Abstract
The TabZIP15 gene encoding a 396 amino acid (aa) polypeptide in the fungus Trichoderma asperellum ACCC30536 was cloned and characterised. The protein includes a basic region motif (NR-x2-QR-x2-R) and has a pillar-like structure. The 25 basic region/leucine zipper transcription factors (TFs) identified in the T. asperellum genome were divided into YAP (14 TFs), ATF2 (5), GCN4 (2), Zip1 (2), BRLZ (1) and u1 (1) subfamilies based on conserved domains. T. asperellum was cultured in minimal media (MM) control, C-Hungry and N-Hungry medium (to simulate nutrient competition and interaction with pathogens, respectively), and differential expression analysis showed that 14 TabZIP genes (including TabZIP15) were significantly altered under both conditions; TabZIP23 responded strongly to N-Hungry media and TabZIP24 responded strongly to C-Hungry media. However, only YAP genes TabZIP15, TabZIP12 and TabZIP2 were significantly upregulated under both conditions, and expression levels of TabZIP15 were highest. T. asperellum was also cultured in the presence of five fungal pathogenic toxins, and RT-qPCR results showed that TabZIP15 was significantly upregulated in four of the five toxin stress conditions (MM + Rhizoctonia solani, MM + Fusarium oxysporum, MM + Alternaria alternata and MM + Cytospora chrysosperma).
Similar content being viewed by others
Introduction
Members of the fungal genus Trichoderma are important biological control agents, and their biological control mechanisms have been investigated in detail1,2,3. Species in the Trichoderma genus can readily adapt to their environment, propagates and grow, hence their use worldwide. Genome sequencing programs have targeted Trichoderma4 and the genomes of 16 Trichoderma species have been published (https://mycocosm.jgi.doe.gov/mycocosm/home/), laying the foundation for molecular biological analysis.
Basic region/leucine zipper (bZIP) transcription factors (TFs) are one of the largest and most diverse TF families, and proteins with bZIP domains are present in all eukaryotes5. These proteins possess two distinctive structural features located on a contiguous alpha helix; (1) a basic region of ~ 16 amino acid (aa) residues comprising a nuclear localisation signal followed by an invariant N-x7-R/K motif that is responsible for interacting with DNA and nuclear import, and (2) a heptad repeat of leucines or other bulky hydrophobic amino acids positioned exactly nine amino acids from the C-terminus that creates an amphipathic helix which regulates dimerisation5. The DNA binding and dimerisation mechanism in fungi has been studied in Saccharomyces cerevisiae, and the results revealed that Met4 and Met28 form a heteromeric complex with a centromere binding factor to activate transcription6,7.
The mechanism of bZIP TFs has been analysed in various fungus species8. Recent studies showed that members of the bZIP TF family play diverse regulatory roles in filamentous fungi. Eight bZIP TFs in Ustilaginoidea virens are involved in stress tolerance and pathogenicity9. In Aspergillus spp., bZIP TFs including AtfA, NapA, AflR, RsmA and Apyap1 have been demonstrated to be the key factors responding to oxidative, osmotic, environmental and drug stresses10,11,12,13. In other fungi, such as Botrytis cinerea, Neurospora crassa, Fusarium graminearum and Magnaporthe oryzae, bZIP TFs are known to be involved in responding to oxidative stress and pathogenicity14,15. These findings suggest that bZIP TFs play an important role in stress responses in several fungi, and a similar regulatory function can be inferred in other fungal species. However, most of the research on fungal bZIP proteins has focused on pathogens, which bZIP proteins in biological control agents such as Trichoderma spp. have received little attention. Phytopathogen toxins are metabolites in pathogens that can harm plants and other fungi. To better understand the detoxification mechanism of Trichoderma that is triggered in response to phytopathogen toxins, the roles of bZIP TFs should be investigated under different stress conditions.
Herein, the TabZIP15 TF gene of Trichoderma asperellum ACCC30536 was cloned, and characterised alongside another 24 similar bZIP TFs in the Joint Genome Institute (JGI) database. The basic biochemical characteristics of these bZIP TFs were investigated, and expression levels were analysed based on RT-PCR data obtained for T. asperellum under three conditions at 72 h. Furthermore, transcription levels of TabZIP15 were measured by RT-qPCR following exposure to toxins produced by five pathogenic fungi (Rhizoctonia solani, Fusarium oxysporum, Sclerotinia sclerotiorum, Alternaria alternata, and Cytospora chrysosperma). The results provide theoretical support for the analysis and development of TFs from T. asperellum.
