Abstract
Saprophytic bacteria and plants compete for limited nutrient sources. Bacillus subtilis grows well on steamed soybeans Glycine max to produce the fermented food, natto. Here we focus on bacterial responses in conflict between B. subtilis and G. max. B. subtilis cells maintained high growth rates specifically on non-germinating, dead soybean seeds. On the other hand, viable soybean seeds with germinating capability attenuated the initial growth of B. subtilis. Thus, B. subtilis cells may trigger saprophytic growth in response to the physiological status of G. max. Scanning electron microscope observation indicated that B. subtilis cells on steamed soybeans undergo morphological changes to form apertures, demonstrating cell remodeling during saprophytic growth. Further, transcriptomic analysis of B. subtilis revealed upregulation of the gene cluster, yesOPQR, in colonies growing on steamed soybeans. Recombinant YesO protein, a putative, solute-binding protein for the ATP-binding cassette transporter system, exhibited an affinity for pectin-derived oligosaccharide from plant cell wall. The crystal structure of YesO, in complex with the pectin oligosaccharide, was determined at 1.58 Å resolution. This study expands our knowledge of defensive and offensive strategies in interspecies competition, which may be promising targets for crop protection and fermented food production.
Similar content being viewed by others
Introduction
Plant seeds, each consisting of an embryo, storage tissues, and protective outer coat, remain dormant, waiting for favorable environmental conditions for germination. Stored nutrient sources, such as carbohydrates, lipids, and proteins, are available to the germinating embryo, and also to seed microbiota, a community of bacteria and fungi surrounding the seeds1,2. Plants have evolved sophisticated mechanisms to prevent phytopathogen colonization and infection. However, once ungerminated seeds undergo senescence, decreased antimicrobial activity of dead seeds and, thus, increased accessibility to nutrients may permit microbial growth using seed decomposition products, i.e., saprophytic growth3,4,5. The molecular basis for recognition of seed physiological status is required to understand the competition between microorganisms and plants.
Bacillus subtilis is a Gram-positive, spore-forming, saprophytic bacterium ubiquitously isolated from soil, water, air, decaying plant materials, and fermented foods6,7,8,9. During production of natto, a traditional fermented soybean food in Japan, soybean seeds are soaked and steamed before inoculation with B. subtilis. This process, which kills seeds, is regarded as essential for enhancing B. subtilis growth. Therefore, B. subtilis cells growing on soybean seeds may be a useful model to study cell responses and molecular recognition at saprophytic growth onset. B. subtilis is widely used in emerging research fields as a Gram-positive model organism10,11; however, its physiological responses and behaviors on soybean seeds remain unclear.
Saprophytic growth requires nutrient uptake by decomposer organisms from decaying plant material. Bacterial ATP-binding cassette (ABC)-family transporters translocate a variety of substrates into cells, using energy from ATP hydrolysis12,13,14. A typical ABC transporter consists of three components: integral membrane proteins that form pores, nucleotide-binding proteins (NBPs) that bind and hydrolyze ATP, and solute-binding proteins (SBPs) that recognize and deliver target compounds into cells. SBPs are critical elements for substrate specificity and high affinity of ABC transporter systems15,16,17,18. For instance, a conformational change of SBP in pathogenic bacteria is critical for selective recognition and import of mammalian host glycosaminoglycans19,20.
Pathogenic and symbiotic microorganisms secrete numerous enzymes to penetrate and degrade plant cell walls to provide nutrition21,22,23,24,25. Thus, cell wall decomposition products are potential targets for saprophytic bacteria, which may use them to monitor plant physiological status. Primary plant cell walls consist of a cellulose-hemicellulose framework embedded in an inter-fibrillar matrix of pectins26,27. Complex and diverse pectic polysaccharides include homogalacturonan (HG), rhamnogalacturonan type I (RG-I), and rhamnogalacturonan type II (RG-II). HG is a linear polymer of α-1,4-linked galacturonic acid. RG-I is composed of a backbone, based on disaccharide-repeating units of rhamnose and galacturonic acid, with side chains of galactans, arabinans, and arabinogalactans. RG-II contains HG as a backbone, with complex side chains composed of a variety of sugars. Although studies on HG degradation have progressed28,29,30, bacterial decomposition of RG-I and RG-II has been reported in a limited number of papers so far31,32,33.
This report is the first on the competition between saprophytic B. subtilis and soybean Glycine max on the surfaces of soybean seeds. This article further focuses on the physiological and transcriptomic traits of B. subtilis, and provides structural insight into SBP-dependent recognition of RG-I-derived trisaccharide during the saprophytic growth.
