Abstract
To estimate regional Alzheimer disease (AD) pathology burden clinically, analysis methods that enable tracking brain amyloid or tau positron emission tomography (PET) with magnetic resonance imaging (MRI) measures are needed. We therefore developed a robust MRI analysis method to identify brain regions that correlate linearly with regional amyloid burden in congruent PET images. This method was designed to reduce data variance and improve the sensitivity of the detection of cortical thickness–amyloid correlation by using whole brain modeling, nonlinear image coregistration, and partial volume correction. Using this method, a cross-sectional analysis of 75 tertiary memory clinic AD patients was performed to test our hypothesis that regional amyloid burden and cortical thickness are inversely correlated in medial temporal neocortical regions. Medial temporal cortical thicknesses were not correlated with their regional amyloid burden, whereas cortical thicknesses in the lateral temporal, lateral parietal, and frontal regions were inversely correlated with amyloid burden. This study demonstrates the robustness of our technique combining whole brain modeling, nonlinear image coregistration, and partial volume correction to track the differential correlation between regional amyloid burden and cortical thinning in specific brain regions. This method could be used with amyloid and tau PET to assess corresponding cortical thickness changes.
Similar content being viewed by others
Introduction
The 2018 National Institute on Aging and Alzheimer's Association Research Framework emphasizes basing the diagnosis of Alzheimer's disease (AD) on underlying pathologic processes and supporting the clinical diagnosis with the identification of biomarkers grouped into the categories of β amyloid burden, pathologic tau burden, and neurodegeneration; this is known as the AT(N) model1. Among the imaging biomarkers, amyloid positron emission tomography (PET) ligand binding (biomarker of Aβ plaques, “A”) is increasingly used clinically, whereas cortical tau PET ligand binding (biomarker of fibrillar tau, “T”) is often used for research. Volumetric structural magnetic resonance imaging (MRI) assessment of neurodegeneration (“N”) is even more widely available and is often the initial biomarker available to a clinician1.
The ability of structural MRI to precisely measure regional brain volumes has yielded insights identifying patterns of atrophy related to AD (“N” in the AT[N] model). A “cortical signature” of AD pathology has been reported: more pronounced atrophy in the limbic and heteromodal association cortices, including the hippocampus, posterior and polar temporal lobe, parietal lobe, and portions of the frontal lobe2,3. Nonetheless, even as cortical thinning in AD is postulated to be a result of neuronal damage from a combination of neurofibrillary tangles, neuritic plaques, and other pathology2,4, the burden of neuritic plaques does not conform to the AD signature areas5. Furthermore, cortical thinning is also influenced by other factors including age6,7,8, cognitive reserve9, and stage of disease10,11,12.
To better use MRI regional cortical thickness as a biomarker for tracking clinical AD progression while accounting for these above factors, a robust method for coregistration of a PET image (amyloid or tau) to the corresponding MR image is required. An important challenge here is the size scale of the voxels (approximately 1 mm for MRI and 6–8 mm for PET), which creates uncertainty in coregistration. Another challenge involves the nonlinear distortions common with MR images that are more pronounced in the periphery of the imaged volume; these distortions can be several millimeters in size and can be significant compared to normal cerebral cortex, which ranges from 1.0 to 4.5 mm in thickness. Imprecise coregistration can lead to cortex in one modality being compared to white matter in the other modality. It is therefore not surprising that although multiple studies have tracked MRI cortical changes to cross-sectional and longitudinal PET amyloid burden and assessed the relationship between these factors and cognitive decline13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28, two key questions related to precise quantification of regional cortical information remain: (1) Is the relationship between PET amyloid burden and cortical thickness linear or nonlinear13,17 and (2) does this relationship vary across individual cortical regions? To answer these questions, precise measurement of spatially coincident cortical thickness and amyloid is required, for which we have developed improved image processing tools; nevertheless, methodological challenges remain an obstacle to accurate correlation of amyloid and tau PET signal with regional cortical thickness.
To this end, we designed a novel image processing method to reduce data variance and improve the sensitivity of detection of the cortical thickness–amyloid correlation in individual cortical regions. The key elements of the method, detailed in the next section, involve whole brain modeling, nonlinear image coregistration, modified partial volume correction, and 3 T imaging. While these technical advances have been applied separately in previous work by others, this study is the first to demonstrate the utility of combining these methods. We next used our novel method to investigate the spatial patterns of correlations between regional amyloid burden and regionally coincident cortical thinning. This research is unique in applying this method fully to all brain regions, in particular the cortex, and also in including cerebrospinal fluid (CSF) regions as controls (as there should be no relationship between CSF standardized uptake value ratios [SUVR] and CSF volumes).
Previous research in prodromal AD found a pronounced correlation between tau PET ([18F]flortaucipir) and cortical thickness in temporal and parietal regions, whereas the correlation between amyloid PET ([18F]flutemetamol) and cortical thickness was less region specific4. We therefore used our novel image processing method to assess concomitant amyloid PET and MRI images from a cross-sectional cohort of memory clinic patients to test the hypothesis that the degree of correlation between amyloid burden and cortical thickness is highest in the temporoparietal neocortical regions where early AD pathology is noted (also described by tau PET). We also hypothesized that evaluation of the relationship between regional amyloid burden and cortical thickness would allow us to estimate the accuracy of using structural MRI metrics to infer the burden of amyloid and tau as measured using amyloid and tau PET.
