Abstract
Wildlife management in rapid changing landscapes requires critical planning through cross cutting networks, and understanding of landscape features, often affected by the anthropogenic activities. The present study demonstrates fine-scale spatial patterns of genetic variation and contemporary gene flow of red panda (Ailurus fulgens) populations with respect to landscape connectivity in Kangchenjunga Landscape (KL), India. The study found about 1,309.54 km2 area suitable for red panda in KL—India, of which 62.21% area fell under the Protected Area network. We identified 24 unique individuals from 234 feces collected at nine microsatellite loci. The spatially explicit and non-explicit Bayesian clustering algorithms evident to exhibit population structuring and supported red panda populations to exist in meta-population frame work. In concurrence to the habitat suitability and landscape connectivity models, gene flow results supported a contemporary asymmetric movement of red panda by connecting KL—India in a crescent arc. We demonstrate the structural-operational connectivity of corridors in KL—India that facilitated red panda movement in the past. We also seek for cooperation in Nepal, Bhutan and China to aid in preparing for a comprehensive monitoring plan for the long-term conservation and management of red panda in trans-boundary landscapes.
Similar content being viewed by others
Introduction
Habitat mapping and modelling corridors across species distribution are cardinal for prioritization of conservation strategies1,2. Landscape connectivity demonstrates feasibility for wildlife to move through fragmented habitats and therefore maintaining corridors in fragmented landscapes are vital to ensure natural gene flow and the long-term survival of the species1,3. Further, heterogeneity and rapid changes imposed in the landscape often accelerate restriction in the species movement between suitable patches4,5. This restricted movement may lead to genetic consequences including disruption of gene flow, inflation of inbreeding and loss of rare alleles supporting local adaptation and genetic fitness6,7. This phenomenon may induce multifaceted challenges in small populations inhabiting in trans-boundary landscapes (TBL). Prioritizing species conservation across TBL is challenging due to differences in the national interest, policies, local communities, funds allocation and political will8.
The Kangchenjunga Landscape (KL) is one of the six TBL in the Hindu Kush Himalayan region, sharing boundary among Nepal, India and Bhutan9. The red panda (Ailurus fulgens), a magnificent iconic species of this landscape, is endemic to temperate conifer and cool broadleaf forest with dense bamboo undergrowth of preferring altitude range 2,300 to 4,000 m of Central and Eastern Himalayan biotic province10,11,12. Red Panda was taxonomically classified to occur in two subspecies based on the morphology and distribution—Ailurus fulgens fulgens distributed in Nepal, India, Bhutan, Myanmar, and China (Tibet and western Yunnan province) and Ailurus fulgens styani, occurred from the Sichuan and Yunnan provinces of China and Nujiang river was believed to be a biogeographic barrier for the separation of two subspecies13,14. However, a recent study by Hu et al.15 demonstrated the presence of two phylogenetic species of red panda, i.e. the Himalayan red panda (Ailurus fulgens) and Chinese red panda (Ailurus styani), with high depth sequencing data and revealed that the Yalu Zangbu river might be the potential boundary for species divergence. Further, Himalayan red panda reported to be distributed in India, Nepal, Bhutan, northern Myanmar, and Tibet and western Yunnan Province of China, while the Chinese red panda distributed in the Yunnan and Sichuan provinces of China15. The anthropogenic factors including habitat loss, poaching for pelt, jhoom cultivation and conversion of forest to non-forest land use have caused rapid decline of red panda12,14,16. Nearly 50% of red panda habitat has been lost in the last three generations, bringing it as ‘Endangered’ in the Red list category12. In India, the red panda is protected under Schedule-I of the Wildlife (Protection) Act, 1972 and most of its populations reside in small and isolated protected areas (< 500 sq. km), thereby increasing high risk of local extirpation of red pandas due to genetic inbreeding and loss of heterozygosity15,17,18,19. Earlier studies available on red panda from KL—India have addressed population status, distribution, and abundance11,20,21, habitat preferences and diet composition22,23. However, with the emergence of landscape genetics, it is now feasible to explicitly quantify the effects of landscape features on the spatial patterns of genetic variation, population structure, gene flow, and adaptation24,25,26. Thus, population genetics integrated with landscape ecology and remote sensing data can be used to aid delineating shift, if any, in the identified corridors that maintain connectivity between habitat patches and facilitate biotic processes such as dispersal and gene flow27,28,29,30. In this view, the detailed population genetic assessment of red panda with respect to landscape connectivity and anthropogenic activities is imperative to prioritize the management strategies for ensuring long term population viability of red panda in Himalayas. The present study is aimed to address the fine-scale spatial patterns of genetic differentiation and gene flow among the habitat clusters supporting red panda population in KL—India.
Results
Forty-eight candidate maxent models were generated (Table S3) and model with lowest Delta AICc informative AUC (0.911 ± 0.098) was selected, which predicted species distribution better than the random model (Table S1). The generated binary map discriminated between suitable and unsuitable habitat of red panda based on 10-percentile training presence thresholds (Fig. 1b). The jackknife test revealed that bioclimatic variable siwb_bio19, precipitation of coldest quarter contributed 45.6% and maximum temperature of the warmest month contributed 29.4% to predict habitat suitability. The land use land cover (LULC) contributed 12.9% followed by canopy height 11.8% and isothermality found to be less important and contributed only 0.3% (Table 1). The Jackknife test for regularized training gain in the present model showed that the variable with the highest gain, when used in isolation, was precipitation of coldest quarter, which therefore appeared to have the most useful information by itself. The environmental variable that decreased the gain the most when it was omitted, was Max Temperature of Warmest Month, which therefore appeared to have the most information that was not present in the other variables (Fig. S1).
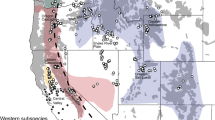
Study area map, red panda species distribution and landscape connectivity model. (a) Map of Kangchenjunga landscape (KL), India with overlaid sampling locations [SNP—Singalila National Park and NVNP—Neora Valley National Park in north West Bengal; BRS—Barsey Rhododendron Wildlife Sanctuary and KNP—Kanchenjunga National Park in West Sikkim (WS); PWLS—Pangolakha Wildlife Sanctuary and KAS—Kyongnosola Alpine sanctuary in the East Sikkim (ES)], (b) predicted habitat suitability model of red panda in KL—India, (c) model based on genetic divergence of red panda in KL—India. (d) Landscape connectivity model based on ensemble approach by combining both the genetic divergence and environmental conductance in KL—India.