Results and discussion
Growth of B. subtilis on live and dead soybeans
Natto, a traditional Japanese fermented food, is manufactured from steamed soybean seeds7,9. Saprophytic B. subtilis cells grow on dead soybeans, using decomposition products as nutrients. Steaming may cause not only loss of viability but also denaturation of proteins; it may alter other soybean components as well. Therefore, what exactly triggers saprophytic growth is an open question. Heat-related impacts to soybean components were reduced by adopting physiological senescence, induced at different temperatures, by prolonged seed storage. Six-month storage at 4 °C maintained viability, but six-month storage at an ambient temperature severely limited seed germination (Supplementary Fig. S1A). Thus, seeds stored under the former and the latter conditions were used as live and dead soybeans, respectively.
Bacillus subtilis cells were found to grow vigorously on non-germinating dead soybean seeds, but not on live seeds (Supplementary Video S1, Fig. 1A). Thus, seed germination and bacterial growth inhibition may be closely linked. We first discovered this phenomenon using B. subtilis natto-producing strain NBRC 1644934. Quantitative analysis of colony-forming units (CFUs) indicated that B. subtilis initially proliferated on both live and dead soybeans. However, growth attenuated specifically on live soybeans when bacterial cell numbers reached 106 CFUs per seed (Fig. 1B). B. subtilis laboratory standard strain 16835 exhibited a similar growth profile on live and dead seeds (Fig. 1C). Thus, live soybeans can severely limit B. subtilis growth on their surfaces. Invading bacteria and germinating plant embryos may compete for nutrients in soybean seeds. B. subtilis cells gain access to seed nutrients when the seeds lose germination ability. B. subtilis growth on soybean seeds is a promising model for bacterial-plant interactions in seed microbiota.
Saprophytic growth of B. subtilis on soybean surfaces. (A) Soybean seeds inoculated with B. subtilis NBRC 16449 cells at 24 h after inoculation. Live (left) or dead (right) soybeans were used. Bars, 1 cm. (B) Growth curves of B. subtilis NBRC 16449 cells on soybean seeds. (C) Growth curves of B. subtilis 168 cells on soybean seeds. Each data point represents the mean ± standard deviations (SD) from three independent experiments. Asterisks indicate significant differences from the data of live soybeans (p < 0.05; t-test)
Morphological traits of B. subtilis during saprophytic growth
To understand the strategy of B. subtilis for saprophytic growth on dead soybeans during natto production, morphological and transcriptional responses were examined. Scanning electron microscope (SEM) analysis revealed cell morphological changes of B. subtilis NBRC 16449 during saprophytic growth on steamed seeds (Fig. 2A–C). The early logarithmic growth phase, 5 h after inoculation, was characterized by elongated, rod-shaped cells, 6–10 μm long. As the growth progressed, most cells displayed short-rod forms, approximately 2 μm long, with distinctive aperture-like structures (Fig. 2C). B. subtilis NBRC 16449 (Supplementary Fig. S2) and 168 (Fig. 2D–F) strains exhibited apertures on soybean seed-mimicking solid medium (Supplementary Fig. S1B). No apertures were observed in liquid medium (Fig. 2G–I), suggesting that a solid substrate is needed for this culture-specific response. Similar aperture-like structures are previously reported in Clostridium sporogenes during formation of endospores36. B. subtilis cell membranes are subject to engulfment during spore development under nutrient-depleted conditions37,38, and saprophytically growing B. subtilis may undergo similar morphological remodeling. How solid-state culture stimulates the starvation-triggered sporulation pathway is of great interest from a signal transduction viewpoint.
SEM observation of B. subtilis cell morphology. (A) B. subtilis NBRC 16449 cells, 5 h after inoculation on steamed soybean seeds. (B,C) B. subtilis NBRC 16449 cells, 48 h after inoculation on steamed soybean seeds. (D) B. subtilis 168 cells, 4 h after inoculation on soybean-mimicking solid medium blocks. (E,F) B. subtilis 168 cells, 216 h after inoculation on soybean-mimicking solid medium blocks. (G) B. subtilis 168 cells, 4 h after inoculation in liquid medium. (H, I) B. subtilis 168 cells, 239 h after inoculation in liquid medium. Bars, 1 μm (A,B,D,E,G,H) or 0.2 μm (C,F,I).
Transcriptomic profile of B. subtilis during saprophytic growth
Comprehensive understanding of saprophytism-specific recognition and responses was sought using the transcriptomic profiles of B. subtilis NBRC 16449 cells from steamed soybeans and from soybean seed-mimicking solid medium, using RNA-Seq analysis. Reads were mapped to the genomic sequence of B. subtilis subsp. natto BEST195 strain39,40. Expression of 93 and 107 genes among a total of 4271 analyzed genes was over tenfold upregulated and downregulated, respectively, in B. subtilis grown on steamed soybeans. Upregulated transcripts included several ABC transporter components and their neighboring genes (Fig. 3A). The gene cluster yesOPQR was highly expressed, as previously reported in B. subtilis 168 cells grown on RG-I, a component of plant cell wall pectin32. Based on sequence homology, yesO and yesPQ genes encode a putative SBP and transmembrane permeases for the ABC-family sugar transport system, respectively. Deleting yesOPQ has a negative impact on utilization of RG-I as a carbon source41. YesR is a cytosolic hydrolase that acts on unsaturated rhamnogalacturonan disaccharide derived from RG-I31. Another upregulated gene, yurJ, may encode a functional NBP for YesOPQ and other ABC transporters41,42. Thus, B. subtilis may recognize and use plant cell wall-derived RG-I as a carbon source by inducing yesOPQR and yurJ genes during saprophytic growth (Fig. 3B).