Methods
Overview of approach
Achieving accurate correlation of cortical atrophy and amyloid burden throughout the brain depends on obtaining a near-exact coregistration of PET to MRI, such that the cortical ribbon from each modality shares considerable overlap (≥ 50%). Thus, if the mean cortical thickness is 2–3 mm, the accuracy of coregistration needs to be on order of 1 mm. To achieve this accuracy, three sources of potential error must be addressed. The first is the considerable difference in spatial resolution between the modalities (approximately 1 mm for MRI and 6–8 mm for PET). The second involves the markedly different tissue contrast characteristics of the two modalities; although many algorithms can accurately coregister two scans using the same modalities, the accuracy is understandably lower when coregistering scans from different modalities, such as PET and MRI. The third source of error involves subtle but widespread spatial distortions of MRI. While not apparent to the naked eye, these distortions can alter measured distances in the brain by more than 1 mm when compared with measurements obtained with less-distorted modalities such as CT. In this study, we addressed the first two errors by constructing a PET model derived from the brain region segmentation of the MRI data, which should achieve similar spatial resolution and permit coregistration between like modalities (PET to PET). To address the third error, we used a higher order coregistration (both affine and nonlinear) between the modeled PET and PET. Additionally, the final values of cortical amyloid burden were adjusted for partial volume correction (PVC), a goodness-of-fit parameter was calculated, and final cortical amyloid values for all cortical parcels were compared to cortical thickness in a patient cohort.
Patients
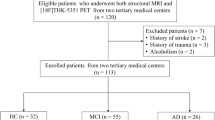
This retrospective study was approved by the Cleveland Clinic Institutional Review Board, and all protected health information requirements were strictly followed and all methods were carried out in accordance with relevant guidelines and regulations. Informed consents were also waived by the Cleveland Clinic Institutional Review Board, in this retrospective study with minimal risk. Using a patient registry from the Cleveland Clinic Lou Ruvo Center for Brain Health, a tertiary memory clinic, we identified 75 patients who underwent both a brain florbetapir PET scan and a head MRI scan (following the ADNI protocol available at adni.loni.usc.edu) between January 2013 and January 2017; both scans were performed as part of a diagnostic evaluation of neurocognitive deficits. Of these 75 patients, 55 had a consensus diagnosis of AD dementia, 5 had mild cognitive impairment (MCI) likely from AD (all amyloid positive), and 15 were amyloid negative (as determined from the radiologist’s report on the amyloid PET scan) and had diagnoses of subjective memory complaints that were likely non-neurodegenerative and were included in the analysis to establish an amyloid-negative baseline. The clinical consensus diagnoses were made by a team of behavioral neurologists, neuropsychologists, and neuroradiologists and were based on clinical criteria for AD dementia29 and for MCI due to AD30. APOE ε 4 genetic information was not available for all patients and was not included in the analysis.
MRI acquisition
Volumetric T1-weighted images were obtained at 3 T using Siemens MRI scanners (Skyra, Prisma, or Trio, depending on the site; Siemens Medical Systems, Erlangen, Germany). The volumetric sequences were obtained using the ADNI MPRAGE protocol with the following acquisition parameters: repetition time/echo time/inversion time = 2300/2.98/900; flip angle = 9°; bandwidth = 240 Hz/pixel; 240 × 256 matrix; 160 slices; voxel size = 1 × 1 × 1.2 mm; scan time = 9:14; and 32-channel head coil. Images were acquired in the sagittal plane with 1.2-mm slice spacing and in-plane voxel size of 1.0 × 1.0 mm.
PET acquisition
All PET images were obtained using a combined PET/computed tomography (CT) system (Siemens Biograph mCT 20 or 40, depending on the site) and were analyzed retrospectively. The voxel sizes were 0.78 × 0.78 × 2.0 (thick) mm for CT images and 2.12 × 2.12 × 2.03 (thick) mm for PET images. The Biograph 20 scanner used a 16-cm axial field of view, with reconstruction performed using a 3D OSEM algorithm (6 iterations, 21 subsets, 2-mm Gaussian filter, without resolution modeling, and with time-of-flight information). The Biograph 40 scanner used a 21-cm axial field of view, with reconstruction performed using a 3DOSEM algorithm (6 iterations, 24 subsets, 3-mm Gaussian filter, with resolution modeling, and without time-of-flight information). The amyloid PET radiotracer used was 18F florbetapir, administered intravenously at a prescribed dose of 10.0 mCi (delivered doses ranged from 8.1 to 12.0 mCi). A 15-min image acquisition was obtained approximately 50 min after injection. Standard data corrections were applied (randoms, deadtime, normalization, decay, scatter, and CT-derived attenuation correction).
MRI segmentation
For all patients, FreeSurfer 6.0 processing31,32 was performed on the volumetric T1-weighted sequence, which provided segmentation of each hemisphere’s 34 cortical parcels, 34 subcortical white matter parcels, and 97 remaining parcels of deep gray matter structures, other white matter structures, brainstem, cerebellum, and ventricles. A small number of voxels remained as unsegmented structures with common labels. Because this confounded later modeling, these small areas were relabeled to match those of the most common nearest neighbor segmentations, using custom software written in IDL (Interactive Data Language, Harris Geospatial Solution, Boulder, CO, USA). A total of 165 volumes for a model were fit to the PET images. A binary mask was formed around the parcellated brain using a 3-voxel dilation. All subsequent coregistrations between PET and MRI data were conducted with the MRI space used as the reference.