Habitat suitability and landscape connectivity
The study covered 10 protected areas (PAs) that included Singalila National Park (SNP), Senchal Wildlife Sanctuary (SWLS), and Neora valley National Park (NVNP) located in Northern West Bengal and Barsey Rhododendron Sanctuary (BRS), Maenam Wildlife Sanctuary (MWLS), FambongLho Wildlife Sanctuary (FLWLS), Pangolakha Wildlife Sanctuary (PWLS), Kangchenjunga National Park (KNP), Shingba Rhododendron Sanctuary (SRS), and Kyongnosla Alpine Sanctuary (KAS) located in Sikkim State of India. Our results predicted a total of 1,309.54 km2 (12.78%), comprised of 1,097.26 km2 in Sikkim and 212.28 km2 in North West Bengal as suitable habitat for red panda with largest suitable habitats in KNP (493.37 km2) (Table 2). The habitat suitability demonstrated that from West to East, SNP was connected with BRS and BRS was connected with KNP which is the only National Park in Sikkim. The MWLS in south Sikkim was connected with KNP on its northern border. On eastern part of the landscape NVNP and PWLS were connected, however, large amount of habitat suitability was observed outside PA on the eastern part of Sikkim.
The ensemble approach by combining both the genetic divergence and environmental conductance depicted the landscape connectivity and its functionality to support movement corridors of red panda in KL—India. The predicted genetic divergence among the identified clusters was in coherence to the habitat suitability model (Fig. 1b,c). The landscape genetics model by an ensemble conductance surface, lumping both the genetic divergence and environmental suitability demonstrated high-density currents radiated from SNP and NVNP in the North West Bengal (Fig. 1d). Further, to decipher the direction of gene flow among the clusters, we estimated the contemporary migration rate using BayesAss (Fig. S4). The landscape connectivity model and the contemporary migration rate, jointly inferred the permeability of floating currents, indicating structural connectivity in KL—India (Fig. 1d), well supported by the asymmetric gene flow from West to East (Fig. S4). The landscape connectivity model did not show horizontal connection from West to East in KL—India. Instead, the current flow forming a crescent arc to connect KL—India from the West to East with high-density currents radiating from SNP and NVNP of North West Bengal. The current formed bottleneck position of corridor in the KNP of north Sikkim. On the eastern part of the KL, the cumulative current flow was narrow running in longitudinal extent between PAs (Fig. 1d).
Individual identification and assessment of genetic variability
Allelic Drop out (ADO) ranged from 0 to 0.294 while we did not observe any significant amount of False allele (FA) (Table 3). Four loci i.e. CRP357, CRP385, CRP409 and CRP367 exhibited relatively high frequencies of null alleles that may consequently affected the Hardy Weinberg Equilibrium (HWE) and Inbreeding coefficient (FIS) (Table 3). Among 234 faecel samples collected from KL, we obtained positive PCR amplification in 98 faeces (38%) with nine microsatellite loci. Of which, 37 samples (approx. 40%) yielded mixed profiles on capillary electrophoresis, plausibly due to the red panda behaviour, being arboreal and group defecation in piles at same places. We finally obtained 57 unambiguous genotypes and with a select panel of seven loci with a cumulative PIDsib 2.91 × 10−3, we identified 24 unique genotypes—two individuals originated from SNP, 11 from WS, six from ES and five from NVNP (Table 3). We uploaded multilocus genotype data of 24 unique individuals on DRYAD and available on https://doi.org/10.5061/dryad.2280gb5nz. For population genetic analysis, we used nine microsatellite data that exhibited ≥ 90% amplification success. The exhibited mean observed number of alleles (Na)—6 ± 0.58, observed heterozygosity (HO)—0.493 ± 0.06 and expected heterozygosity (HE) 0.664 ± 0.027 (Table 3). Three loci, CRP357, CRP385 and CRP367 deviated significantly from HWE and only one pair of loci out of 36 pair-wise comparisons were in significant Linkage disequilibrium (LD) (P < 0.0012) after Bonferroni correction. HWE deviation of these markers might be due to the multiple reasons, like inflated frequencies of null alleles, Wahlund effect, and inbreeding due to consanguineous and assortative mating31. The mean FIS was 0.276, indicating a significant inbreeding and loss of heterozygosity (Table 3).
Population genetic structure
The STRUCTURE analysis detected three clusters based on the mean ln P (k) and Delta K (Fig. 2, Fig. S3). We showed probabilities of population assignment at varying K (Fig. S3). At K-2, a majority of individuals from WS (West Sikkim) showed a distinct cluster but no geographical assignment was observed among the individuals originated from SNP, ES (East Sikkim), and NVNP. At K-3 individuals from SNP showed a distinct isolated cluster while individuals from ES and NVNP assigned to one another cluster. However, a further increase in the K did not reveal any structuring in population. The results indicated that NVNP and ES clusters were relatively connected meta-populations as found shared ancestry in one-another populations while the other two clusters SNP and WS shared ancestry with additional genetic influxes of an unknown population which plausibly be the North Sikkim (NS) and or the historic/uncaptured gene flow from Nepal, which could not be addressed due to unavailability of samples (Fig. 2a). In congruity, GENELAND also testified to the similar clustering patterns, NVNP and ES grouped into a single cluster and SNP and WS in a distinct clusters (Fig. 2b). The sPCA revealed a west–east differentiation from SNP to NVNP in the allele frequencies, indicating samples of NVNP and ES in one cluster. SNP in the other and WS was assigned as an intermediate population with more affinity to SNP (Fig. 2c, Fig. S2). The DAPC also identified three major clusters, providing strong signals to support red panda populations to exist in metapopulations framework (Fig. 2d). The non-Bayesian methods of population assignment in coherence to Bayesian clustering methods supported strong population genetic structure with asymmetric gene flow among the habitat patches in KL—India.
Population genetic structure of red panda population in KL—India. (a) Population assignment using STRUCTURE at K3; (b) map of estimated cluster membership showing spatial distribution of the three inferred genetic clusters through GENELAND; (c) spatial PCA showing clusters in spatially distributed populations; (d) Eigen values of PCA estimation showing three clusters in DAPC, each identified by individual colours and inertia eclipses.