Affinity between YesO and RG-I oligosaccharides
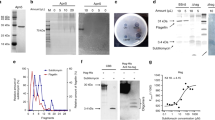
YesO is a putative SBP involved in RG-I assimilation, but its binding substrate is unknown. Recombinant YesO protein and the decomposition product of RG-I backbones (oligo-RG-I) were prepared to address this knowledge gap. Comparisons between YesO of B. subtilis laboratory strain 168 and its orthologs from Bacillus sonorensis, Bacillus licheniformis DSM 13, Paenibacillus sp. Aloe-11, and Paenibacillus macerans revealed low conservation of the N-terminus (Supplementary Fig. S3). Thus, N-terminus-deleted YesO, of approximately 45 kDa, was overexpressed in Escherichia coli, and purified to homogeneity (Fig. 4A,B). To obtain oligo-RG-I, RG-I backbones were treated with exolyase YesX (Fig. 4C). Thin-layer chromatography (TLC) analysis of reaction products showed that released oligosaccharides consisted mainly of unsaturated rhamnogalacturonan disaccharide, as reported previously32. The interaction between YesO and oligo-RG-I was analyzed by measuring YesO fluorescent intensity in the presence or absence of oligo-RG-I (Fig. 4D). Adding oligo-RG-I decreased the fluorescent intensity from tryptophan residues of YesO by 3–7%. Based on the plot, the dissociation constant (Kd) was 1.6 μM. No significant decrease of fluorescent intensity was observed when other disaccharides, such as sucrose, maltose, cellobiose, chitobiose, and digalacturonic acid, were used as a ligand (Supplementary Fig. S4). YesO appears to recognize oligo-RG-I as a substrate selectively.
Interaction between oligo-RG-I and YesO. (A) SDS-PAGE analysis of purified YesO, followed by CBB staining. (B) Elution profile of YesO via gel filtration chromatography. Bovine serum albumin (BSA, 66 kDa, light gray) and chymotrypsin (Chy, 25 kDa, dark gray) were used for comparison. (C) TLC analysis of oligo-RG-I. Note that unsaturated rhamnogalacturonan disaccharide (di) showed two distinctive signals, corresponding to ammonium ion-unbound and bound forms32. Traces of oligosaccharides (oligo) were also detected. Rhamnose (Rha) and galacturonate (GalUA) were used for comparison. The profile of TLC plate is not an image cropped from different parts of the same plate or from different plates. (D) Fluorescent spectrum analysis. The decrease in fluorescent intensity (Δfluorescence), caused by increasing ligand concentrations, was plotted after modification based on volume change in the cuvette. Each data point represents the mean ± SD from three independent experiments. (E) Overall structure of ligand-free YesO protein. The assigned secondary structures of α-helices (α1–α15) and β-sheets (β1–β8) are indicated. (F) Overall structure of YesO/oligo-RG-I. Orange, N-domain; green, C-domain. (G) Superimposition of YesO and YesO/oligo-RG-I. Magenta, YesO; cyan, YesO/oligo-RG-I. The ball model shows ΔGalUA-Rha-Gal (green, carbon atom; red, oxygen atom). The rotation axis and angle for the Venus-flytrap conformational change are shown. (H) Residues interacting with ΔGalUA-Rha-Gal via hydrogen bonds and C–C contacts. Electron density map (2Fo–Fc) of ΔGalUA-Rha-Gal in YesO/oligo-RG-I is shown in blue mesh contoured with 1.4 σ. Hydrogen bonds are represented by dotted lines with labeled distances. The residues of the N- and C-domains are colored orange and green, respectively. The residues interacting only by C–C contacts (Trp52, Pro176, Ile179, Phe180, Met236, Tyr252, Phe259, and Met287) are pale-colored.
