Abstract
Epidermodysplasia verruciformis (EV) is a genodermatosis characterized by the inability of keratinocytes to control cutaneous β-HPV infection and a high risk for non-melanoma skin cancer (NMSC). Bi-allelic loss of function variants in TMC6, TMC8, and CIB1 predispose to EV. The correlation between these proteins and β-HPV infection is unclear. Its elucidation will advance the understanding of HPV control in human keratinocytes and development of NMSC. We generated a cell culture model by CRISPR/Cas9-mediated deletion of CIB1 to study the function of CIB1 in keratinocytes. Nine CIB1 knockout and nine mock control clones were generated originating from a human keratinocyte line. We observed small changes in gene expression as a result of CIB1 knockout, which is consistent with the clearly defined phenotype of EV patients. This suggests that the function of human CIB1 in keratinocytes is limited and involves the restriction of β-HPV. The presented model is useful to investigate CIB1 interaction with β-HPV in future studies.
Similar content being viewed by others
Introduction
Epidermodysplasia verruciformis (EV) (OMIM 226400) is a primary immunodeficiency to cutaneous human papillomaviruses of the genus β-HPV1,2,3. Furthermore, patients with clinical symptoms of EV associated with inborn or acquired immunodeficiency by T-cell defects have been reported. This clinical picture is referred to as “atypical EV”1. Patients with congenital EV exhibit plane warts with onset in early childhood and have an increased risk of developing non-melanoma skin cancer (NMSC)3,4,5. More than 500 patients have been described to date6. Bi-allelic pathogenic variants in TMC6 or TMC8 (also known as EVER1 and EVER2, respectively) have been identified in 22 families (Table 1). Recently, we identified homozygous loss-of-function variations in CIB1 in six EV-families7. Majority of the bi-allelic loss-of-function variants in these three genes are either nonsense, frameshift, or splice site variants. A homozygous three amino acid in-frame duplication in TMC8 was recently described in one family8. Loss of protein expression has been shown in patients with a frameshift in TMC89 and in patients with CIB1 variants7.
CIB1, TMC6, and TMC8 form a complex and CIB1 has been shown to be degraded in case of one missing interaction partner7. The CIB1-TMC6-TMC8 complex acts as restriction factor for β-HPV via an unknown mechanism. Effects based on known functions of these three proteins were disproven in patients’ keratinocytes7. CIB1 has been shown to influence cell migration and adhesion in animal models10. These processes normally work in CIB1 deficient patients’ keratinocytes7. It has been hypothesized that TMC6 and TMC8 influence intracellular zinc levels, which control activity of transcription factors such as AP-1, which is important for the HPV life cycle11,12. However, changes in intracellular zinc homeostasis were not observed in TMC6, TMC8, or CIB1 deficient keratinocytes derived from EV patients7. Finally, TMC8 was hypothesized to switch the cellular response to TNF-α from pro-apoptotic to pro-survival combined with NF-κB activation13,14. Such changes could not be confirmed in keratinocytes from EV patients or in in vitro studies7. Additionally, no influences on the transcriptome could be identified in the patients' cells by RNA-seq or qPCR. Due to the heterogeneity of patients' samples, small differences may be lost in the background. The presented keratinocyte model aimed to overcome this diversity and to identify effects of CIB1 deficiency on the expression of downstream genes.
Materials and methods
Cell culture and validation of keratinocyte line
The human keratinocyte line NKc2115 was maintained in CnT-PR medium (CELLnTEC Advanced Cell Systems, Bern, Switzerland). The mouse fibroblast line 3T316 was maintained in DMEM (Lonza, Basel, Switzerland) supplemented with 10% FCS (Thermo Fisher Scientific, Waltham, MA, USA). All cell lines were cultured at 37 °C in a 5% CO2 atmosphere.
CIB1, TMC6, and TMC8 genes of NKc21 were analysed by Sanger sequencing after PCR amplification from isolated gDNA. Sequences of primers for TMC6 and TMC817 were kindly provided by Gérard Orth and Michel Favre (Department of Virology, Institute Pasteur, Paris, France). Primer sequences for CIB1 are listed in Table S1. Expression of TMC6, TMC8, and CIB1 was verified by RT-PCR on RNA isolated from NKc21 using primers CIB1_9, CIB1_10, TMC6_1, TMC6_2, TMC8_1, and TMC8_2 (Table S1). Additionally, gDNA was analyzed by an Illumina HumanOmniExpressExome-8 BeadChip v1.3 SNP array. SNP array data was visualized using GenomeStudio and copy number analysis performed using CNV-Partition (both Illumina, San Diego, CA, USA).
Transfection of keratinocytes
Transfection of keratinocytes was performed using Xfect transfection reagent (Takara Bio, Kusatsu, Japan) as previously described18. In short, 1 day after seeding 70,000 cells per well in 6-well plates, the medium was replaced by fresh medium and a mixture of 330 ng of each CRISPR/Cas9 plasmid (total of 660 ng DNA), 0.2 µl Xfect polymer, and 100 µl Xfect buffer was added to the keratinocytes after incubation at room temperature for 10 min. After four hours at 37 °C, the medium was replaced by 3 ml fresh CnT-PR.