Gene flow and detection of migrants
In AMOVA, we found relatively lower genetic variance within population which indicated gene flow within population was higher than the between groups. Red panda populations in KL—India exhibited 58% variance within population than 12% between populations and 30% among group with high Fst (0.321). Though Fst value was not statistically significant but observed signal was an indication of the red panda population to exist in metapopulation framework. Pairwise FST based gene flow revealed SNP and NVNP populations were highly differentiated (FST 0.352) and NVNP and ES did not qualify to be separate populations (FST − 0.052) (Fig S4).
The contemporary migration assessed by the BAYESASS resulted to favour asymmetric movement of red panda from West to East in KL—India (Fig. S4). We detected significant asymmetric migration of individuals from SNP to WS (5.55%) and WS to ES (5.48%) but did not obtain a rational backward gene flow. In contrary, we detected significant bidirectional migration between ES to NVNP (5.91%) and NVNP to ES (6.05%).
Discussion
Habitat suitability and corridor connectivity
The red panda population in KL has experienced massive habitat loss and fragmentation in the past11,12,16,32 which must have deteriorated connecting corridors in this landscape. For habitat suitability, predictors such as precipitation, temperature and vegetation with high weightage appeared reasonably correct as vegetation cover, and climatic factors are directly linked to the species diet, survival and reproductive necessities. A few earlier studies addressed the species distribution modelling (SDM) of Red panda in Himalaya16,20,33,34,35. These studies considered different variables like elevation, slope, aspect and distance to water etc. for predicting red panda habitat. These variables may facilitate in old growth forest dominated by Betula utilis, Rhododendron spp. and Abies spp. with dense bamboo cover in the understory and high densities of fallen logs and tree stumps at ground level. Further, the growth of bamboo understory in the temperate forests is highly influenced by the rainfall and temperature36 and red pandas as being the canopy-dwelling species prefer temperate forests in the central and eastern Himalayas with dense bamboo undergrowth. Therefore, rainfall, temperature and vegetation cover played a significant role in predicting red panda habitat.
Earlier, Choudhury11 estimated 1,700 km2 potential habitat of red panda in Sikkim and 200 km2 in North West Bengal by undertaking exploratory field surveys based on one time efforts. Ziegler37 estimated 684 km2 suitable forest for red panda in Sikkim with limited efforts confining to PWLS only. However, Ghose and Dutta38 estimated 1,200 km2 suitable forest for red panda in Sikkim and 250 km2 in North West Bengal. These authors collected primary data by undertaking field surveys through transect and trail monitoring39 from Sikkim and North West Bengal and also recorded indirect evidences such as droppings, browsing marks, nest sites, pugmarks, skins or pelts, etc. Ghose and Dutta38 also collected secondary data through questionnaires and public interviews in the villages around the potential red panda habitats. Our field surveys and efforts were relatively comparable to Ghose and Dutta38 and we obtained an estimated suitable habitat 1,097.26 km2 in Sikkim and 212.28 km2 in North West Bengal, which represented a relative decline the habitat suitability in last ten years when compare to Ghose and Dutta38. In northern region, red panda habitat was relatively fragmented and circuitscape also supported lack of horizontal connectivity in KL—India. However, the identified corridor favoured red panda movement from West to East in KL India, forming a crescent arc (Fig. S4). The model demonstrated that the identified corridor in the east, south Sikkim and eastern part of north Sikkim was relatively narrow and passing through the non-PAs (Fig. 1d). We propose corridor passing through the non-PAs to be monitored for the emerged developmental activities and enriched by bamboo plantation to avoid further loss of red panda habitat in KL—India.
Genetic diversity and inbreeding
Red panda population of KL—India being a trans-boundary population, must be carrying ancestral genetic attributes shared with Nepal and Bhutan populations. The genetic assessment suggested a relatively low genetic diversity in red panda population of KL—India (HE = 0.66), when compared to other red panda populations i.e. HE = 0.71940 and HE = 0.77241. The observed low genetic diversity of the Himalayan red panda might be due to historical bottlenecks15. The observed loss of suitable habitat by the emerged anthropogenic activities in last few decades might have disrupted the contiguous gene flow and confined red panda in isolated patches, also a contributing factor for inbreeding and loss of heterozygosity in KL—India.
Population genetic structure and gene flow
Explicit Bayesian and non-Bayesian clustering methods to a great extent, showed similar patterns clustering red panda into at least three populations/meta-populations in KL—India. Indisputably, the NVNP and ES was found to represent one single population, However, SNP was another distinct cluster where WS was assigned as an intermediate population with more affinity to SNP. SNP and NVNP that lacks horizontal connection, were two distinct clusters (FST = 0.352). These populations earlier reported to exist in relatively high density (1 individual per 1.67 sq. km in SNP20; 32 animals in SNP and 34 in NVNP42). Individuals from these populations showed asymmetric gene flow and contributed migration to Sikkim population from either-side. The WS population was rationally distinct from SNP (FST 0.142), however ES and NVNP, to a large extent represent a single population based on the multiple evidences, e.g. a negative FST − 0.052 and a high migration rate from one another population (ES to NVNP—5.91% and NVNP to ES—6.05%; Fig. S4). In concurrence to this, AMOVA also supported a high variance (within group) in red panda populations. The observed scenario of contemporary gene flow patterns in KL—India suggested that corridors connecting these populations were functional in the recent past. The pragmatic asymmetric migration detected in the last two generations exhibited asymmetric migration of individuals in KL—India from West to East, SNP to WS and WS to ES. The ES and NVNP had bidirectional migration, also an indicator to support them for being a single population. The observed asymmetric gene flow among populations was due to the landscape heterogeneity and habitat suitability. The circuitscape results evident to prove that SNP and NVNP were two relatively dense populations in KL—India, where individuals might be moving from more stable, relatively high-density population to neighbouring low-density populations43. The high migration rate from SNP and NVNP maintained the demographic connectivity in the landscape. Both, the significant genetic differentiation in some areas and the presence of a weak population structure in other regions in KL—India may be explained by the corridor connectivity model (Fig. 1d), which coincides to the results of BAYESASS and supports current flow from West to East forming a crescent arc in the KL—India.
Functional connectivity and conservation priorities
The present study unveils the facts that landscape features have shaped the current distribution patterns of red panda in KL—India. The results demonstrated that NVNP and PWLS were connecting North West Bengal–Sikkim border. But there has been a large gap between PWL to KAS which extended up to the SRS. Further, the suitable habitat was patchy and connecting corridors were narrow between those habitat patches. Habitat on the eastern part of the study landscape was more vulnerable to destruction due to the anthropogenic activities. The FLWLS had only a small suitable area with poor connectivity to the other populations. Ziegler37 conducted a survey in FLWLS but failed to confirm the presence of red panda. There have been recent historic records of red panda from SWLS in West Bengal44 but intensive surveys need to be conducted to assess the current status of red panda in SWLS, which is also poorly connected to SNP.