Binding mode of YesO to oligo-RG-I
Oligo-RG-I recognition by YesO was analyzed with X-ray crystallography. The crystal structures of YesO and its complex form with oligo-RG-I (YesO/oligo-RG-I) were determined at 1.97 Å and 1.58 Å resolution, respectively. Data collection and refinement statistics are summarized in Supplementary Table S1. Ligand-free YesO structure (Fig. 4E) was similar to a previously determined structure (PDB ID 4R6K). YesO is an α/β protein with 15 α helices (residues 23–39, 52–65, 76–86, 92–96, 111–113, 135–140, 152–166, 177–187, 205–219, 253–262, 312–318, 328–336, 340–354, 368–383, and 381–405) and 8 β sheets (residues 15–20, 45–55, 71–73, 122–126, 130–133, 248–252, 267–270, and 288–290). Note that amino acid residue numbers of our YesO are 20 fewer than those of the full-length YesO to adjust to PDB ID 4R6K. The substrate-binding cleft is formed between two globular N- and C-domains (Fig. 4F), as typically observed among SBPs of ABC transport systems15,16,17,18. YesO is categorized as a Class II SBP, based on a β2–β1–β3–β8–β4 topology of the β sheet core in the N-domain. N- and C-domains are connected through two short hinges (residues Val126-Leu129 and Leu281-Lys284), assigning YesO to Cluster D SBPs. Comparison of ligand-free YesO with YesO/oligo-RG-I revealed a 26.2°-closed conformation upon binding of a ligand molecule (Fig. 4G), suggesting a Venus-flytrap conformational change43.
Although our prepared oligo-RG-I was mostly disaccharide (Fig. 4C), X-ray crystallography of YesO/oligo-RG-I indicated an interaction between YesO and the trisaccharide, ΔGalUA-Rha-Gal, composed of unsaturated galacturonic acid and rhamnose from the main chain and galactose from the side chain (Fig. 4H). This result reproduced another independent YesO/oligo-RG-I crystal structure. The molecular docking simulation suggested that rhamnogalacturonan tetrasaccharide is too large to fit in the ligand-binding cleft (Supplementary Fig. S5). Thus, YesO may selectively recognize ΔGalUA-Rha-Gal trisaccharide as a ligand. ΔGalUA-Rha-Gal binds to the YesO cleft via hydrogen bonds and C–C contacts (Table 1). There were 19 hydrogen bonds (ΔGalUA, 10; Rha, 2; Gal, 7) and 44 C–C contacts (ΔGalUA, 22; Rha, 17; Gal, 5). ΔGalUA-interacting residues, Arg25, Asn127, Asn254, and Ser286, are highly conserved among the known YesO orthologs, highlighting the importance of ΔGalUA in the YesO ligand.
Recognition of ΔGalUA-Rha-Gal during saprophytic growth
In the present study, B. subtilis yesOPQR was identified as an upregulated gene cluster during saprophytic growth on dead soybean surfaces (Fig. 3B). YesO was found to act as an SBP for uptake of RG-I-derived ΔGalUA-Rha-Gal trisaccharide. YesPQ are putative transmembrane permeases, and YesR catalyzes intracellular degradation of unsaturated rhamnogalacturonan disaccharide31. Expression of the yurJ gene, encoding a putative NBP for YesOPQ40, was also induced on dead soybeans. Thus, YesOPQR/YurJ-dependent assimilation of ΔGalUA-Rha-Gal is likely to play a major role in saprophytic growth. Assuming that plant cell wall degradation is closely associated with wounding, pathogen attack, or cell death, induction of the yesOPQR and yurJ genes might be one of the initial responses for saprophytic growth on dead plant surfaces.
Our hypothesis is that ΔGalUA-Rha-Gal is used as a nutrient or signaling molecule, or both. ΔGalUA-Rha-Gal is degraded into assimilable monosaccharides by an unidentified intracellular β-1,4-galactosidase and YesR. Since the soybean surface is a low-nutrient environment, pectin-derived oligosaccharides may be the carbon source that supports saprophytic growth. Further, ΔGalUA-Rha-Gal may also serve as a signaling molecule to trigger saprophytism. Some bacterial two-component regulatory systems adopt SBPs for signal recognition18. Our recent study demonstrated that SPH1118, an SBP in Sphingomonas sp. strain A1, is responsible not only for pectin assimilation but also for chemotaxis toward pectin44. Assuming such alternative functions of YesO, recognizing ΔGalUA-Rha-Gal may trigger signal transduction for proliferation or adaptation under saprophytic conditions.
Based on the YesO/oligo-RG-I three-dimensional structure, YesO traps ΔGalUA-Rha-Gal trisaccharide, although most of the oligo-RG-I used in this study was disaccharide. How is ΔGalUA-Rha-Gal generated via pectin degradation? B. subtilis cells secrete a series of enzymes for RG-I degradation, such as YesWX lyases to cleave α-1,4-glycosidic linkages between rhamnose and galacturonic acid residues in the RG-I backbone32, GanAB galactosidases to degrade galactan branches45, and AbfA and Xsa arabinofuranosidases to catalyze hydrolysis of terminal arabinofuranoside bonds of arabinogalactan or arabinan46. YesWX-dependent degradation of RG-I backbones, with a single galactose residue at each branch point, may release ΔGalUA-Rha-Gal trisaccharide (see red areas in Fig. 3B).