Preprocessing and initial coregistration of PET, CT, and MRI
The head frame was first removed from CT images because it confounded coregistration algorithms. Because we used a combined PET/CT scanner, the native PET and CT images were oriented in a common physical space; however, because of the large difference in voxel sizes, the native PET image was upsampled to match the image space of the CT scan. A rigid coregistration was performed between the MRI T1-weighted image and CT images using FLIRT from FSL33,34, with 6 degrees of freedom and a normmi search cost. The resulting transformation matrix was applied to the native PET image, thereby producing a new image (hereafter referred to as the “PET image”) that was coarsely coregistered to the MR image. Because of the intrinsic higher spatial resolution of the MR image, the MR image was used as a reference image in all subsequent analyses, and all other transformations were made on the PET image.
Construction of a PET model from segmented MRI
A PET model was constructed using the entire parcellation from FreeSurfer output of MR images. This enabled us to make comparisons of the same modality (PET images with modeled PET), thus allowing for fine-tuned coregistration using higher order techniques. The model was constructed by first assigning the mean PET activity in the corresponding location of the PET image for each cortical parcel. However, a simple correspondence is not possible because of partial volume averaging caused by the relatively larger point spread function of PET, wherein the activity originating from one parcel will “bleed in” to adjacent parcels (an effect that will be corrected at a later stage). The typical full width at half maximum (FWHM) of this spread can range from 6 to 8 mm, much larger than the typical cortical thickness of 2–3 mm. Thus, this blurring was modeled independently for all segmented parcels using a Gaussian convolution with a FWHM of either 6 or 8 mm, assuming each parcel had a uniform distribution of amyloid signal. These values were chosen to reflect the range of observed resolution from PET images, which combines the intrinsic resolution of the hardware and some degree of superimposed additional blurring due to any patient motion. A modeled PET image could then be constructed if the value of uniform amyloid signal was provided for each parcel. PET and MRI could then be compared using the surrogate of a PET model, and a more refined coregistration could be performed by removing the nonlinear distortions from the MRI scan.
Fine-tuned coregistration
To fine-tune the coregistration between the PET image and the PET model, two methods were used: an affine (degrees of freedom = 12) using FLIRT, and a nonlinear coregistration using Advanced Normalization Tools35,36,37. The FLIRT routine used a cost function of leastsq, with a constrained search in all three dimensions of − 20 to 20. The Advanced Normalization Tools allowed us to bring the gyration and sulcation patterns into more precise agreement.
Partial volume correction
Because the point spread function of PET is larger than the typical thickness of cortex, amyloid activity measured in one region necessarily includes activity partially spreading or averaging from adjacent regions. If the spread function is known, it should be possible to account for this spread and compute PET maps without any spread. A number of PVC techniques have been described for this use38; these methods increase the accuracy of cortical amyloid measurements but at the cost of increased noise. For this study, we adopted the geometric transfer matrix method, which was first described by Rousset et al.39 and Labbe et al.40. In short, given a set of N parcels covering the entire brain and a spread function characterized by a parameter such as the FWHM, a square matrix can be constructed (in our case, 165 × 165) for which the partial spread or loss from any parcel to all other parcels is computed. The matrix equation can then be solved to obtain the intrinsic (i.e., nonspread) values at each parcel. Routine least-squares methods can be used to invert the matrix, but we found that these results were often unstable, resulting in parcels with nonphysical values (e.g., negative or unrealistically high). In effect, a superior fit could be obtained in some brain parcels at the expense of a negative SUVR in other parcels. A solution was to use a constrained least-squares routine that could place lower and upper limits on the results, for which we used the lsqlin routine in MATLAB (MathWorks, Natick, MA, USA), using a lower limit constraint of 0 SUVR in any parcel (i.e., cannot be negative). The final result was a set of 165 numbers representing the mean amyloid activity in all parcels. Because there is no practical calibration for the absolute amyloid PET radiotracer uptake in the brain, all values were normalized to a common region to generate SUVRs, and for this research, the brainstem was used. Alternative calculations (not presented) tested other regions such as the cerebellum, which is commonly used, but goodness-of-fit parameters were marginally better using the brainstem. All cortical parcels have a corresponding subcortical white matter parcels; therefore, the mean ratio of cortical activity to subcortical white matter activity was computed. All calculations were performed using IDL.
Measuring goodness of fit
To quantify the accuracy of coregistration methods, a normalized root-mean-square difference between two images was used. In particular, for an image comparing N voxels, the following equation was used:
where the mean is computed over all voxels. The fit was computed for all models over the entire brain or only the cortex. A value close to 0 would indicate a good fit; a value close to 1 would indicate a poor fit. Goodness-of-fit results are presented in Supplementary Information.
Application to patients and statistical analysis
Three different models were applied to images from all patients: (1) rigid coregistration with no PVC; (2) affine coregistration with PVC; and (3) nonlinear coregistration with PVC. Each model yielded best-fit values for normalized amyloid activity in the 165 parcels used in this study. Of these, 68 cortical parcels could be compared with the mean cortical thickness values provided by FreeSurfer. Computing these values for each patient then allowed us to further analyze the relationship between cortical amyloid and thickness. Each parcel could be evaluated for significance of correlation (described below), and the overall performance of each model could be judged by the number of parcels with a significant correlation. A drawback to this measure is its assumption that there is an underlying relationship between SUVR and cortical thickness; there may be some parcels for which no such relationship actually exists. Thus, although this may not be the best technique for evaluating any of the three described methods individually, it is likely a reasonable way to compare the relative merits of the three methods against each other.