Though, there are several PAs on the eastern part of KL—India, there is a need to extend PA network in East Sikkim, South Sikkim and eastern part of North Sikkim. Moreover, the territorial forest divisions must take adequate measures, as much of suitable habitat of red panda exists outside the PA. The present study has laid down the foundation to extend the PAs boundaries and by effectively implementing the eco-sensitive zone planning adopted by the Ministry of Environment and Forest, 2011 of Government of India45. We propose buffer zone may be declared around the PAs and community conservation area to protect important wildlife corridors. Since, the red panda across the range has experienced habitat loss, fragmentation and population decline due to change in the land use pattern and anthropogenic activities in past few decades. Any significant change in the climatic isotherm might result in vacating the site and or shifting the species to other sites based on varying extent of species resilience and inherent adaptive plasticity46,47. Thus, red panda being ecological specialist, serves a good model to test the composite impact of landscapes, historical climate change and contemporary human activities on the possible shift in the ranges.
Further, studying wildlife using genome-wide markers (e.g. GWAS—15,48; SNPs—15,49) is fascinating to evaluate fine scale population genetic structure and investigating loci under natural selection facilitating populations to adapt in the changing climatic conditions50. However, to check the immediate effects of landscape features on the genetic variability and population contiguity, the assessment of wild populations using microsatellites is still most cost effective and widely applied way to genetic monitor of free ranging populations30,51. Further, consolidating landscape connectivity through mapping corridors, validating movement through genetics and expanding natural PAs is fundamental to make red panda conservation a long-term success across the distribution range. We also seek for the possible collaboration in Nepal, Bhutan and China to aid in preparing a comprehensive monitoring plan for red panda conservation in TBL and to evaluate the ongoing conservation efforts across the political boundaries.
Materials and methods
Study area
KL—India, situated in the Central Himalayan biotic province with spanning between 26° 21′ 40.49ʺ–28° 7′ 51.25ʺ N and 87° 30′ 30.67ʺ–90° 24′ 31.18ʺ E. The landscape is highly rugged with mountainous terrain including the world’s 3rd highest mountain peak, the Mount Kangchenjunga (8586 m asl). The habitat types ranging from tropical, subtropical, warm temperate, cool temperate, subalpine, and alpine forest types52,53. The study landscape is bestowed with 10 PAs i.e. Singalila National Park (SNP), Senchal Wildlife Sanctuary (SWLS), and Neora valley National Park (NVNP) located in North West Bengal and Barsey Rhododendron Sanctuary (BRS), Maenam Wildlife Sanctuary (MWLS), FambongLho Wildlife Sanctuary (FLWLS), Pangolakha Wildlife Sanctuary (PWLS), Kanchenjunga National Park (KNP), Shingba Rhododendron Sanctuary (SRS), and Kyongnosla Alpine Sanctuary (KAS) located in Sikkim State of India. Together these PAs, although most of them are fairly small in size, contribute approximately 3,112.21 km2 area (Fig. 1a).
Study design and occurrence data
Firstly, we stratified the study area based on the forest types, topography to cover all the logistically possible habitat patches in KL—India. Secondly, we adopted a landscape approach, instead of prioritizing PAs for sampling and we aimed to cover maximum reported habitats of red panda in KL—India. Thirdly, for recording the species presence, we adopted a three-pronged approach i.e. transects/trail surveys, questionnaire surveys, and also the remote camera traps following Pradhan et al.20, Buckland et al.39 and Joshi et al.54. Further, it was not logistically feasible to cover the entire area systematically due to the rugged terrain, unpredictable weather and poor resources availability. Hence, representative sampling was carried out in KL—India that represented 109 spatially distinct locations, 12 locations from camera traps, seven direct sightings 12 presence locations from reliable interviews and 78 distinct sites where we collected 234 faecal samples of red panda. For model building, we used 56 spatially independent locations after correlation testing among the location following Kramer-Schadt55. Informed consent was obtained from all participants who responded to the questionnaire interviews and all methods including human participation in the study was in accordance to the relevant guidelines and regulations of Zoological Survey of India (ZSI), Kolkata. All experimental protocols and methods were approved by the Research and Academic committee (RAC) of ZSI, Kolkata that has a ethics committee/institutional review board. All field surveys and sampling were conducted with taking necessary permits issued by the Forest Department of West Bengal and Sikkim.
Ecological modelling
Selection of environmental variables
Species are sensitive to habitats as well as the climatic isotherms56 while, any significant change in climatic isotherm to a species having special requirements might result in local extirpation or shifting the species to other ranges46,47. Therefore, it is imperative to use variables for SDM which define the likely habitat of a species based on the field observation. Considering these facts, we selected 26 variables out of which 19 variables represented climatic isotherm, four-topography and three variables represented land covers. These variables were also related to the habitat selection of red panda16,35,57. The selected variables represented the present environmental conditions and habitat covariates to facilitate red panda distribution patterns in KL—India. The biotic predictors (19) were obtained from the WorldClim data base at 30 arc second scale58; https://www.worldclim.org/ (Table S2) and Bioclime v 1.4 dataset was used for the model building. The vegetation classification was carried out following the methodology developed by Forest Survey of India (ISFR 2017). ArcGIS 10.6 (ESRI 2018) was used to develop topographic variables from digital elevation model. For obtaining the forest canopy classes, the satellite data (Landsat 8) was downloaded from US Geological Survey (https://earthexplorer.usgs.gov/). The forest canopy was classified into four categories viz., very dense, moderately dense, open forest and scrubland59. Moreover, considering the fact that the species is a canopy dweller11 the Canopy height data was obtained from Oak Ridge National Laboratory (https://webmap.ornl.gov/ogc) for understanding the influence on the species habitat suitability. Since, different variables like climate, landscape, topography and anthropogenic influences used in present study were compiled from various resources present in different resolution scales. Hence, to bring uniformity in the selected data, we re-sampled all raster on the same resolution scale (1 km resolution).