In conclusion, this report is the first to describe the competition between saprophytic B. subtilis and soybean G. max on soybean seeds surfaces. Live soybeans prevent bacterial invasion, while B. subtilis YesO recognizes pectin-derived ΔGalUA-Rha-Gal trisaccharide to establish saprophytic growth on dead soybean seeds. This study will help elucidate the diversity of bacterial-plant interactions, particularly for seed microbiota.
Materials and methods
Materials and microorganisms
Soybean seeds of Suzumaru (G. max) were purchased from Mamehei (Japan). RG-I (from potatoes) was purchased from Megazyme (Ireland). B. subtilis laboratory standard strain 168 was obtained from the National Bio Resource Project (NBRP), Japan. B. subtilis strain NBRC 16449 was obtained from the NITE Biological Research Center (NBRC), Japan. E. coli strain BL21-Gold(DE3) (Agilent, USA) was used for recombinant protein overexpression. E. coli strain DH5α was used for cloning and plasmid maintenance. Bacterial cells were routinely grown in LB medium (1% tryptone, 0.5% yeast extract, 1% sodium chloride, pH 7.2) at 37 °C.
Growth test
Soybean seeds were stored at 4 °C and at room temperature for 6 months to prepare live and dead soybeans, respectively. The seeds were washed and soaked in pure water at room temperature for 3.5 h. The seed surfaces were sterilized by 20-min ultraviolet exposure. This exposure did not produce severe effects on living soybean seeds’ germination capability (Supplementary Fig. S1A). Soybean seed-mimicking solid LB medium blocks (solidified by 3% agar, 300 μl each; Supplementary Fig. S1B) were prepared aseptically in microtubes. Thirty seeds or blocks were used for each growth test.
Bacillus subtilis was precultured at 37 °C in liquid LB medium overnight, washed, and resuspended in 0.85% sodium chloride. Approximately 104 cells per seed or solid medium block were inoculated and incubated at 37 °C. To assay CFUs, B. subtilis cells on the sampled seeds or blocks were collected in 0.85% sodium chloride, spread on LB agar plates, and incubated at 37 °C overnight.
SEM analysis
Bacillus subtilis cells were grown on steamed soybean seeds, on soybean mimicking solid LB medium blocks, and in liquid LB medium at 37 °C. Steamed soybeans were prepared by washing and soaking live seeds with pure water at room temperature for 3.5 h, followed by autoclaving at 121 °C for 20 min. For protein fixation, samples (soybean seeds or collected cells) were mixed with 4% formaldehyde and incubated for 1 h. Subsequently, for lipid fixation, samples were washed with 10 mM potassium phosphate buffer or pure water three times and immersed in 1% osmium oxide for 1–2 h. Fixed samples were washed with 10 mM potassium phosphate buffer or pure water three times, dehydrated sequentially with 50%, 70%, 90%, and 100% ethanol, and immersed in t-butyl alcohol at 4 °C. Freeze-dried samples were coated with a platinum-palladium layer under argon gas and observed with a field emission (FE)-SEM SU8230 (Hitachi, Japan) using a 1.5 kV electron beam.
RNA-Seq analysis
Bacillus subtilis NBRC 16449 cells were grown on steamed soybean seeds or soybean-mimicking solid LB medium blocks to the logarithmic phase. Cells on the seeds or blocks were collected in 0.85% sodium chloride, treated with RNAprotect bacteria reagent (Qiagen, Netherlands), and frozen immediately in liquid nitrogen. Total RNA was prepared using the hot phenol method47 by Nihon Gene Research Laboratories, Inc. (Japan). RNA-Seq analysis was performed by Macrogen Japan Corp. (Japan). In brief, paired-end sequencing of the constructed library was performed using HiSeq 2500 (Illumina, USA). Raw data were filtered using Trimmomatic v0.3248 to remove adapter sequences and bases with a Phred score below 30. Trimmed reads were aligned against the B. subtilis subsp. natto BEST195 reference genome (DDBJ accession no. DRA000001)39,40, using Bowtie as the alignment algorithm49. Differential expression analysis was conducted using HTSeq50. RNA-Seq data, generated from this study, were deposited in the NCBI Gene Expression Omnibus (GEO) and are accessible through GEO series accession number GSE109523.
Expression and purification of recombinant YesO protein
To construct an expression system for YesO that lacks the weakly conserved N-terminus (Supplementary Fig. S3), the yesO gene of B. subtilis strain 168 was amplified by genomic PCR, using primers 5′-GGCATATGACACTCAGAATCGCGTGGTGGGGC-3′ and 5′GGCTCGAGTCAATTATTCCTCTCTAATATCTCATT-3′ (with underlined restriction sites), and cloned into the NdeI-XhoI site of pET21b(+) (Novagen, USA). The resultant pET21b(+)-yesO plasmid was introduced into E. coli BL21-Gold(DE3) cells.