Materials and methods
Strains and materials
T. asperellum ACCC30536 was obtained from the Agricultural Culture Collection of China. The five phytopathogens were R. solani, F. oxysporum, S. sclerotiorum, A. alternata and C. chrysosperma, and they were stored at the Laboratory of Forestry Protection, Northeast Forestry University, Harbin, China. T. asperellum was cultured on potato dextrose agar (PDA) slant culture medium at 28 °C for 7 days and stored at 4 °C. A. alternata, S. sclerotiorum, R. solani, C. chrysosperma and F. oxysporum were inoculated in 200 mL of 1/4, 1, 1/2, 1 and 1/2 strength PD medium and cultured in shake flasks for 10 days at 28 °C with shaking at 200 rpm. The medium was filtered using 0.2 μm filters (Pall Corporation, MI, USA) to remove spores, and the filtered fermentation liquid from each pathogen was combined, mixed, and stored in tubes at − 20 °C.
Cloning and analysis of TabZIP15 transcription factor from T. asperellum
Primers for cloning TabZIP15 (TabZIP15-1, 5′-GCGAATCCGGATGAGTTCAC-3′; TabZIP15-2, 5′-TCGCCGCACCCTATACTTTT-3′) were designed using Primer Premier 6.0 software (PREMIER Biosoft, Vancouver, Canada)16. Extraction of T. asperellum DNA was performed as described previously17, and thermal cycling for PCR was performed at 94 °C for 5 min, followed by 35 cycles at 94 °C for 25 s, 56 °C for 30 s, and 72 °C for 30 s, and followed by 72 °C for 7 min. The PCR product was purified and ligated into the pGEM-T vector (A3600; Promega, Madison, USA) and sequenced (Shanghai Sangon Co., Shanghai, China). Conserved domain prediction and identification of other homologous proteins was performed using the Blastp tool18 from the National Center for Biotechnology Information (NCBI). Multiple sequence alignment was conducted using the Clustal X program (https://www.ebi.ac.uk/Tools/clustalw2/)19, and three-dimensional structure prediction was carried out by SWISS-MODEL ( https://swissmodel.expasy.org/)20.
Characteristic analysis and construction of phylogenetic tree of 25 bZIPs in the T. asperellum genome
Full-length T. asperellum bZIP protein (TabZIP protein) sequences were obtained from the T. asperellum CBS 433.97 v1.0 JGI database (https://genome.jgi.doe.gov/)21, and conserved domains were predicted using the NCBI tool Blastp. Proteins without conserved bZIP domains were omitted. The molecular weight (MW) and isoelectric point (pI) of each bZIP protein was calculated using ExPASy (https://www.expasy.org/)22, and gene structural information for exons and introns was obtained from the JGI database (https://genome.jgi.doe.gov/)21 and analysed by the Gene Structure Display Server 2.0 website (https://gsds.cbi.pku.edu.cn/index.php)23. A phylogenetic tree was constructed via the maximum likelihood method with 1,000 bootstrap replicates using the MEGA 7.0 program24. Motifs were searched using the MEME suite (http://meme-suite.org/tools/meme)25 with the site distribution set as ‘zoops’, and the number of motifs set as ‘3’. Conserved domains and motifs were analysed by the MEME suite and TBtools26.
