends. NeoBiota 42, 19 (2019).
Dunbar, J., Schulte, J., Lyons, K., Fort, A. & Dugon, M. New Irish record for Steatoda triangulosa (Walckenaer, 1802), and new county records for Steatoda nobilis (Thorell, 1875), Steatoda bipunctata (Linnaeus, 1758) and Steatoda grossa (CL Koch, 1838). Ir. Nat. J. 36, 39–43 (2018).
Nolan, M. Three Spiders (Araneae) New to Ireland: Bolyphantes alticeps Oonops domesticus and Steatoda nobilis. Ir. Nat. J. 26, 200–202 (1999).
Türkeş, T. & Mergen, O. The comb-footed spider fauna of the central Anatolia region and new records for the Turkish fauna (Araneae: Theridiidae). Serket 10, 112–119 (2007).
Zamani, A., Mirshamsi, O., Jannesar, B., Marusik, Y. M. & Esyunin, S. L. New data on spider fauna of Iran (Arachnida: Araneae) Part II. Zool. Ecol. 25, 339–346 (2015).
Faúndez, E. I., Carvajal, M. A. & Aravena-Correa, N. P. On a bite by Steatoda nobilis (Thorell, 1875) (Araneae: Theridiidae) on a human being, with comments on its handling during the 2020 SARS-COV-2 pandemic. Rev. Iber. Aracnol. 51, 178–180 (2020).
Faúndez, E. I., Carvajal, M. A., Darquea-Schettini, D. & González-Cano, E. Nuevos registros de Steatoda nobilis (Thorell, 1875)(Araneae: Theridiidae) de Sudamérica. Rev. Iber. Aracnol. 33, 52–54 (2018).
Faúndez, E. I. & Téllez, F. Primer registro de una mordedura de Steatoda nobilis (Thorell, 1875) (Arachnida: Araneae: Theridiidae) en Chile. Arquivos Entomol. 15, 237–240 (2016).
Taucare-Ríos, A., Mardones, D. & Zúñiga-Reinoso, Á. Steatoda nobilis (Araneae: Theridiidae) in South America: a new alien species for Chile. Can. Entomol. 148, 479–481 (2016).
Vetter, R. S., Adams, R. J., Berrian, J. E. & Vincent, L. S. The European spider Steatoda nobilis (Thorell, 1875)(Araneae: Theridiidae) becoming widespread in California. Pan-Pac. Entomol. 91, 98–101 (2015).
Porras-Villamil, J. F., Olivera, M. J., Hinestroza-Ruiz, Á. C. & López-Moreno, G. A. Envenomation by an arachnid (Latrodectus or Steatoda): Case report involving a woman and her female dog. Case Rep. 6, 33–43 (2020).
Warrell, D., Shaheen, J., Hillyard, P. & Jones, D. Neurotoxic envenoming by an immigrant spider (Steatoda nobilis) in southern England. Toxicon 29, 1263–1265 (1991).
Hambler, C. The’Noble false widow’spider Steatoda nobilis is an emerging public health and ecological threat. PCI. Zool. https://doi.org/10.31219/osf.io/axbd4 (2020).
Petrov, B. & Lazarov, S. Steatoda triangulosa (Walckenaer, 1802) feeding on a European blind snake. Newsltr. Br. Arachnol. Soc. 81, 9–10 (2000).
Zamani, A. Predation on Cyrtopodion scabrum (Squamata: Gekkonidae) by Steatoda paykulliana (Araneae: Theridiidae). Riv. Arachnol. Ital. 8, 12–15 (2016).
Dunbar, J. P. et al. Venomics approach reveals a high proportion of Lactrodectus-Like toxins in the venom of the noble false widow spider Steatoda nobilis. Toxins 12, 402 (2020).
Garb, J. E. & Hayashi, C. Y. Molecular evolution of α-latrotoxin, the exceptionally potent vertebrate neurotoxin in black widow spider venom. Mol. Biol. Evol. 30, 999–1014 (2013).
Gendreau, K. L. et al. House spider genome uncovers evolutionary shifts in the diversity and expression of black widow venom proteins associated with extreme toxicity. BMC Genom. 18, 178 (2017).
Isbister, G. K. & Gray, M. R. Effects of envenoming by comb-footed spiders of the genera Steatoda and Achaearanea (family Theridiidae: Araneae) in Australia. J. Toxicol. Clin. 41, 809–819 (2003).