Escherichia coli cells harboring the plasmid were cultured in LB liquid medium with 100 μg ml−1 ampicillin at 30 °C. When optical density at 600 nm (OD600) reached 0.3, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture at a final concentration of 0.1 mM. After further incubation at 16 °C for 2 days, cells were washed, suspended in 20 mM Tris–HCl (pH 7.5), and disrupted with an ultrasonicator 201 M (Kubota, Japan). The supernatant obtained by centrifugation (20,000 × g, 4 °C, 20 min) was used for YesO purification.
Cell extracts were applied to an anion exchange chromatography system with a TOYOPEARL DEAE-650M column (Tosoh, Japan). Proteins were eluted using a linear gradient of sodium chloride (0–250 mM) in 20 mM Tris–HCl (pH 7.5). After separation with 12.5% SDS-PAGE, fractions containing the target protein were combined and purified by gel filtration chromatography using HiLoad 16/60 Superdex 200 pg resin (GE Healthcare, USA) equilibrated with 200 mM sodium chloride in 20 mM Tris–HCl (pH 7.5). After separation with 12.5% SDS-PAGE, fractions containing the target protein were combined.
Preparation of oligo-RG-I
Substrate RG-I backbones were prepared from commercially available RG-I, as previously reported32. A reaction mixture, consisting of 1% RG-I backbone, 50 mM Tris–HCl (pH 7.5), 20 mM manganese(II) chloride, and lysates of YesX-expressing E. coli cells32, was incubated at 30 °C for 3 h. The mixture was boiled for 5 min and centrifuged (7200 × g, room temperature, 5 min). The supernatant was passed through a Centriprep centrifugal filter (3 kDa NMWL; Merck Millipore, USA) to obtain decomposition products. Resultant oligo-RG-I was detected by TLC analysis, as reported previously32.
Fluorescence spectrum analysis
Fluorescence of protein, with a peak around 340 nm, is partially quenched by adding a binding ligand51. Fluorescent intensities of YesO, with increasing concentrations of oligo-RG-I, were measured by spectrophotometer FP-6500 (JASCO, Japan). Measurement parameters during reactions were: excitation band width, 1 nm; emission band width, 10 nm; response, 2 s; sensitivity, high; excitation wavelength, 280 nm; start to end emission wavelength, 300–500 nm; data pitch, 1 nm; and scan speed, 100 nm min−1. The reaction mixture contained 0.14 μM YesO, 20 mM Tris–HCl (pH 7.5), and 0–40 μM ligands. Decreases in fluorescent intensity by adding ligands were plotted, and the dissociation constant (Kd) was determined based on nonlinear regression with one site-specific binding model51.
X-ray crystallography
Ligand-free YesO and YesO/oligo-RG-I were crystallized by sitting drop vapor diffusion, using the JBScreen crystallization kit (Jena Bioscience, Germany). In 96-well sitting drop plates, 1 μl of 15 mg ml−1 YesO (without or with 1 mM oligo-RG-I) and 1 μl of reservoir solution were mixed. The mixture was kept at 20 °C until crystals grew sufficiently. Ligand-free YesO was crystallized in the drop, consisting of 100 mM malonate-imidazole-boric acid (MIB) buffer (pH 6.0), 25% polyethyleneglycol (PEG)-1500, and 1 mM digalacturonate. YesO/oligo-RG-I was crystallized in the drop, consisting of 100 mM Tris–HCl (pH 8.5), 20% PEG-1000, and 25% glycerol.
Each single crystal was picked up using a nylon loop from the drop, soaked in a reservoir solution containing 20% ethylene glycol, and instantly frozen using cold nitrogen gas. Synchroton radiation X-ray irradiated the crystals at 1.00 Å wavelength, and X-ray diffraction data were collected using a MAR225HE CCD detector (Rayonix, USA) at the BL-26B1 and BL-38B1 beamlines in SPring-8 (Japan). Diffraction data were processed using the HKL-2000 program52.
The crystal structures of YesO and YesO/oligo-RG-I were determined through molecular replacement with the Molrep program53 in the CCP4Interface package, using the ligand-free YesO structure (PDB ID 4R6K) as a reference model. Structure refinement was conducted using the phenix.refine54 and REFMAC553 programs. At each refinement cycle, the model was adjusted manually with the WinCoot program55. Images of protein structures were prepared using the PyMOL molecular graphics system (Schrödinger, USA). The coordinates and structure factors (accession codes 5Z6C for ligand-free YesO and 5Z6B for YesO/oligo-RG-I) have been deposited in the PDB.
Code availability
Accession codes RNA-Seq data were deposited in the NCBI GEO and are accessible through GEO series accession number GSE109523. Protein structures of YesO and YesO/oligo-RG-I were deposited in the PDB under accession codes 5Z6C and 5Z6B, respectively.
References
Nelson, E. B. Microbial dynamics and interactions in the spermosphere. Annu. Rev. Phytopathol. 42, 271–309 (2004).
Shade, A., Jacques, M. A. & Barret, M. Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr. Opin. Microbiol. 37, 15–22 (2017).