Differential expression of bZIP family genes and expression of TabZIP15 genes in the presence of five pathogenic toxins
Minimal media (MM) contained 15 g/L NaH2PO4, 5 g/L (NH4)2SO4, 600 mg/L CaCl2·2H2O, 600 mg/L MgSO4·7H2O, 5 mg/L FeSO4, 2 mg/L CoCl2, 1.6 mg/L MnSO4, 1.4 mg/L ZnSO4 and 0.1% (w/v) glucose. MM + A comprised MM plus 5% (v/v) fermentation liquid from A. alternata. MM + S comprised MM plus 5% (v/v) fermentation liquid from S. sclerotiorum. MM + R comprised MM plus 5% (v/v) fermentation liquid from R. solani. MM + C comprised MM plus 5% (v/v) fermentation liquid from C. chrysosperma. MM + F comprised MM plus 5% (v/v) fermentation liquid from F. oxysporum, C-Hungry was MM without a carbon source, and N-hungry was MM without a nitrogen source. Differential expression of 25 bZIP family genes was analysed using T. asperellum ACCC30536 transcriptome data obtained previously27, and the heatmap was drawn by TBtools26. Expression of TabZIP15 genes was analysed by RT-qPCR under five pathogenic toxin stress conditions. Spores from T. asperellum were inoculated into 200 mL 1/4 strength PD broth medium at a final concentration 1 × 104 spores/mL and cultured at 28 °C with continuous shaking at 200 rpm for 48 h. The resulting mycelia were filtered, washed, transferred to MM for 2 h, then transferred into medium containing different pathogenic toxins for 72 h, three replicates were set. Mycelia were collected at 0, 4, 8, 12, 24, 48, and 72 h. At each time point, the biomass was calculated (averaged from three replicates), and 25 mL mycelium cultures were collected and stored at − 80 °C. Total RNA was extracted from mycelia28 using TRIzol reagent (Invitrogen, Carlsbad, USA), digested with DNaseI (Promega, Madison, USA), and reverse-transcribed into cDNA using a PrimeScript RT Kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions.
Expression levels of TabZIP15 in T. asperellum were measured by RT-qPCR, and calculated according to the 2−ΔΔCt method29 using cDNA as template, with actin, α-tubulin and β-tubulin as reference genes. Three RT-qPCR replicates were performed per cDNA sample. Primers for RT-qPCR (Table 1) were designed using Primer Premier 6.0 software (PREMIER Biosoft, Vancouver, Canada)16, and figures were drawn using Origin 8.0 (OriginLab Corp, Northampton, Massachusetts, USA)30.
Statistical analysis
All experimental data were subjected to analysis of variance (ANOVA) and independent sample t tests using SPSS19.031. Statistical significance of differences between groups was determined by Turkey’s multiple range tests (p < 0.05).
Results
Characteristics of 25 TabZIP TFs in the T. asperellum genome
The 25 TabZIP genes encode proteins ranging from 177 to 753 aa in length, with an average length of 408 aa. The molecular weight of TabZIP proteins ranges from 19.63 to 80.93 kDa, with an average of 44.7 kDa. The pI ranges from 4.66 to 10.05, and the average pI is 6.65. TabZIP TF genes in T. asperellum are distributed on scaffold 1 to scaffold 41, and some chromosomes have more than one TabZIP TF gene (four, three and three TFs are in scaffold 5, scaffold 2 and scaffold 9, respectively; Table 2). The number of exons in the 25 TabZIP genes ranges from one to six, with most having two or three exons. Eight genes possess untranslated exon regions (Fig. 1).
Phylogenetic analysis, conserved domains, and motif prediction
Phylogenetic relationships between the 25 TabZIP proteins were explored, and sequences were divided into seven clades (Fig. 2a). The number of conserved domains in TabZIP TFs ranges from one to four (Fig. 2b), with seven bZIP TFs (TabZIP10, TabZIP17, TabZIP18, TabZIP22, TabZIP23, TabZIP24 and TabZIP25) having more than one conserved domain. Fourteen bZIP TFs have a bZIP-YAP conserved domain, five have a bZIP-ATF2 conserved domain, two have a bZIP-GCN4 conserved domain, two have a bZIP-Zip1 conserved domain, one has a BRLZ conserved domain, and one has a bZIP-u1 conserved domain. The 25 bZIP TFs were searched for three motifs (Fig. 2c), Motif 1 (E[KR][RK][RK][AI][QR]NR[VA]A[QA]R[KA][FYC]R[QE][RK][KR]KER[LI][EK][ED]LEEE[LV]); Motif 2 ([LM][ES]A[EQ]NN[NQ]LRMEYXQLREE[IV]GQ[LV]K[NS][DL]L[LM][AE]HTECNDDNINXWI); and Motif 3 (L[ED]ER[FLM][ET][FGH][IM][LMR][ELV][CKN][ATV][LRS][AEV]AGF[DHK][SD][IF]D[ADS][ML][AIV][KLS][QA]YY[TC][AEG][DNT][FL][GRS][EHY][DA]S[VF][IL][AHS][NQW][EA]Q[RK][LNR]SR[HMN]). We found that 10 bZIP TFs have two motifs, but interestingly, none of the bZIP TFs have three motifs and bZIP TFs in the same clade have similar numbers and types of motifs (Fig. 2).