Roberts, M. J. Spiders of Britain & Northern Europe (HarperCollins Publishers, New York, 1995).
Metcalf, J. L. et al. A microbial clock provides an accurate estimate of the postmortem interval in a mouse model system. Elife 2, e01104 (2013).
Giordano, N. et al. Erythema nodosum associated with Staphylococcus xylosus septicemia. J. Microbiol. Immunol. Infect. 49, 134–137 (2016).
Pain, M., Hjerde, E., Klingenberg, C. & Cavanagh, J. P. Comparative genomic analysis of Staphylococcus haemolyticus reveals keys to hospital adaptation and pathogenicity. Front. Microbiol. 10, 2096 (2019).
Premkrishnan, B. N. et al. Complete genome sequence of Staphylococcus haemolyticus type strain SGAir0252. Genome Announc. 6, e00229-e1218 (2018).
Ladhani, S. & Bhutta, Z. Neonatal Pseudomonas putida infection presenting as staphylococcal scalded skin syndrome. Eur. J. Clin. Microbiol. Infect. Dis. 17, 642–644 (1998).
Mustapha, M. M. et al. Draft genome sequences of four hospital-associated Pseudomonas putida isolates. Genome Announc. 4, e01039-e11016 (2016).
Horino, T., Inotani, S., Matsumoto, T., Ichii, O. & Terada, Y. Reactive arthritis caused by Rothia mucilaginosain an elderly diabetic patient. JCR J. Clin. Rheumatol. 8, 228–234 (2019).
Poyer, F. et al. Rothia mucilaginosa bacteremia: A 10-year experience of a pediatric tertiary care cancer center. Pediatr. Blood. Cancer 66, e27691 (2019).
Kozlov, S. A. et al. Latarcins, antimicrobial and cytolytic peptides from the venom of the spider Lachesana tarabaevi (Zodariidae) that exemplify biomolecular diversity. J. Biol. Chem 281, 20983–20992 (2006).
Ventola, C. L. The antibiotic resistance crisis: part 1: causes and threats. Pharmacol. Ther. 40, 277 (2015).
Lane, D. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics (eds Stackebrandt, E. & Goodfellow, M.) 115–175 (Wiley, West Sussex, 1991).
Caporaso, J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. PNAS 108, 4516–4522 (2011).
Senthilraj, R., Prasad, G. S. & Janakiraman, K. Sequence-based identification of microbial contaminants in non-parenteral products. Braz. J. Pharm. 52, 329–336 (2016).
Wayne, P. A. Clinical and Laboratory Standards Institute; CLSI. Performance Standard for Antimicrobial Disk Susceptibility Tests; Approved standard-Eleventh edition. CLSI document M02-A11 (2012).
EUCAST, T. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 7.1, http://www.eucast.org, (2017).
Acknowledgements
The authors thank Eoin MacLoughlin for assistance during field work. We thank Maria Condon for access to property for sampling. The authors also thank Dr Maria Barrett for assistance with preparing samples for sequencing. We are also very grateful to the two anonymous referees whose critical review of this manuscript was extremely valuable. This project was funded through the Irish Research Council under a Government of Ireland Postgraduate Scholarship held by Neyaz Khan (GOIPG/2017/910), the NUI Galway Ryan Award for Innovation held by Michel Dugon and a NUI Galway College of Science PhD scholarship held by John Dunbar.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: M.M.D., J.P.D., V.O.F., C.L.A., A.B., N.A.K. Performed the experiments: J.P.D., N.A.K., C.L.A., P.B., J.M., S.A. Analysed the data: A.B., V.O.F., M.M.D., J.P.D., N.A.K., C.L.A. Wrote the paper: J.P.D., N.A.K. Sourcing spiders and venoms: J.P.D. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dunbar, J.P., Khan, N.A., Abberton, C.L. et al. Synanthropic spiders, including the global invasive noble false widow Steatoda nobilis, are reservoirs for medically important and antibiotic resistant bacteria. Sci Rep 10, 20916 (2020). https://doi.org/10.1038/s41598-020-77839-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-77839-9
This article is cited by
-
Effects of cadmium and diethylhexyl phthalate on skin microbiota of Rana chinensis tadpoles
Environmental Science and Pollution Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.