Analysis of the ventricles formed a useful control for testing the PVC method, as the ventricular size would not be expected to have any correlation with soluble amyloid contained within the CSF (i.e., there should not be a significant correlation between ventricular SUVR and ventricular volume).
Statistical analyses were performed using the IMSL_MULTIREGRESS routine in IDL to create multiple linear regression models and the IMSL_PARTIAL_COV routine to calculate partial correlation coefficients. The first statistical analysis performed a simple linear regression to calculate the Pearson correlation coefficient between the cortical thickness and amyloid uptake, yielding a correlation coefficient ρ and its associated p value. A second statistical analysis used a multivariable linear regression including the clinical variables of education, age, sex, and Montreal Cognitive Assessment (MoCA) score with the model assuming
yielding an overall correlation coefficient and regression coefficients with associated p values for the 5 dependent variables. Following multiple comparisons across 68 FreeSurfer cortical regions (using both hemispheres), the false discovery rate (FDR) was controlled using the Benjamini–Hochberg procedure41, using an FDR of 0.1 to yield p < 0.018. An independent sample t-test was used to compare continuous variables, and Fisher’s exact test was used for categorical variables. A difference with a two-tailed probability of p < 0.05 was considered statistically significant (Null hypothesis was that the degree of correlation between amyloid burden and cortical thickness is the same in all regions).
A pipeline diagram of the analysis method is provided in the supplementary material as Supplementary Fig. 2.
Results
Patients
A summary of demographic features of the 75 patients included in the final data analysis is provided in Table 1. These patients were subcategorized into 3 groups roughly reflecting the severity of dementia, as demarked by MoCA scores: 0–10 (n = 11); 11–19 (n = 31); and 20–30 (n = 33).
Image coregistration
Figure 1 shows imaging data from a single patient and demonstrates the utility of higher order coregistration and PVC. The best correspondence was seen between nonlinear coregistered PET and the PET model using PVC, particularly in the details of cortical overlap. Additional details comparing our novel MR image processing to previous methods are provided in Supplementary Information, which includes results about goodness-of-fit measurements. For example the goodness of fit parameter (Eq. 1) across the entire brain assuming a FWHM of 8 mm was 0.163 for nonlinear coregistered PET with PVC, whereas 0.388 for a rigid coregistration with no PVC.
Sample images of PET data, coregistration, and PET models. Images show (A) PET data after rigid coregistration of native PET data to MRI space; (B) initial MRI-based model of PET data derived from FreeSurfer parcels, using values from (A) as inputs, with no PVC; (C) nonlinear coregistration of native PET data to MRI space; and (D) final MRI-based PET model using input values from (C) and incorporating PVC. The correspondence between (A) and (B) was grossly adequate but insufficient for details close to the cortical ribbon. Differences are best appreciated along the margins of features (e.g., near the apex, as shown in the orange circles). The correspondence between (C) and (D) was greatly improved with the use of nonlinear coregistration and PVC (see blue circles). The colored display reflects the SUVR, with the normalized magnitude indicated by the color bar, scaled between the maximum value (which was close to SUVR = 3.0) and zero. MRI magnetic resonance imaging, PET positron emission tomography, PVC partial volume correction, SUVR standardized uptake value ratio.
Association between cortical amyloid and cortical thickness
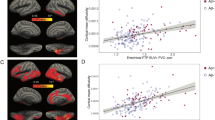
Figure 2 shows scatter plots of regional amyloid uptake versus cortical thickness for two cortical parcels: the middle temporal cortex and the pericalcarine cortex. In each case, the group with the lowest MoCA score demonstrated the highest amyloid uptake. In the middle temporal lobe, this was associated with significant cortical thinning; in the pericalcarine cortex, there was no change in cortical thickness. These two plots are exemplary of the ends of the relationship spectrum, i.e., one plot shows a strong relationship between cortical thickness and amyloid uptake, whereas the other shows no relationship. In particular these examples show a range of Pearson coefficient between a high correlation (R = 0.69; p = 7 × 10–12) and no correlation (R = 0.07; p = 0.53).
Scatter plot demonstrating cortical thickness and cortical amyloid uptake as measured by SUVR. Left: a region of high Pearson correlation (R = 0.69; p = 7 × 10–12) in the middle temporal gyrus. Right: a region of low Pearson correlation (R = 0.07; p = 0.53) in the pericalcarine cortex. The patients are color-coded into three groups reflecting the degree of degenerative disease (stratified by MoCA score), with crosshairs for each group representing means and standard deviations. Red data are from patients with MoCA score ≤ 10; blue data are from patients with MoCA score of 11–19; and yellow data are from patients with MoCA score ≥ 20. MoCA Montreal Cognitive Assessment, SUVR standardized uptake value ratio.
For each brain parcel, a simple linear regression model and multivariable linear regression were used to measure the association between cortical amyloid and cortical thickness (Table 2). In the simple regression model, the temporal lobe had the greatest number of regions with highly significant correlations (three regions), followed by the parietal lobe (also with three regions, but less significant compared with the temporal lobe). In the multivariable model, the variable with the strongest correlation was the MoCA score (with 28 parcels out of 34 being significant), followed by education (20 parcels) and cortical thickness (11 parcels). All brain regions except the entorhinal, parahippocampal, and posterior cingulate regions showed significant correlation of MoCA with amyloid SUVR. Analysis of the lateral ventricles showed no significant correlation between ventricular SUVR and ventricular volume when using a nonlinear coregistration with a constrained PVC (r = 0.04, p = 0.66). A significant correlation resulted when using either a rigid coregistration without PVC (r = − 0.72, p < 0.001) or nonlinear coregistration with an unconstrained PVC (r = − 0.38, p < 0.001) (see Supplementary Information and Supplementary Fig. 2).