Phylogenetic relationships, conserved domain prediction, and motif composition of the 25 bZIP transcription factors (TFs) indentified in T. asperellum (TabZIP TFs). (a) Phylogenetic relationships among the 25 TabZIP TF amino acids; (b) Conserved domain prediction using the NCBI database. The x-axis shows amino acid numbers; (c) Motif prediction. The x-axis shows amino acid numbers.
Differential expression of 25 TabZIPs in the three transcriptomes
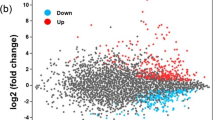
Expression of the 25 TabZIP genes in 3 transcriptomes was measured (Fig. 3a; Supplementary Table S1). Interestingly, 13, 14 and 11 TabZIP genes were highly expressed (Log2RPKM > 6, Reads Per Kilobase per Million mapped reads) in MM, C-Hungry and N-Hungry media, respectively. Genes were clustered based on their expression, and TabZIP15, TabZIP13 and TabZIP23 were highly expressed in all three conditions, while TabZIP18, TabZIP24 and TabZIP 25 were highly expressed in MM and N-Hungry media, and TabZIP6 and TabZIP20 were highly expressed in MM and C-Hungry media.
In the differential expression analysis, a two-fold (Log2 value > 1) change in expression level relative to MM was considered significant. The results showed that six TabZIP genes were significantly upregulated and six were significantly downregulated in C-Hungry media (Fig. 3b). Meanwhile, four TabZIP genes were significantly upregulated and 11 were significantly downregulated in N-Hungry media (Fig. 3b). Additionally, 14 genes were significantly altered in N-Hungry and C-Hungry media compared with MM media, but only three genes (TabZIP15, TabZIP12 and TabZIP2) were significantly upregulated in both N-Hungry and C-Hungry media, while TabZIP6, TabZIP11, TabZIP20 and TabZIP25 were the only genes significantly downregulated in these two conditions. During the stress response process, upregulated genes may act as positive response factors. Thus, we focused on TabZIP15, TabZIP12 and TabZIP2, and TabZIP15 displayed the highest expression, hence this gene was subjected to further analysis.
Characteristics of TabZip15 TF in T. asperellum
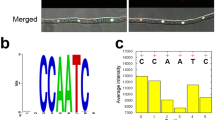
The TabZIP15 DNA sequence (2062 bp) includes six exons and five introns. The TabZIP15 cDNA (1,188 bp) encodes a 396 aa polypeptide with a calculated molecular weight of 45.05 kDa, and a pI of 5.54. TabZIP15 is a member of the bZIP-YAP family of TFs (Fig. 4a). Multiple sequence alignment revealed a highly conserved basic region motif (NR-x2-QR-x2-R), and three conserved hydrophobic amino acids in the sequence following the basic region (Fig. 4b). Three-dimensional structure prediction showed that the TabZIP15 protein is a cylindrical bundle of α-helices (Fig. 4c).
Characteristics of bZIP transcription factor TabZIP15. (a) Conserved domains in TabZIP15 predicted by NCBI Blastp. (b) Multiple sequence alignment of TabZIP15 and another 17 TFs. Except for TabZIP15, all names are NCBI accession numbers. Asterisk (*) represent identity, colon (:) represent high similarity, and periods (.) represent low similarity. Basic regions are marked by blue squares and hydrophobic amino acids are marked by red circles. The sequence logo was created based on the sequences of the basic region, and the height of letters in the logo represent the sequence conservation at that position. (c) Predicted three-dimensional structure of the TabZIP15 protein.
Differential expression of TabZIP15 under five fungal toxin stress conditions
Expression of TabZIP15 was investigated in the presence of five fungal pathogenic toxins (Fig. 5). In MM media, expression of TabZIP15 was downregulated at 4 h and 8 h, then upregulated at 12 h, and the peak expression level (25.54) occurred at 24 h. Expression of TabZIP15 was upregulated at 4 h in MM + A, at 4 h in MM + C, at 8 h in MM + F, at 4 h in MM + R, and at 12 h in MM + S, with peak expression levels at 48 h (26.55-fold), 48 h (26.98-fold), 48 h (24.74-fold), 12 h (26.98-fold) and 24 h (24.42-fold), respectively. Analysis of differential expression (relative to controls) showed that TabZIP15 was highly expressed when T. asperellum was exposed to toxins from A. alternata, C. chrysosperma, F. oxysporum and R. solani, but expression in MM + S media did not differ significantly from that in MM media.