Prusky, D., Alkan, N., Mengiste, T. & Fluhr, R. Quiescent and necrotrophic lifestyle choice during postharvest disease development. Annu. Rev. Phytopathol. 51, 155–176 (2013).
Fatima, U. & Senthil-Kumar, M. Plant and pathogen nutrient acquisition strategies. Front. Plant Sci. 6, 750 (2015).
Poole, P., Ramachandran, V. & Terpolilli, J. Rhizobia: From saprophytes to endosymbionts. Nat. Rev. Microbiol. 16, 291–303 (2018).
Earl, A. M., Losick, R. & Kolter, R. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 16, 269–275 (2008).
Murooka, Y. & Yamashita, M. Traditional healthful fermented products of Japan. J. Ind. Microbiol. Biotechnol. 35, 791–798 (2008).
Fira, D., Dimkić, I., Berić, T., Lozo, J. & Stanković, S. Biological control of plant pathogens by Bacillus species. J. Biotechnol. 285, 44–55 (2018).
Kimura, K. & Yokoyama, S. Trends in the application of Bacillus in fermented foods. Curr. Opin. Biotechnol. 56, 36–42 (2019).
Kalamara, M., Spacapan, M., Mandic-Mulec, I. & Stanley-Wall, N. R. Social behaviours by Bacillus subtilis: Quorum sensing, kin discrimination and beyond. Mol. Microbiol. 110, 863–878 (2018).
Liu, Y., Liu, L., Li, J., Du, G. & Chen, J. Synthetic biology toolbox and chassis development in Bacillus subtilis. Trends Biotechnol. 37, 548–562 (2019).
Davidson, A. L., Dassa, E., Orelle, C. & Chen, J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–364 (2008).
Murata, K., Kawai, S., Mikami, B. & Hashimoto, W. Superchannel of bacteria: Biological significance and new horizons. Biosci. Biotechnol. Biochem. 72, 265–277 (2008).
Locher, K. P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016).
Fukami-Kobayashi, K., Tateno, Y. & Nishikawa, K. Domain dislocation: A change of core structure in periplasmic binding proteins in their evolutionary history. J. Mol. Biol. 286, 279–290 (1999).
Berntsson, R. P., Smits, S. H., Schmitt, L., Slotboom, D. J. & Poolman, B. A structural classification of substrate-binding proteins. FEBS Lett. 584, 2606–2617 (2010).
Maqbool, A. et al. The substrate-binding protein in bacterial ABC transporters: Dissecting roles in the evolution of substrate specificity. Biochem. Soc. Trans. 43, 1011–1017 (2015).
Scheepers, G. H., Nijeholt, J. A. L. & Poolman, B. An updated structural classification of substrate-binding proteins. FEBS Lett. 590, 4393–4401 (2016).
Oiki, S., Mikami, B., Maruyama, Y., Murata, K. & Hashimoto, W. A bacterial ABC transporter enables import of mammalian host glycosaminoglycans. Sci. Rep. 7, 1069 (2017).
Oiki, S., Kamochi, R., Mikami, B., Murata, K. & Hashimoto, W. Alternative substrate-bound conformation of bacterial solute-binding protein involved in the import of mammalian host glycosaminoglycans. Sci. Rep. 7, 17005 (2017).
Juge, N. Plant protein inhibitors of cell wall degrading enzymes. Trends Plant Sci. 11, 359–367 (2006).
Oldroyd, G. E., Murray, J. D., Poole, P. S. & Downie, J. A. The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45, 119–144 (2011).
Glass, N. L., Schmoll, M., Cate, J. H. & Coradetti, S. Plant cell wall deconstruction by ascomycete fungi. Annu. Rev. Microbiol. 67, 477–498 (2013).
Kubicek, C. P., Starr, T. L. & Glass, N. L. Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu. Rev. Phytopathol. 52, 427–451 (2014).
Garron, M. L. & Cygler, M. Uronic polysaccharide degrading enzymes. Curr. Opin. Struct. Biol. 28, 87–95 (2014).
Caffall, K. H. & Mohnen, D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344, 1879–1900 (2009).
Lampugnani, E. R., Khan, G. A., Somssich, M. & Persson, S. Building a plant cell wall at a glance. J. Cell Sci. 131, jcs207373 (2018).
Charnock, S. J., Brown, I. E., Turkenburg, J. P., Black, G. W. & Davies, G. J. Convergent evolution sheds light on the anti-β-elimination mechanism common to family 1 and 10 polysaccharide lyases. Proc. Natl. Acad. Sci. USA 99, 12067–12072 (2002).
Abbott, D. W. & Boraston, A. B. Structural biology of pectin degradation by Enterobacteriaceae. Microbiol. Mol. Biol. Rev. 72, 301–316 (2008).
Hugouvieux-Cotte-Pattat, N., Condemine, G. & Shevchik, V. E. Bacterial pectate lyases, structural and functional diversity. Environ. Microbiol. Rep. 6, 427–440 (2014).