Figure 3 shows an inflated surface image of both hemispheres, with a colored overlay indicating the partial regression coefficient. The threshold of significance for each region was 0.23, derived from an FDR of 0.1 and yielding a p-value of 0.037. The regions of significance corresponding to AD signature regions included the medial and lateral parietal lobes, temporal lobes, and prefrontal lobes (left more than right). Of note, regions outside of the AD signature regions, including the fusiform gyri and lateral occipital regions, were also significant. Averaged over all parcels, the earliest stages of the disease (subjective memory complaints, MCI) were associated with increased amyloid SUVR but stable cortical thickness, whereas the later stages of the disease (AD dementia) were associated with correlated change (increased amyloid in parallel with cortical thinning) (Fig. 4). The average cortical SUVR and cortical thickness values for each of the three patient groups in Fig. 4A are shown in Table 3. AD signature regions of the parietal and temporal lobes were associated with the most change in cortical thickness across AD stages, while the frontal lobes were associated with a smaller degree of change (Fig. 4D).
Cortical surface overlay showing the partial correlation coefficient between cortical amyloid and cortical thickness. Partial correlation coefficient is shown for each cortical parcel after adjustments were made for age, education, sex, and MoCA score. Displayed are the medial and lateral views of each hemisphere. Using a FDR of 0.1 yielded a p value threshold of 0.037, corresponding to a correlation coefficient threshold of 0.23. FDR false discovery rate, MoCA Montreal Cognitive Assessment.
Patterns of amyloid–cortical thickness values extended to all cortical parcels. (A) Scatter plot for all 34 cortical parcels (each an average of both hemispheres). Each parcel was subanalyzed separately classified by the MoCA score of the patient (red: MoCA score 0–10; blue: MoCA score 11–19; yellow: MoCA score 20–30). Each dot is an average of the cortical thickness and amyloid (SUVR) for each parcel, for all patients in the group indicated by the color. Overlaid crosshairs show the means and standard deviations for the three groups. (B) Scatter plot of the same data points from (A) but with overlaid lines connecting the points belonging to the same parcel. (C) Similar scatter plot as shown in (A) and (B) but with each parcel normalized to a common cortical amyloid SUVR (1.0) and cortical thickness value (0.0) for the high MoCA score group (yellow circles). The parcels were grouped into 2 categories (thicker or thinner) depending on how the high MoCA group (yellow circles) compared with the middle MoCA group (blue circles). This shows the trend of decreased cortical thickness from high MoCA to low MoCA groups. (D) Surface image of an inflated brain using data from (C), showing the spatial variation from a simple dichotomization from (C) (i.e., between high MoCA to medium MoCA. The regions of less thinning generally occur in the frontal parenchyma, Heschl’s gyrus, and calcarine sulcus. MoCA Montreal Cognitive Assessment, SUVR standardized uptake value ratio.
The number of regions with significant correlations also varied depending on the coregistration and PVC methods used, with more accurate coregistration yielding more brain regions demonstrating statistical significance (Table 4). In general, the left hemisphere showed more significant correlations than the right (17 versus 13, using a nonlinear coregistration with constrained least-square fit PVC), and there was an increasing number of significant relationships with increasing goodness of fit (30 for nonlinear coregistration with constrained least-square fit PVC, compared to 17 for rigid coregistration with no PVC).
Discussion
In this cross-sectional study, we used a novel MRI-PET image processing method to investigate in better detail the cortical thickness as measured by MRI with the corresponding amyloid burden as measured by amyloid PET. We found that increasing accuracy of coregistration methods yields more brain regions of significant correlation between the MRI and PET measures of interest. Linear relationships were notable in the fusiform gyri and lateral occipital regions in our study as reported in prior neuropathology studies5, rather than in the medial temporal or the AD cortical signature regions. Our results suggest that the sigmoidal nature of cortical thinning reported across AD stages previously42,43 in fact varies depending on regional brain amyloid burden. Additionally, age, sex, and education altered the linear relationship between amyloid and cortical thickness, suggesting that these variables (in addition to stage of disease) also relate to the correlation between cortical thickness and amyloid burden in AD signature regions, thus limiting cortical thickness utility to track disease progression.
Hippocampal volume derived from structural MRI is a robust and commonly used metric of neurodegeneration (“N”) in the AT(N) model. Although hippocampal volume is often used as a clinical biomarker of AD in clinical trials, our results highlight the severe limitations of inferring corresponding amyloid burden (“A”) using hippocampal volume or cortical thickness in AD signature regions as a proxy for inferring amyloid severity burden (such as with amyloid PET or CSF amyloid assays) across the AD spectrum. Our results demonstrated linear correlation between cortical thickness and amyloid burden in the fusiform gyrus and lateral occipital cortices but not the medial temporal regions regardless of disease severity.