Expression of the TabZIP15 gene in the presence of five fungal pathogenic toxins. Differential expression was measured between treatments, and minimal media (MM) alone served as a control. Error bars represent standard deviation. In each condition, different letters denote a statistically significant difference according to one-way analysis of variance (ANOVA; p < 0.05). Independent-sample t tests were performed on pairs of samples at the same timepoint (*p < 0.05; **p < 0.01, n = 3). (a) MM + A (Alternaria alternata). (b) MM + C (Cytospora chrysosperma). (c) MM + F (Fusarium oxysporum). (d) MM + R (Rhizoctonia solani). (e) MM + S (Sclerotinia sclerotiorum).
Discussion
Members of the Trichoderma genus have been widely used as biological control agents, and the biocontrol mechanism has been investigated3. bZIP proteins regulate various biological processes10,11,12,13 and are present in all eukaryotes5. Thus, bZIP TFs may regulate biological control processes in T. asperellum. Herein, 25 TabZIP TFs were identified in T. asperellum, and multiple sequence alignment revealed relatively low sequence similarity (data not shown). Seventeen sequences sharing homology with TabZIP15 were identified using Blastp (Fig. 4), and multiple sequence alignment showed that these sequences share short conserved regions (~ 30 aa), as expected for bZIP proteins that typically include 18 conserved residues followed by a leucine zipper32,33. Thus, the sequences of bZIP TFs are diverse, and this may explain why they perform diverse functions33. The bZIP TFs in T. asperellum were divided into seven clades, with TabZIP15, TabZIP9 and TabZIP10 in the same clade (Fig. 2a). The close phylogenetic relationship indicates that these three proteins may perform similar functions and share similar expression profiles. Consistently, motif prediction showed that TabZIP15, TabZIP19 and TabZIP21 are the only proteins possessing motif 3, and they share a similar motif arrangement (Fig. 2c). This further suggests that TabZIP15, TabZIP19 and TabZIP21 may perform similar functions or share similar expression profiles.
Next, we explored the expression of these 25 TabZIP genes in C-Hungry and N-Hungry media using an RNA sequencing (RNA-Seq), which simulated Trichoderma nutrient competition and interaction with the pathogens27, TabZIP19 and TabZIP21 have similar motif arrangements to TabZIP15, hence they may be expected to share similar expression modes, but the RNA-Seq data showed that they did not have similar expression modes. Similarly, TabZIP9 and TabZIP10 are in the same clade as TabZIP15, and they share similar phylogenetic relationships, hence they may be expected to share similar expression modes, but again, RNA-Seq data showed that they did not have similar expression modes (Fig. 3).
The YAP protein and its homologs have been analysed in several fungal pathogens, and they all perform similar functions in stress response9,15,34. In T. asperellum, 14 of the 25 bZIP TFs have a conserved bZIP-YAP domain (Fig. 2b). The presence of a large number of YAP bZIPs may explain the robust ability of T. asperellum to adapt to environmental changes, but their exact functions need to be further investigated. In Claviceps purpurea, the transcription factor CPTF1, a homolog of ATF in mammals, is related to oxygen stress responses35. In T. asperellum, five bZIP proteins (TabZIP4, TabZIP7, TabZIP8, TabZIP13 and TabZIP18) have a conserved bZIP-ATF2 domain, and they may perform similar functions, but this should be further investigated (Fig. 2b). The TabZIP18 protein includes Aft1-HRR, Aft1-HRA and Aft1-OSA domains, and is a homolog of UvbZIP89, AtfB36 and Aft137. Aft1 proteins are reportedly involved in the regulation of sexual development and various stress responses, and they activate meiotic recombination, which suggests that TabZIP18 may also play a key role in these processes.