Habitat modelling and landscape connectivity
We employed maximum entropy algorithm for species distribution modelling (MaxEnt 3.4.1), using presence data only60,61,62. Bias file was prepared in ENMeval; R package63 following the methodology suggested by Dudik et al.64. We omitted highly correlated variables with a threshold of 0.865,66. Although we selected initially 26 variables but final model was predicted using 19 variables which passed on the multicollinearity test67 and other related variables with a threshold of 0.8 were excluded to get rid of the over fit model (Table S6)66,68. For modelling, we have used 70% of the total locations as training and remaining 30% for testing the significance60,66,69. The Akaike’s information criteria (AICc) values were used to determine the best fit model with the lowest values in ENMeval. The resultant habitat was classified into suitable, with probabilities 0.584–1 and unsuitable below 0.584 based on the 10-percentile training presence thresholds following Radosavljevic and Anderson70 (Table S4). The Jackknife was used to analyse the contribution of each variable provided to the MaxEnt model. We tested the model with the receiver operated characteristics area under curve (AUC) where, AUC value > 0.75 indicated high discrimination performance71. We also estimated the true skill statistic (TSS—0.742) that compensates for the shortcomings of kappa while keeping all of its advantages following Allouche et al.72 We generated a raster surface of genetic divergence using Genetic landscape GIS Toolbox Ver. 10.173 by generating the inverse distance weighted interpolation within the study area boundary74. To evaluate the operational-structural connectivity by predicted movement corridors of red panda in KL—India, we used an ensemble approach by combining both the genetic divergence and environmental conductance (SDM output) in the final circuit model following Mateo-Sánchez et al.75 and Roffler et al.76 in Circuitscape 4.077.
Population genetic analyses
DNA extraction, PCR amplification and microsatellite genotyping
We collected 234 red panda feces, i.e. 87 feces from North West Bengal and 147 from Sikkim in KL—India (Fig. 1a). All samples were stored in 70% ethanol and DNA was extracted using QIAamp Fast DNA Stool Mini Kit (QIAGEN Germany) following manufacturer's instructions. Nine polymorphic STRs, i.e. Aifu01, Aifu05 from Liang78 and CRP357, CRP385, CRP381, CRP367, CRP409, CRP240 and CRP260 from Yang79 were amplified into two multiplex PCRs. Forward primer of all nine microsatellite loci was fluorescently labelled at 5′ with one of the four dyes, FAM, VIC, NED and PET (Table S5). The PCRs were carried-out in 10 µl reaction volume following QIAGEN Multiplex PCR Kit (Qiagen, Germany). The thermal cycle profile was: initial denaturation at 95 °C for 15 min, followed by 40 cycles of PCR and a final step of 72 °C for 30 min. The annealing temperature (Ta) for multiplex 1 (CRP381, CRP367, CRP409, CRP240 and CRP260) was 55 °C and multiplex 2 (Aifu01, Aifu05, CRP357, CRP385) was 57 °C (Table S5). The PCR products were resolved on an ABI 3730 Genetic analyzer (Applied Biosystems, Foster City, CA, USA) and allele scoring was done using Gene Mapper 4.1 (Applied Biosystems, Foster City, CA, USA).
Genotyping error and individual identification
We genotyped each sample four times to minimize genotyping errors and a heterozygote was ascertained only if there were different alleles in at least three independent attempts. In addition, we also followed an independent allele scoring method where two different researchers perform allele scoring individually and only the consensus genotypes were used for further analysis. The genotyping errors arising due to null allele and the presence of stutters, scoring errors were assessed using MICRO CHECKER 2.2.280. Maximum likelihood allele dropout (ADO) and false allele (FA) error rates were quantified using PEDANT version 1.0 involving 10,000 search steps for enumeration of per allele error rates81. To avoid ambiguity in ascertaining unique genotypes, we limited the number of loci used based on their high success rate (> 90%), presence of no or minimum genotyping errors and exhibiting an informative PID value (probability of obtaining identical genotypes between two samples by chance). The locus wise and cumulative probability of identity for unrelated individuals (PID) and siblings (PID sibs) were calculated following identity analysis module in GenAlEx version 682.
Genetic diversity and inbreeding
The genetic diversity estimates were accounted by calculating the number of allele (Na), observed (Ho) and expected (He) heterozygosity using GENALEX 682. For the Hardy–Weinberg equilibrium (HWE) test, we followed the probability test approach using the program GENEPOP version 4.2.183. Wright’s inbreeding coefficient (FIS) was estimated following Weir & Cockerham84 using GENEPOP83. Linkage disequilibrium (LD) was tested using GENEPOP83 to determine the extent of distortion from independent segregation of loci following 10,000 dememorizations, 500 batches and 10,000 iterations per batch after Bonferroni correction85.
Population genetic structure
We attempted three different clustering methods to capture the most possible population genetic structure of red panda in KL—India i.e. Fst based Analysis of Molecular Variance (AMOVA), explicit Bayesian and non-Bayesian clustering algorithms. We used Arlequin 3.5.2.186 to estimate the proportions of the total genetic variation, arose from, within and between populations using AMOVA. Among different Bayesian clustering methods, individuals were assigned exclusively on the basis of their multi-locus genotypes (e.g. STRUCTURE), and also using the both, multi-locus genotypes and geo-referenced information (e.g. GENELAND). Non-Bayesian multivariate ordination analyses, i.e. discriminant analysis of principle components (DAPC) and spatial principle component analysis (sPCA) were also used, to compare population assignment with the Bayesian clustering outputs87 since they were not based on any model assumptions.
In Bayesian analysis, STRUCTURE 2.3.4 software88 was used to determine the number of genetic clusters (K) following 20 iterations (20,000 burn-in; 200,000 Markov chain Monte Carlo replicates in each run) with NOPRIOR with admixed and correlated allele frequencies. We considered there were K populations (1 to 10), with repeating each analysis for 10 times at each K value. The most probable cluster was calculated via estimating the distribution of Delta K89 using STRUCTURE HARVESTER v.0.6890. GENELAND v 4.0.391 was run through an extension of R v.3.0.1 with the correlated allele frequency and spatial uncertainty model. We allowed K to vary between 1 and 10 following 20 independent runs, each with 100,000 iterations, and a thinning of 1,000. The inferred spatial clusters were georeferenced in ArcGIS 10.6. Two non-Bayesian clustering methods i.e., DAPC and sPCA, were run to assign the possible clusters in Adegenet v1.3.4 package of R92.