Most previous studies of AD used a global measure of amyloid burden, and early studies reported variations in the regions of significant correlation4,13,14,15,16. In an earlier study that used local measures of amyloid and atrophy in cognitively normal elderly participants, a correlation was identified in the parietal regions, including the precuneus, but not in the medial temporal regions17. Local measures of amyloid and cortical atrophy in our study of a memory clinic cohort, using a novel image coregistration technique, again found that cortical thickness in the AD signature regions did not demonstrate the strongest correlation with amyloid.
There are several possible reasons for these results. First, our study population displayed a wide range of AD severity, ranging from subjective cognitive impairment to AD dementia. Second, there were differences between our study and previous research in MRI processing and analytical techniques. Finally, atrophy in the AD signature regions may be sensitive to multiple factors beyond amyloid burden alone. Indeed, recent research demonstrating the appearance of neurodegeneration in the absence of Aβ and tau has challenged the Aβ- and tau-centric model of AD pathophysiology44,45,46.
Metrics derived from structural MRI are widely used in memory clinics worldwide; thus, the strength of our study is that it was performed in a clinical cohort of patients with memory changes. Accordingly, the robust cortical atrophy patterns and correlation with amyloid burden are likely to be replicated in typical clinical populations. Additionally, rather than using a global measure of amyloid burden, we used a novel image analysis method to improve upon limitations in methods previously used for these analyses, including improved coregistration of MRI and PET images.
The study design was cross-sectional; no longitudinal data were obtained. Given that there was temporal progression of all patients from high MoCA (20–30) to middle (11–19) to low (0–10) scores, with each group having differences in mean cortical thickness and mean SUVRs, longitudinal trends may be inferred. A cross-sectional cohort of patients with a wide range of MoCA scores may be helpful in exploring longitudinal behavior in a scatter plot of SUVR versus cortical thickness. In the context of this study, the data suggest that a decrease in cognitive scores (high to mid-range MoCA score) is correlated with increased cortical SUVR but little change in cortical thickness; the change from mid-range to low MoCA score is revealed by both increased cortical SUVR and decreased cortical thickness. In this cross-sectional evaluation of different stages of the disease, amyloid SUVR changes are noted without significant cortical thinning. True longitudinal data would be required to verify this interpretation.
The limitations of this study reflect the challenges involved in characterizing cortical thickness, which can be affected by multiple variables. Variabilities in the manifestation of amyloid burden and atrophy in different disease subtypes limit the current results to the common amnestic form of AD, which was typical of the amyloid-positive patients in this cohort. Because we used local correlations in our analysis, these results, like those from previous studies, are not sensitive to network effects, whereby amyloid accumulation in a given region contributes to cortical atrophy in a distinct non-coincident region. Tau PET was not available for our study cohort; although neurofibrillary tangles may contribute to atrophy in some brain regions, this factor could not be assessed in this study. However, the image analysis methods used in this study can be used to improve the analysis of tau burden and cortical thickness in studies using tau PET.
In addition to neurodegeneration related specifically to Alzheimer pathology, the aging brain is subject to other acquired injuries, most notably vascular disease, and these injuries likely contribute at least additively and possibly synergistically to the neurodegeneration of AD47. Patients with vascular dementia were excluded from this study, and so this factor could not be characterized. Study patients may have differed in other possibly confounding factors affecting atrophy, including the presence and severity of metabolic conditions such as diabetes48 and nutritional status. Finally, the presence of hippocampal sclerosis49 and mixed pathology such as combinations of AD, Lewy body pathology, and vascular pathology50 are also known to affect rates of cognitive decline, but these conditions could not be reliably identified in our cohort without neuropathology confirmation. It would be helpful to further evaluate these findings in a larger autopsy-confirmed cohort of individuals to overcome the likelihood of a false positive (type 1) error.
In conclusion, with the use of novel imaging analysis methods to enable better coregistration for regional cortical analysis, we identified a robust regional cortical atrophy signature that linearly tracks regional amyloid SUVRs in patients with AD. We found that fusiform gyrus and lateral occipital cortical thickness, not medial temporal cortical thickness, had the strongest linear relationship with regional amyloid SUVRs. Our novel image processing methods could also be used for tau PET to better assess the correlation between regional tau burden and cortical thickness. These analyses could contribute to improved understanding of the imaging correlates of AD pathology.
Data availability
The datasets analyzed during the current study are available from the corresponding author upon reasonable request pending IRB and institutional approval.
Abbreviations
- AD:
-
Alzheimer’s disease
- CSF:
-
Cerebrospinal fluid
- CT:
-
Computed tomography
- FDR:
-
False discovery rate
- FWHM:
-
Full-width at half-maximum
- MCI:
-
Mild cognitive impairment
- MoCA:
-
Montreal Cognitive Assessment
- MRI:
-
Magnetic resonance imaging
- PET:
-
Positron emission tomography
- PVC:
-
Partial volume correction
- SUVR:
-
Standardized uptake value ratio
References
Jack, C. R. Jr. et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 14, 535–562 (2018).
Dickerson, B. C. et al. The cortical signature of Alzheimer’s disease: Regionally specific cortical thinning relates to symptom severity in very mild to mild AD dementia and is detectable in asymptomatic amyloid-positive individuals. Cereb. Cortex 19, 497–510 (2009).
Bakkour, A., Morris, J. C. & Dickerson, B. C. The cortical signature of prodromal AD: Regional thinning predicts mild AD dementia. Neurology 72, 1048–1055 (2009).
Ossenkoppele, R. et al. Associations between tau, Aβ, and cortical thickness with cognition in Alzheimer disease. Neurology 92, e601–e612 (2019).