Our transcriptome data showed that 16 of the 25 bZIP TF genes were significantly differentially expressed in C-Hungry medium (simulating competition with pathogens) or N-Hungry medium (simulating interaction with pathogens; Fig. 2b). Of these, 15 genes appear to be involved in nutrient competition and 15 appear to be related to interactions with pathogens. The 15 nutrient competition-related genes include nine YAP, four ATF2, one Zip1 and one u1 genes, while the 15 pathogen interaction-related genes include eight YAP, four ATF2, one Zip1, one u1 and one BRLZ genes. These results suggest that YAP and ATF2 proteins in T. asperellum may be involved in various stress responses, similar to other homologs9,15,34,35,36,37. One BRLZ gene was significantly downregulated in N-Hungry media (interaction with pathogens), and a homolog of this protein is reportedly involved in fungal development and differentiation38. Meanwhile one u1 and one Zip1 were significantly downregulated in both N-Hungry and C-Hungry media. A homolog of Zip1 was reportedly induced under stress conditions39, but u1 proteins have not been investigated, and further studies are needed. Thirteen genes were highly expressed in MM media (Log2RPKM > 6), and they may be involved in normal physiological regulation (Fig. 2a). Three YAP genes (TabZIP15, TabZIP12 and TabZIP2) were significantly upregulated in both C-Hungry and N-Hungry media (simulating nutrient competition and interactions with pathogens, respectively), which suggests that they may be important for resistance against pathogens. TabZIP12 and TabZIP2 were both significantly upregulated. TabZIP12 expression levels were upregulated 22.31- and 21.85-fold in C-Hungry and N-Hungry media respectively, while TabZIP2 expression levels were upregulated 22.14- and 21.58-fold in C-Hungry and N-Hungry media respectively. However, the RPKM value for TabZIP12 in MM and C-Hungry media was 45 and 224, respectively; compared with 22 and 97 for TabZIP2. The RPKM value for TabZIP12 in MM and N-Hungry media was 45 and 163, respectively, compared with 22 and 66 for TabZIP2. Thus, the RPKM values for both genes were not high, and can be easily influenced by experimental error40, but fold-changes in expression were relatively large, hence further investigation is clearly needed.
The most highly expressed gene (TabZIP15) was chosen for further analysis, and its expression was measured in the presence of five pathogenic toxins by RT-qPCR. The results confirmed our hypothesis (Fig. 5); TabZIP15 was strongly upregulated in four of the five pathogenic toxin conditions, indicating an important function in resistance to pathogens. Interestingly, TabZIP15 was also highly expressed in MM media, which suggests that it may be involved in other functions beyond resistance to pathogens.
Herein, we identified 25 TabZIP genes in T. asperellum, which were divided into seven clades based on phylogenetic analysis, and six conserved bZIP domains were detected in the aa sequences. Analysis of the transcriptome data showed that expression of 16 TabZIP genes was significantly altered under C-Hungry and N-Hungry media conditions. The TabZIP15 gene was identified as a potential biocontrol factor, and its characteristics were further analysed. The RT-qPCR results showed that the YAP gene TabZIP15 was significantly upregulated in the presence of four of the five pathogenic toxins (those from A. alternata, C. chrysosperma, F. oxysporum and R. solani). Our results reveal some of the properties of bZIP TFs in T. asperellum, identify potential biological control-related bZIP TFs, and will guide further research in this area.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files), if there is any other request, the datasets are available from the corresponding author on reasonable request.
References
Lea, A. et al. The Gpr1-regulated Sur7 family protein Sfp2 is required for hyphal growth and cell wall stability in the mycoparasite Trichoderma atroviride. Sci. Rep. 8, 12064. https://doi.org/10.1038/s41598-018-30500-y (2018).
Shoresh, M., Yedidia, I. & Chet, I. Involvement of jasmonic acid/ethylene signaling pathway in the systemic resistance induced in cucumber by Trichoderma asperellum T203. Phytopathology 95(1), 76–84. https://doi.org/10.1094/PHYTO-95-0076 (2005).
An-Le, H. E. et al. Soil application of Trichoderma asperellum GDFS1009 granules promotes growth and resistance to Fusarium graminearum in maize. J. Agric. Sci. 018(003), 599–606. https://doi.org/10.1016/S2095-3119(18)62089-1 (2019).
Mukherjee, P. K., Horwitz, B. A., Herreraestrella, A., Schmoll, M. & Kenerley, C. M. Trichoderma research in the genome era. Annu. Rev. Phytopathol. 51(51), 105–129. https://doi.org/10.1146/annurev-phyto-082712-102353 (2013).
Sornaraj, P., Luang, S., Lopato, S. & Hrmova, M. Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: a molecular model of a wheat bZIP factor and implications of its structure in function. Biochem. Biophys. Acta 1860(1), 46–56. https://doi.org/10.1016/j.bbagen.2015.10.014 (2016).