Gene flow and migration rate
We assessed Fst based gene flow in last 150–200 years93 among the different sub-populations of red panda in KL—India using Arlequin v 3.5.286. The contemporary and asymmetric migration rate in last two generations were estimated using BayesAss 1.343. We used 9 × 106 iterations, with a burn-in of 106 iterations, 1,000 number of permutations and a sampling frequency of 2000 to ensure that the model's starting parameters were sufficiently randomized. We also estimated the first-generation migrants between all pairs of subpopulations of red panda in KL—India using GENECLASS 2.094, an approach described by Paetkau95 for likelihood computation (Lhome/Lmax), with 1,000 simulations at an assignment threshold (alpha) of 0.01 and 0.05.
Data availability
The multilocus genotype data of 24 unique individuals is available on DRYAD (https://doi.org/10.5061/dryad.2280gb5nz) and all other relevant data is included in the manuscript.
Reference:s
Margules, C. R. & Pressey, R. L. Systematic conservation planning. Nature 405(6783), 243–253 (2000).
Lindenmayer, D. B. & Fischer, J. Habitat Fragmentation and Landscape Change: An Ecological and Conservation Synthesis (Island Press, Washington, 2013).
Hilty, J. A., Lidicker, W. Z. Jr. & Merenlender, A. M. Corridor Ecology: The Science and Practice of Linking Landscapes for Biodiversity Conservation (Island Press, Washington, 2012).
Katayama, N. et al. Landscape heterogeneity–biodiversity relationship: Effect of range size. PLoS ONE https://doi.org/10.1371/journal.pone.0093359 (2014).
Doherty, T. S. & Driscoll, D. A. Coupling movement and landscape ecology for animal conservation in production landscapes. Proc. R. Soc. B Biol. Sci. 285(1870), 2017–2272 (2018).
Frankham, R. Challenges and opportunities of genetic approaches to biological conservation. Biol. Conserv. 143(9), 1919–1927 (2010).
Haddad, N. M. et al. Habitat fragmentation and its lasting impact on Earth’s ecosystems. Sci. Adv. 1(2), e1500052 (2015).
Lambertucci, S. A. et al. Apex scavenger movements call for transboundary conservation policies. Biol. Conserv. 170, 145–150 (2014).
Gurung, J. et al. Evolution of a transboundary landscape approach in the Hindu Kush Himalaya: Key learnings from the Kangchenjunga landscape. Glob. Ecol. Conserv. 17, 1–15 (2019).
Wei, F., Feng, Z., Wang, Z., Zhou, A. & Hu, J. Use of the nutrients in bamboo by the red panda (Ailurus fulgens). J. Zool. 248(4), 535–541 (1999).
Choudhury, A. An overview of the status and conservation of the red panda Ailurus fulgens in India, with reference to its global status. Oryx 35(3), 250–259 (2001).
Glatston, A., Wei, F., Than, Z., & Sherpa, A. Ailurus fulgens. The IUCN red list of threatened species, 4 (2015).
Roberts, M. S. & Gittleman, J. L. Ailurus fulgens. Mamm. Species 222, 1–8 (1984).
Wei, F., Feng, Z., Wang, Z. & Hu, J. Current distribution, status and conservation of wild red pandas Ailurus fulgens in China. Biol. Conserv. 89, 285–291 (1999).
Hu, Y. et al. Genomic evidence for two phylogenetic species and long-term population bottlenecks in red pandas. Sci. Adv. 9, 5751 (2020).
Thapa, A. et al. Predicting the potential distribution of the endangered red panda across its entire range using MaxEnt modeling. Ecol. Evol. 8, 10542–10554 (2018).
Cohen, J. E. & Small, C. Hypsographic demography: The distribution of human population by altitude. Proc. Natl. Acad. Sci. U.S.A. 95, 14009–14014 (1998).
Panthi, S., Wang, T., Sun, Y. & Thapa, A. An assessment of human impacts on endangered red pandas (Ailurus fulgens) living in the Himalaya. Ecol. Evol. 9, 13413–13425 (2019).
Hansen, A. J. & De Fries, R. Ecological mechanisms linking protected areas to surrounding lands. Ecol. Appl. 17(4), 974–988 (2007).
Pradhan, S., Saha, G. K. & Khan, J. A. Ecology of the red Panda Ailurus fulgens in the singhalila national Park, Darjeeling, India. Biol. Conserv. 98, 11–18 (2001).
Chakraborty, R. et al. Status, abundance, and habitat associations of the red panda (Ailurus fulgens) in Pangchen Valley, Arunachal Pradesh, India. Mammalia https://doi.org/10.1515/mammalia-2013-0105 (2015).
Sharma, H. P., Belant, J. L. & Swenson, J. E. Effects of livestock on occurrence of the Vulnerable red panda Ailurus fulgens in Rara National Park, Nepal. Oryx 48(2), 228–231 (2014).
Panthi, S., Coogan, S. C., Aryal, A. & Raubenheimer, D. Diet and nutrient balance of red panda in Nepal. Sci. Nat. 102(9–10), 54 (2015).
Holderegger, R. & Wagner, H. H. Landscape genetics. Bioscience 58(3), 199–207 (2008).
Balkenhol, N., Waits, L. P. & Dezzani, R. J. Statistical approaches in landscape genetics: An evaluation of methods for linking landscape and genetic data. Ecography 32(5), 818–830 (2009).
Manel, S. et al. Perspectives on the use of landscape genetics to detect genetic adaptive variation in the field. Mol. Ecol. 19(17), 3760–3772 (2010).
Cushman, S. A., McKELVEY, K. S. & Schwartz, M. K. Use of empirically derived source-destination models to map regional conservation corridors. Conserv. Biol. 23(2), 368–376 (2009).
Brodie, J. F. et al. Evaluating multispecies landscape connectivity in a threatened tropical mammal community. Conserv. Biol. 29(1), 122–132 (2015).
Zeller, K. A. et al. Are all data types and connectivity models created equal? Validating common connectivity approaches with dispersal data. Divers. Distrib. 24(7), 868–879 (2018).
Menchaca, A. et al. Population genetic structure and habitat connectivity for jaguar (Panthera onca) conservation in Central Belize. BMC Genet. 20(1), 1–13 (2019).
Hedrick, P. Large variance in reproductive success and the Ne/N ratio. Evolution 59(7), 1596–1599 (2005).
Mallick, J. K. Status of Red Panda Ailurus fulgens in Neora Valley National Park, Darjeeling District, West Bengal, India. Small Carnivore Conserv. 43(30), e36 (2010).