Arnold, S. E., Hyman, B. T., Flory, J., Damasio, A. R. & Van Hoesen, G. W. The topographical and neuroanatomical distribution of neurofibrillary tangles and neuritic plaques in the cerebral cortex of patients with Alzheimer’s disease. Cereb. Cortex 1, 103–116 (1992).
Salat, D. H. et al. Thinning of the cerebral cortex in aging. Cereb. Cortex 14, 721–730 (2004).
Lemaitre, H. et al. Normal age-related brain morphometric changes: Nonuniformity across cortical thickness, surface area and gray matter volume?. Neurobiol. Aging 33(617), e1-9 (2012).
Busovaca, E. et al. Is the Alzheimer’s disease cortical thickness signature a biological marker for memory?. Brain Imaging Behav. 10, 517–523 (2016).
Pillai, J. A. et al. Alzheimer’s Disease Neuroimaging Initiative. Higher education is not associated with greater cortical thickness in brain areas related to literacy or intelligence in normal aging or mild cognitive impairment. J. Clin. Exp. Neuropsychol. 34, 925–935 (2012).
Ridgway, G. R. et al. Early-onset Alzheimer disease clinical variants: Multivariate analyses of cortical thickness. Neurology 79, 80–84 (2012).
Lehmann, M. et al. Global gray matter changes in posterior cortical atrophy: A serial imaging study. Alzheimers Dement. 8, 502–512 (2012).
Suarez-González, A. et al. Effect of age at onset on cortical thickness and cognition in posterior cortical atrophy. Neurobiol. Aging 44, 108–113 (2016).
Jack, C. R. Jr. et al. Alzheimer’s Disease Neuroimaging Initiative. Serial PIB and MRI in normal, mild cognitive impairment and Alzheimer’s disease: Implications for sequence of pathological events in Alzheimer’s disease. Brain 132, 1355–1365 (2009).
Chetelat, G. et al. Australian Imaging Biomarkers and Lifestyle Research Group. Relationship between atrophy and beta-amyloid deposition in Alzheimer disease. Ann. Neurol. 67, 317–324 (2010).
Whitwell, J. L. et al. Does amyloid deposition produce a specific atrophic signature in cognitively normal subjects?. Neuroimage Clin. 2, 249–257 (2013).
Doherty, B. M. et al. Amyloid burden, cortical thickness, and cognitive function in the Wisconsin Registry for Alzheimer’s Prevention. Alzheimers Dement. (Amst) 1, 160–169 (2015).
Becker, J. A. et al. Amyloid-β associated cortical thinning in clinically normal elderly. Ann. Neurol. 69, 1032–1042 (2011).
Jack, C. R. Jr. et al. 11C PiB and structural MRI provide complementary information in imaging of Alzheimer’s disease and amnestic mild cognitive impairment. Brain 131, 665–680 (2008).
Chételat, G. et al. AIBL Research Group. Accelerated cortical atrophy in cognitively normal elderly with high β-amyloid deposition. Neurology 78, 477–484 (2012).
Mattsson, N. et al. Alzheimer’s Disease Neuroimaging Initiative. Emerging β-amyloid pathology and accelerated cortical atrophy. JAMA Neurol. 71, 725–734 (2014).
Villeneuve, S. et al. Cortical thickness mediates the effect of β-amyloid on episodic memory. Neurology 82, 761–767 (2014).
Su, Y. et al. Dominantly Inherited Alzheimer Network. Partial volume correction in quantitative amyloid imaging. Neuroimage 107, 55–64 (2015).
Li, C. et al. Alzheimer’s Disease Neuroimaging Initiative. The relationship of brain amyloid load and APOE status to regional cortical thinning and cognition in the ADNI cohort. J. Alzheimers Dis. 59, 1269–1282 (2017).
d’Oleire Uquillas, F., et al. Interactive versus additive relationships between regional cortical thinning and amyloid burden in predicting clinical decline in mild AD and MCI individuals. Neuroimage Clin. 17, 388–396 (2017).
Fan, L. Y. et al. The relation between brain amyloid deposition, cortical atrophy, and plasma biomarkers in amnesic mild cognitive impairment and Alzheimer’s disease. Front. Aging Neurosci. 10, 175 (2018).
Ten Kate, M. et al. MRI predictors of amyloid pathology: Results from the EMIF-AD Multimodal Biomarker Discovery study. Alzheimers Res. Ther. 10, 100 (2018).
Knopman, D. S. et al. Joint associations of β-amyloidosis and cortical thickness with cognition. Neurobiol. Aging. 65, 121–131 (2018).
Schwarz, C. G. et al. A comparison of partial volume correction techniques for measuring change in serial amyloid PET SUVR. J. Alzheimers Dis. 67, 181–195 (2019).
McKhann, G. M. et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 7, 263–269 (2011).
Albert, M. S. et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 7, 270–279 (2011).
Fischl, B. & Dale, A. M. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc. Natl. Acad. Sci. USA 97, 11050–11055 (2000).
Fischl, B. et al. Automatically parcellating the human cerebral cortex. Cereb. Cortex 14, 11–22 (2004).
Jenkinson, M., Bannister, P., Brady, M. & Smith, S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17, 825–841 (2002).
Jenkinson, M., Beckmann, C. F., Behrens, T. E., Woolrich, M. W. & Smith, S. M. FSL. Neuroimage 62, 782–790 (2012).