Thomas, D., Jacquemin, I. & Surdin-Kerjan, Y. MET4, a leucine zipper protein, and centromere-binding factor 1 are both required for transcriptional activation of sulfur metabolism in Saccharomyces cerevisiae. Mol. Cell. Biol. 12(4), 1719–1727. https://doi.org/10.1128/mcb.12.4.1719 (1992).
Kuras, L., Cherest, H., Surdin Kerjan, Y. & Thomas, D. A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J. 15(10), 2519–2529. https://doi.org/10.1002/j.1460-2075.1996.tb00609.x (1996).
Pi Syk, S. et al. The Aspergillus nidulans metZ gene encodes a transcription factor involved in regulation of sulfur metabolism in this fungus and other Eurotiales. Curr. Genet. 61(2), 115–125. https://doi.org/10.1007/s00294-014-0459-5 (2015).
Yin, W., Cui, P., Wei, W., Lin, Y. & Luo, C. Genome-wide identification and analysis of the basic leucine zipper (bZIP) transcription factor gene family in Ustilaginoidea virens. Genome https://doi.org/10.1139/gen-2017-0089 (2017).
Kazutoshi, S. et al. Aspergillus oryzae atfA controls conidial germination and stress tolerance. Fungal Genet. Biol. 46(12), 887–897. https://doi.org/10.1016/j.fgb.2009.09.004 (2009).
Yin, W. et al. An Aspergillus nidulans bZIP response pathway hardwired for defensive secondary metabolism operates through aflR. Mol. Microbiol. 83(5), 1024–1034. https://doi.org/10.1111/j.1365-2958.2012.07986.x (2012).
Hong, S. Y., Roze, L. V., Wee, J. & Linz, J. E. Evidence that a transcription factor regulatory network coordinates oxidative stress response and secondary metabolism in aspergilli. Microbiologyopen 2(1), 144–160. https://doi.org/10.1002/mbo3.63 (2013).
Jin, W. B. et al. Illumina identification of RsrA, a conserved C2H2 transcription factor coordinating the NapA mediated oxidative stress signaling pathway in Aspergillus. BMC Genom. 15(1), 1011. https://doi.org/10.1186/1471-2164-15-1011 (2014).
Wang, X. et al. The bZIP transcription factor PfZipA regulates secondary metabolism and oxidative stress response in the plant endophytic fungus Pestalotiopsis fici. Fungal Genet. Biol. 81, 221–228. https://doi.org/10.1016/j.fgb.2015.03.010 (2015).
Montibus, M. et al. The bZIP transcription factor Fgap1 mediates oxidative stress response and trichothecene biosynthesis but not virulence in Fusarium graminearum. PLoS ONE 8(12), e83377. https://doi.org/10.1371/journal.pone.0083377 (2013).
Johnson, M. et al. NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9. https://doi.org/10.1093/nar/gkn201 (2008).
Sambrook, J., Fritsch E. F. & Maniatis, T. Molecular Cloning: A Laboratory Manual (No. Ed 2) 721–722 (Cold Spring Harbor Laboratory Press, 1989).
Madeira, F. et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. https://doi.org/10.1093/nar/gkz268 (2019).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. https://doi.org/10.1093/nar/gky427 (2018).
Nordberg, H. et al. The genome portal of the department of energy joint genome institute: 2014 updates. Nucleic Acids Res. 42, D26–D31. https://doi.org/10.1093/nar/gkt1069 (2014).
Artimo, P. et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, W597–W603. https://doi.org/10.1093/nar/gks400 (2012).
Hu, B. et al. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31(8), 1296–1297. https://doi.org/10.1093/bioinformatics/btu817 (2015).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. https://doi.org/10.1093/molbev/msw054 (2016).
Bailey, T. L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. https://doi.org/10.1093/nar/gkp335 (2009).
Singh, V. K., Mangalam, A. K., Dwivedi, S. & Naik, S. Primer Premier: program for design of degenerate primers from a protein sequence. Biotechniques 24, 318–319. https://doi.org/10.1271/bbb.62.412 (1998).
Chen, C., Rui, X., Hao, C. & He, Y. TBtools, a Toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv https://doi.org/10.1101/289660 (2018).