Zhang, Z. J. et al. Microhabitat separation during winter among sympatric giant pandas, red pandas and tufted deer: The effects of diet, body size, and energy metabolism. Can. J. Zool. 82, 1451–1458 (2004).
Dorji, S., Vernes, K. & Rajaratnam, R. Habitat correlates of the Red Panda in the temperate forests of Bhutan. PLoS ONE 6(10), e26483 (2011).
Zhou, X. et al. The winter habitat selection of red panda (Ailurus fulgens) in the Meigu Dafengding national nature reserve, China. Curr. Sci. 105, 1425–1429 (2013).
Rao, A. N., Zhang, X. P. & Zhu, S. L. Selected papers on recent bamboo research in China (1991).
Ziegler, S. et al. Sikkim-under the sign of the red panda. Z. Des Kölner Zoos 2, 79–92 (2010).
Ghose, D. & Dutta, P. K. Status and Distribution of Red Panda Ailurus fulgens fulgens in India. In Red Panda 357–373 (William Andrew Publishing, Park Ridge, 2011).
Buckland, S. T., Anderson, D. R., Burnham, K. P. & Laake, J. L. Distance Sampling: Estimating Abundance of Biological Populations (Chapman and Hall, London, 1993).
Hu, Y. et al. Genetic structuring and recent demographic history of red pandas (Ailurus fulgens) inferred from microsatellite and mitochondrial DNA. Mol. Ecol. 20(13), 2662–2675 (2011).
Liu, Z., Zhang, B., Wei, F. & Li, M. Isolation and characterization of microsatellite loci for the red panda, Ailurus fulgens. Mol. Ecol. Notes 5(1), 27–29 (2005).
Mallick, J. K. In situ and ex situ conservation of Red Panda in Darjeeling district, West Bengal, India. Animal Divers. Nat. Hist. Conserv. 5, 283–305 (2015).
Wilson, G. A. & Rannala, B. Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163(3), 1177–1191 (2003).
Bahuguna, N. C. & Mallick, J. K. Handbook of the Mammals of South Asia: With Special Emphasis on India, Bhutan, and Bangladesh (Natraj Publishers, Dehradun, 2010).
Deb, S., Ahmed, A. & Datta, D. An alternative approach for delineating eco-sensitive zones around a wildlife sanctuary applying geospatial techniques. Environ. Monit. Assess. 186(4), 2641–2651 (2014).
Angert, A. L. et al. Do species’ traits predict recent shifts at expanding range edges?. Ecol. Lett. 14, 677–689 (2011).
Guo, F., Lenoir, J. & Bonebrake, T. C. Land-use change interacts with climate to determine elevational species redistribution. Nat. Commun. 9, 1315 (2018).
Santure, A. W. & Garant, D. Wild GWAS—Association mapping in natural populations. Mol. Ecol. Resour. 18(4), 729–738 (2018).
Provan, J., Glendinning, K., Kelly, R. & Maggs, C. A. Levels and patterns of population genetic diversity in the red seaweed Chondrus crispus (Florideophyceae): A direct comparison of single nucleotide polymorphisms and microsatellites. Biol. J. Linn. Soc. 108(2), 251–262 (2013).
Allendorf, F. W., Hohenlohe, P. A. & Luikart, G. Genomics and the future of conservation genetics. Nat. Rev. Genet. 11(10), 697–709 (2010).
Waraniak, J. M., Fisher, J. D., Purcell, K., Mushet, D. M. & Stockwell, C. A. Landscape genetics reveal broad and fine-scale population structure due to landscape features and climate history in the northern leopard frog (Rana pipiens) in North Dakota. Ecol. Evol. 9(3), 1041–1060 (2019).
Champion, S. H., & Seth, S. K. A revised survey of the forest types of India. In A revised survey of the forest types of India. (1968).
Wikramanayake, E. D., Dinerstein, E. & Loucks, C. J. Terrestrial Ecoregions of the Indo-Pacific: A Conservation Assessment (Island Press, Washington, 2002).
Joshi, B. D. et al. Field testing of different methods for monitoring mammals in Trans-Himalayas: A case study from Lahaul and Spiti. Glob. Ecol. Conserv. 21, e00824 (2020).
Kramer-Schadt, S. et al. The importance of correcting for sampling bias in MaxEnt species distribution models. Divers. Distrib. 19(11), 1366–1379 (2013).
Lenoir, J. et al. Species better track climate warming in the oceans than on land. Nat. Ecol. Evol. 4, 1044–1059 (2020).
Bista, D. et al. Distribution and habitat use of red panda in the Chitwan-Annapurna landscape of Nepal. PLoS ONE 12(10), e0178797 (2017).
Hijmans, R. J., Cameron, S. E., Parra, J. L., Jones, P. G. & Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. J. R. Meteorol. Soc. 25(15), 1965–1978 (2005).
Negi, S. P. Forest cover in Indian Himalayan states—An overview. Indian J. For. 32(1), 1–5 (2009).
Phillips, S. J., Anderson, R. P. & Schapire, R. E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 190(3–4), 231–259 (2006).
Elith, J. et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 29(2), 129–151 (2006).
Elith, J. et al. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 17(1), 43–57 (2011).
Muscarella, R. et al. ENM eval: An R package for conducting spatially independent evaluations and estimating optimal model complexity for Maxent ecological niche models. Methods Ecol. Evol. 5(11), 1198–1205 (2014).
Dudík, M., Phillips, S. J. & Schapire, R. E. Correcting sample selection bias in maximum entropy density estimation. In Advances in Neural Information Processing Systems (eds Dietterich, T. G. et al.) 323–330 (MIT Press, Cambridge, 2006).
Warren, D. L., Glor, R. E. & Turelli, M. ENMTools: A toolbox for comparative studies of environmental niche models. Ecography 33(3), 607–611 (2010).
Sharma, L. K., Mukherjee, T., Saren, P. C. & Chandra, K. Identifying suitable habitat and corridors for Indian Grey Wolf (Canis lupus pallipes) in Chotta Nagpur Plateau and lower Gangetic planes: A species with differential management needs. PLoS ONE https://doi.org/10.1371/journal.pone.0215019 (2019).
Phillips, S. J., Anderson, R. P. & Schapire, R. E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 190, 231–259 (2006).
Bagaria, P. et al. West to east shift in range predicted for Himalayan Langur in climate change scenario. Glob. Ecol. Conserv. 22, e00926 (2020).