Avants, B. B., Epstein, C. L., Grossman, M. & Gee, J. C. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12, 26–41 (2008).
Li, M. et al. Automatic nonlinear transformation to Talairach stereotaxic space with quality assurance in ISMRM 21st Annual Meeting & Exhibition (2013).
Li, M., Lin, J., Koenig, K. & Lowe, M. Automatic nonlinear transformation of 7T MRI brain image to Talairach stereotaxic space in ISMRM 25th Annual Meeting & Exhibition (2017).
Erlandsson, K. et al. MR imaging-guided partial volume correction of PET data in PET/MR imaging. PET Clin. 11, 161–177 (2016).
Rousset, O. G., Ma, Y. & Evans, A. C. Correction for partial volume effects in PET: Principle and validation. J. Nucl. Med. 39, 904–911 (1998).
Labbe, C. et al. Absolute PET quantification with correction for partial volume effects within cerebral structures. In Quantitative Functional Brain Imaging with Positron Emission Tomography (eds. Carson, R. E., Daube-Witherspoon, M. E. & Herscovitch, P.) 59–66 (Academic Press, Cambridge, 1998).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 57, 289–300 (1995).
Sabuncu, M. R. et al. Alzheimer’s Disease Neuroimaging Initiative. The dynamics of cortical and hippocampal atrophy in Alzheimer disease. Arch. Neurol. 68, 1040–1048 (2011).
Schuff, N. et al. Alzheimer’s Disease Neuroimaging Initiative. Nonlinear time course of brain volume loss in cognitively normal and impaired elders. Neurobiol. Aging 33, 845–855 (2012).
Jagust, W. Imaging the evolution and pathophysiology of Alzheimer disease. Nat. Rev. Neurosci. 19, 687–700 (2018).
Villemagne, V. L., Doré, V., Burnham, S. C., Masters, C. L. & Rowe, C. C. Imaging tau and amyloid-β proteinopathies in Alzheimer disease and other conditions. Nat. Rev. Neurol. 14, 225–236 (2018).
La Joie, R. et al. Prospective longitudinal atrophy in Alzheimer’s disease correlates with the intensity and topography of baseline tau-PET. Sci. Transl. Med. 12, eaau5732 (2020).
Villeneuve, S. et al. Vascular risk and Aβ interact to reduce cortical thickness in AD vulnerable brain regions. Neurology 83, 40–47 (2014).
van Elderen, S. G. et al. Progression of brain atrophy and cognitive decline in diabetes mellitus: A 3-year follow-up. Neurology 75, 997–1002 (2010).
Ala, T. A., Beh, G. O. & Frey, W. H. Pure hippocampal sclerosis: A rare cause of dementia mimicking Alzheimer’s disease. Neurology 54, 843–848 (2000).
Pillai, J. A., Butler, R. S., Bonner-Jackson, A. & Leverenz, J. B. Impact of Alzheimer’s disease, lewy body and vascular co-pathologies on clinical transition to dementia in a national autopsy cohort. Dement. Geriatr. Cogn. Disord. 42, 106–116 (2016).
Acknowledgements
We thank Frank DiFilippo, PhD, for his review of physics aspects regarding nuclear medicine imaging, and Megan Griffiths for editorial support.
Funding
This study received funding (NIA K23AG055685-01), which was used to support J.A.P. He was a program participant of the Research Education Component of the Cleveland Alzheimer’s Disease Research Center supported by NIA P30 AG062428.
Author information
Authors and Affiliations
Contributions
(1) Research Project: (A) Conception, (B) Organization, (C) Execution; (2) Statistical Analysis: (A) Design, (B) Execution, (C) Review and Critique; (3) Manuscript Preparation: (A) Writing the First Draft, (B) Review and Critique. J.A.P.: (1) A, B (2) C (3) A. M.L.: (1) A (2) C, (3) A. A.C.: (1) C (2) C (3) B. J.C.: (2) C (3) B. J.L.C.: (2) C (3) B. S.E.J: (1) A, B, C (2) A, B (3) A.
Corresponding author
Ethics declarations
Competing interests
J.A.P. has received research funding from the National Institutes of Health (NIH), Alzheimer’s Association, and Keep Memory Alive. M.L. has received research funding from NIH. J.C. is a consultant to Acadia, Accera, Actinogen, Agenebio, Alkahest, Allergan, Alzheon, Annovis, Avanir, Axsome, Cassava, Cerevel, Cortexyme, EIP Pharma, Eisai, Foresight, Green Valley, Grifols, Karuna, Nutricia, Otsuka, ReMYND, Resverlogix, Roche, Samus, Samumed, Sunovion, Suven, Third Rock, and United Neuroscience pharmaceutical and assessment companies and has received research funding from the National Institute of General Medical Sciences (Grant: P20GM109025) and Keep Memory Alive. S.E.J. has received research funding from NIH, St Jude, and Biogen; has received speaking fees from Siemens, Radnet, and St Jude’s Hospital; and is a consultant to Eisai and Monteris. A.C. and J.L.C. have no potential conflicts of interest to declare. “Research reported in this publication was supported by the National Institute On Aging of the National Institutes of Health under Award Number K23AG055685. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.”
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pillai, J.A., Larvie, M., Chen, J. et al. Spatial patterns of correlation between cortical amyloid and cortical thickness in a tertiary clinical population with memory deficit. Sci Rep 10, 20717 (2020). https://doi.org/10.1038/s41598-020-77503-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-77503-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.