Itoh, T., Ochiai, A., Mikami, B., Hashimoto, W. & Murata, K. A novel glycoside hydrolase family 105: The structure of family 105 unsaturated rhamnogalacturonyl hydrolase complexed with a disaccharide in comparison with family 88 enzyme complexed with the disaccharide. J. Mol. Biol. 360, 573–585 (2006).
Ochiai, A., Itoh, T., Kawamata, A., Hashimoto, W. & Murata, K. Plant cell wall degradation by saprophytic Bacillus subtilis strains: Gene clusters responsible for rhamnogalacturonan depolymerization. Appl. Environ. Microbiol. 73, 3803–3813 (2007).
Kunishige, Y. et al. Crystal structure of exo-rhamnogalacturonan lyase from Penicillium chrysogenum as a member of polysaccharide lyase family 26. FEBS Lett. 592, 1378–1388 (2018).
Ogawa, Y., Hosoyama, H., Hamano, M. & Motai, H. Purification and properties of γ-glutamyltranspeptidase from Bacillus subtilis (natto). Agric. Biol. Chem. 55, 2971–2977 (1991).
Kunst, F. et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390, 249–256 (1997).
Brunt, J., Cross, K. L. & Peck, M. W. Apertures in the Clostridium sporogenes spore coat and exosporium align to facilitate emergence of the vegetative cell. Food Microbiol. 51, 45–50 (2015).
Higgins, D. & Dworkin, J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol. Rev. 36, 131–148 (2012).
Dworkin, J. Protein targeting during Bacillus subtilis sporulation. Microbiol. Spectr. 2, TBS-0006-2012 (2014).
Itaya, M. & Matsui, K. Conversion of Bacillus subtilis 168: Natto producing Bacillus subtilis with mosaic genomes. Biosci. Biotechnol. Biochem. 63, 2034–2037 (1999).
Nishito, Y. et al. Whole genome assembly of a natto production strain Bacillus subtilis natto from very short read data. BMC Genomics 11, 243 (2010).
Ferreira, M. J., Mendes, A. L. & de Sá-Nogueira, I. The MsmX ATPase plays a crucial role in pectin mobilization by Bacillus subtilis. PLoS ONE 12, e0189483 (2017).
Quentin, Y., Fichant, G. & Denizot, F. Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J. Mol. Biol. 287, 467–484 (1999).
Mao, B., Pear, M. R., McCammon, J. A. & Quiocho, F. A. Hinge-bending in l-arabinose-binding protein. The “Venus’s-flytrap” model. J. Biol. Chem. 257, 1131–1133 (1982).
Konishi, H., Hio, M., Kobayashi, M., Takase, R. & Hashimoto, W. Bacterial chemotaxis towards polysaccharide pectin by pectin-binding protein. Sci. Rep. 10, 3977 (2020).
Watzlawick, H., Heravi, K. M. & Altenbuchner, J. Role of the ganSPQAB operon in degradation of galactan by Bacillus subtilis. J. Bacteriol. 198, 2887–2896 (2016).
Inácio, J. M., Correia, I. L. & de Sá-Nogueira, I. Two distinct arabinofuranosidases contribute to arabino-oligosaccharide degradation in Bacillus subtilis. Microbiology 154, 2719–2729 (2008).
Damm, K. et al. Impact of RNA isolation protocols on RNA detection by Northern blotting. Methods Mol. Biol. 1296, 29–38 (2015).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Ohnishi, M., Yamashita, T. & Hiromi, K. Static and kinetic studies by fluorometry on the interaction between gluconolactone and glucoamylase from Rh. niveus. J. Biochem. 81, 99–105 (1977).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 (1997).
Adams, P. D. et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Acknowledgements
Diffraction data were collected at the BL26B1 and BL38B1 stations of SPring-8 (Hyogo, Japan) with the approval of JASRI (proposal nos. 2016A2574, 2016B2574, 2017B2547, and 2017B2592). This work was supported in part by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (to W.H.), and Research Grant (to W.H.) from Fuji Foundation for Protein Research. The authors would like to thank Enago (https://www.enago.com) for the English language review.
Author information
Authors and Affiliations
Contributions
W.H. designed the study; H.S., A.N., S.O., B.M., and W.H. performed the experiments; H.S., A.N., S.O., B.M., D.W., and W.H. analyzed the data; H.S., D.W., and W.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sugiura, H., Nagase, A., Oiki, S. et al. Bacterial inducible expression of plant cell wall-binding protein YesO through conflict between Glycine max and saprophytic Bacillus subtilis. Sci Rep 10, 18691 (2020). https://doi.org/10.1038/s41598-020-75359-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-75359-0
This article is cited by
-
Adaptation of yeast Saccharomyces cerevisiae to grape-skin environment
Scientific Reports (2023)
-
Disentangling the genetic basis of rhizosphere microbiome assembly in tomato
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.