Yang, X. Q., Kushwaha, S. P. S., Saran, S., Xu, J. & Roy, P. S. Maxent modeling for predicting the potential distribution of medicinal plant, Justicia adhatoda L. in Lesser Himalayan foothills. Ecol. Eng. 51, 83–87 (2013).
Radosavljevic, A. & Anderson, R. P. Making better Maxent models of species distributions: Complexity, overfitting and evaluation. J. Biogeogr. 41(4), 629–643 (2014).
Fielding, A. H. & Bell, J. F. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ. Conserv. 24(1), 38–49 (1997).
Allouche, O., Tsoar, A. & Kadmon, R. Assessing the accuracy of species distribution models: Prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol. 43, 1223–1232 (2006).
Amy, G. et al. Genetic landscapes GIS toolbox: Tools to map patterns of genetic divergence and diversity. Mol. Ecol. Resour. 11, 158–161. https://doi.org/10.1111/j.1755-0998.2010.02904 (2010).
Liszka, T. An interpolation method for an irregular net of nodes. Int. J. Numer. Methods Eng. 20, 1599–1612 (1984).
Mateo-Sánchez, M. C. et al. Estimating effective landscape distances and movement corridors: Comparison of habitat and genetic data. Ecosphere https://doi.org/10.1890/ES14-00387.1 (2015).
Roffler, G. H., Schwartz, M. K. & Pilgrim, K. L. Identification of landscape features influencing gene flow: How useful are habitat selection models?. Evol. Appl. 9(6), 805–817 (2016).
McRae, B. H. & Shah, V. B. Circuitscape User’s Guide (The University of California, Santa Barbara, 2009).
Liang, X. U. et al. Isolation and characterization of 16 tetranucleotide microsatellite loci in the red panda (Ailurus fulgens). Mol. Ecol. Notes 7(6), 1012–1014 (2007).
Yang, A. et al. Twenty-five microsatellite loci of Ailurus fulgens identified by genome survey. Sichuan J. Zool. 38(1), 56–61 (2019).
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P. & Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 4(3), 535–538 (2004).
Johnson, P. C. & Haydon, D. T. Maximum-likelihood estimation of allelic dropout and false allele error rates from microsatellite genotypes in the absence of reference data. Genetics 175(2), 827–842 (2007).
Peakall, R. O. D. & Smouse, P. E. GENALEX 6: Genetic analysis in excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6(1), 288–295 (2006).
Rousset, F. Genepop ’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour. 8(1), 103–106 (2008).
Weir, B. S. & Cockerham, C. C. Estimating F-statistics for the analysis of population structure. Evolution 38(6), 1358–1370 (1984).
Bonferroni, C. E., Bonferroni, C., & Bonferroni, C. E. Teoria statistica delle classi e calcolo delle probabilita’ (1936).
Excoffier, L. & Lischer, H. E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10(3), 564–567 (2010).
Vergara, M. et al. Inferring population genetic structure in widely and continuously distributed carnivores: The stone marten (Martes foina) as a case study. PLoS ONE 10(7), e0134257 (2015).
Pritchard, J. K., Wen, W., & Falush, D. Documentation for STRUCTURE software: Version 2 (2003).
Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 14(8), 2611–2620 (2005).
Earl, D. A. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4(2), 359–361 (2012).
Guillot, G., Santos, F. & Estoup, A. Analysing georeferenced population genetics data with Geneland: A new algorithm to deal with null alleles and a friendly graphical user interface. Bioinformatics 24(11), 1406–1407 (2008).
Jombart, T. An introduction to adegenet 1.3-4. R vignette, R-Forge, The R Project for Statistical Computing (2012).
Landguth, E. L., Cushman, S. A., Murphy, M. A. & Luikart, G. Relationships between migration rates and landscape resistance assessed using individual-based simulations. Mol. Ecol. Resour. 10(5), 854–862 (2010).
Piry, S. et al. GENECLASS2: A software for genetic assignment and first-generation migrant detection. J. Hered. 95(6), 536–539 (2004).
Paetkau, D., Slade, R., Burden, M. & Estoup, A. Genetic assignment methods for the direct, real-time estimation of migration rate: A simulation-based exploration of accuracy and power. Mol. Ecol. 13(1), 55–65 (2004).
Acknowledgements
Authors thank Principal Chief Wildlife Warden and other forest officials, of red panda range States for granting the necessary permission to carry out the study. Authors thank Dr. Liang Zhang for providing primer information for standardizing genotyping reactions.
Funding
The study was supported by the DST—INSPIRE FACULTY SCHEME (Grant No. DST/INSPIRE/04/2016/002246) awarded to Dr Mukesh Thakur and funds received from the NMHS-Large Grant project of the Ministry of Environment, Forest and Climate Change, New Delhi (Grant No. NMHS/2017-18/LG09/02).
Author information
Authors and Affiliations
Contributions
S.D., H.K. and S.B. undertook field survey and collected samples. H.K., M.T., T.M., L.K.S. and R.S. participated in analyzing the field collected data. S.D., S.K.S., S.B. and A.G. generated STR data and undertook genetic analysis in the mentorship of M.T. M.T. and L.K.S. conceptualized the idea and H.K., S.D., S.K.S. and S.B. wrote the primary draft of the manuscript. M.T., L.K.S. and R.S. edited and revised the MS. S.D., H.K., M.T., S.K.S., L.K.S. and R.S. finalized the MS. K.C. supervised the overall activities and provided all the logistic support and administrative approval.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dalui, S., Khatri, H., Singh, S.K. et al. Fine-scale landscape genetics unveiling contemporary asymmetric movement of red panda (Ailurus fulgens) in Kangchenjunga landscape, India. Sci Rep 10, 15446 (2020). https://doi.org/10.1038/s41598-020-72427-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-72427-3
This article is cited by
-
Genetic assessment of captive breeding program of Indian Pangolin: implications for conservation and management
Molecular Biology Reports (2024)
-
Landscape genetics identified conservation priority areas for blue sheep (Pseudois nayaur) in the Indian Trans-Himalayan Region
Scientific Reports (2023)
-
Contextualising Landscape Ecology in Wildlife and Forest Conservation in India: a Review
Current Landscape Ecology Reports (2023)
-
Population genetics of the snow leopards (Panthera uncia) from the Western Himalayas, India
Mammalian Biology (2022)
-
Historical genetic diversity and population structure of wild red pandas (Ailurus fulgens) in Nepal
Mammalian Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.