Ji, S., Liu, Z., Liu, B. & Wang, Y. Comparative analysis of biocontrol agent Trichoderma asperellum ACCC30536 transcriptome during its interaction with Populus davidiana × P. alba var. Pyramidalis. Microbiol. Res. 227, 126294. https://doi.org/10.1016/j.micres.2019.126294 (2019).
Argumedo-Delira, R., González-Mendoza, D. & Alarcón, A. A rapid and versatile method for the isolation of total rna from the filamentous fungus Trichoderma Sp. Ann. Microbiol. 58(4), 761–763. https://doi.org/10.1007/BF03175587 (2008).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3(6), 1101–1108. https://doi.org/10.1038/nprot.2008.73 (2008).
May, R. A. & Stevenson, K. J. Software review of origin 8. J. Am. Chem. Soc. 131, 872. https://doi.org/10.1021/ja809638x (2009).
Nie, N. H., Hull, C. H., Jenkins J. G., Steinbrenner K. & Bent D. H. SPSS (Statistical Package For The Social Sciences) (No. Ed 2) 187–196 (Mcgraw-hill Book Company, 2003).
Banerjee, A. & Roychoudhury, A. Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 254, 3–16. https://doi.org/10.1007/s00709-015-0920-4 (2017).
Wang, L., Zhu, J., Li, X., Wang, S. & Wu, J. Salt and drought stress and ABA responses related to bZIP genes from V. radiata and V. angularis. Gene 651, 152–160. https://doi.org/10.1016/j.gene.2018.02.005 (2018).
Montibus, M., Pinson-Gadais, L., Richard-Forget, F., Barreau, C. & Ponts, N. Coupling of transcriptional response to oxidative stress and secondary metabolism regulation in filamentous fungi. Crit. Rev. Microbiol. https://doi.org/10.3109/1040841X.2013.829416 (2015).
Nathues, E. et al. CPTF1, a CREB-like transcription factor, is involved in the oxidative stress response in the phytopathogen Claviceps purpurea and modulates ROS level in its host Secale cereale. Mol. Plant Microbe Interact. 17(4), 383–393. https://doi.org/10.1094/mpmi.2004.17.4.383 (2004).
Wee, J. et al. The fungal bZIP transcription factor Atfb controls virulence-associated processes in Aspergillus parasiticus. Toxins 9(9), 287. https://doi.org/10.3390/toxins9090287 (2017).
Gao, J., Davidson, M. K. & Wahls, W. P. Distinct regions of ATF/CREB proteins Atf1 and Pcr1 control recombination hotspot ade6–M26 and the osmotic stress response. Nucleic Acids Res. 36(9), 2838–2851. https://doi.org/10.1093/nar/gkn037 (2008).
Thompson, C. R. L., Fu, Q., Buhay, C., Kay, R. R. & Shaulsky, G. A bZIP/bRLZ transcription factor required for DIF signaling in Dictyostelium. Development 131(3), 513–523. https://doi.org/10.1242/dev.00939 (2004).
Guo, L., Ghassemian, M., Komives, E. A. & Russell, P. Cadmium-induced proteome remodeling regulated by Spc1/Sty1 and Zip1 in fission yeast. Toxicol. Sci. 129(1), 200–212. https://doi.org/10.1093/toxsci/kfs179 (2014).
Zhang, W. Z. et al. Two-factor ANOVA of SSH and RNA-seq analysis reveal development-associated Pi-starvation genes in oilseed rape. Planta 250(4), 1073–1088. https://doi.org/10.1007/s00425-019-03201-7 (2019).
Acknowledgements
The authors thank Shida Ji, Huifang Zhang, Ping Zhang, Lei Gao and Jian Diao for providing advice on this article.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC: 31870627), the National High Technology Research and Development Program (the 13th Five-Year Plan Program) (Grant Number 2016YFC0501505) and Start-up Funds of Talent Introduction of Shenyang Agricultural University.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Topic selection was performed by Y.W. Material preparation, data collection and analysis were performed by Z.Y. and Y.Z. Experimental supervision was provided by Z.W., Y.W. and Z.L. Fund was provided by Z.L.. The first draft of the manuscript was written by Z.Y., and all authors commented on subsequent versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yu, Z., Wang, Z., Zhang, Y. et al. Analysis of TabZIP15 transcription factor from Trichoderma asperellum ACCC30536 and its function under pathogenic toxin stress. Sci Rep 10, 15084 (2020). https://doi.org/10.1038/s41598-020-72226-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-72226-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.