Generation of active CRISPR plasmids
Sequences for sgRNAs were chosen based on the minimal off-target activity predicted by an online tool (https://crispr.mit.edu)19,20. Plasmids expressing Cas9 and sgRNA were generated by inserting custom sgRNA sequences into pSpCas9(BB)-2A-GFP (Addgene, Cambridge, MA, USA, #48138) as described20. Plasmids and sgRNA used in this study are listed in Table S2. After treatment with ATP-dependent DNase (Lucigen, Middleton, WI, USA), DH5α chemo-competent E. coli were transformed and selected in the presence of ampicillin (Thermo Fisher Scientific). Correct insertion of the sgRNA was verified by sequencing of isolated plasmids (Nucleospin plasmid kit, Macherey–Nagel, Düren, Germany) using U6 primer.
Validation of CRISPR/Cas9 plasmids
Deletion activity of five combinations of sgRNAs with target sites on both sides of CIB1 was tested. For this, NKc21 were transfected with plasmids encoding Cas9 and the tested sgRNAs. Six days after transfection, gDNA was isolated and the target region was amplified using primer CIB1_1 and CIB1_8 (Table S1) located outside of the intended deletion (Fig. 1A). This PCR yields a 4,130 bp wild-type amplicon and in case of successful deletion smaller additional amplicons (in the size range of 500–700 bp).
Nine keratinocyte clones showed a knockout of CIB1 without any detectable transcript or protein product. (A) Primers CIB1_1–CIB1_8 were used to amplify regions of CIB1 or the whole gene. Locations of primers and the resulting amplicons (blue lines and arrows) respective to CIB1 and target sites of sgRNAs (red arrows) are depicted. (B) Flowchart shows the screening process to identify successfully edited clones. Nine clones out of 73 had a complete homozygous CIB1 deletion. (C) Expression of CIB1 mRNA in various clones and untreated NKc21 was checked by RT-PCR. LEMD3 was used as a positive control. Nine clones (bold numbers) showed no amplification of CIB1 transcript. The uncropped gel pictures can be found in Figure S3. (D) Presence of CIB1 protein was analysed by Western blot in the nine CIB1−/− clones, nine mock transfected clones (m1–m9), and untreated NKc21. Vinculin was used as loading control. All nine CIB1−/− clones showed complete absence of CIB1 protein while all mock transfected control clones and untreated NKc21 keratinocytes retained CIB1 expression. The uncropped blot pictures can be found in Figure S4.
Generation of cell line models
Combinations of plasmids were transfected into NKc21 keratinocytes (Table 2). After 5 days, single GFP-positive cells were sorted into 96-well plates pre-seeded with 3T3 murine fibroblasts previously growth arrested with 4 mg l−1 mitomycin C (StressMarq Biosciences, Cadboro Bay, Canada) as described18. Single cell sorting was performed on a FACS Aria III cell sorter (BD Biosciences, Franklin Lakes, NJ, USA) using 100 µm nozzle and 15 psi pressure. Clones were expanded from single cells and serially transferred to 24-well plates, T25, and T75 cell culture flasks. During this process, a few cells of each clone were transferred to 20 µl dilution buffer by scratching with a pipet tip. To lyse cells, 0.5 µl DNA release additive was added and the mixture incubated at room temperature for 5 min and at 95 °C for 2 min. These samples were used to screen clones for successful homozygous knockout of CIB1 using Phire Tissue Direct PCR kit according to manufacturer’s instructions (Thermo Fisher Scientific) with primer combinations CIB1_1/CIB1_8, CIB1_3/CIB1_4, and CIB1_5/CIB1_6 (Table S1). Clones with successful CIB1 knockout were harvested and split into four samples. Two samples were stored in liquid nitrogen for subsequent cell culture experiments. The other two samples were pelleted by centrifugation at 400 g at room temperature for 10 min. One pellet was frozen at − 20 °C for DNA isolation, the other was resuspended in 100 µl RA1 buffer and 2 µl TCEP from the NucleoSpin RNA XS kit (Macherey–Nagel) for RNA isolation.
For generation of control clones, NKc21 were transfected with mock plasmids encoding Cas9 and sgRNA without target sequences in the human genome (mock-sgR-1, mock-sgR-2, and mock-sgR-3; Table S2). Single cell sorted clones were grown and harvested in the same way as the CIB1 knockout clones.
Evaluation of clones
Isolation of gDNA from keratinocytes was performed using the NucleoSpin tissue kit (Macherey–Nagel) according to the manufacturer’s instructions for cultured cells. Isolation of RNA from keratinocytes was performed according to the manufacturer’s instructions for RNA purification from cultured cells using the NucleoSpin RNA XS kit, but without addition of poly-A carrier RNA to not impair RNA-seq. Concentrations of isolated nucleic acids were determined using a DS-11 spectrophotometer (DeNovix Wilmington, DE, USA) and a Qubit fluorometer (Thermo Fisher Scientific). CIB1 deletion was validated on isolated gDNA with PCR using Taq polymerase (Qiagen, Venlo, Netherlands) and the same primer combinations as for the Direct PCR during screening. Additionally, the amplicons covering the targeted region (primers CIB1_1/CIB1_8) were analysed by Sanger sequencing (Microsynth, Balgach, Switzerland) to detect the exact junction sites of deletion. To prove the lack of CIB1 RNA expression, RNA isolated from the clones and the original NKc21 keratinocytes was reverse-transcribed using Verso cDNA synthesis kit (Thermo Fisher Scientific) according to manufacturer’s instructions using a 3:1 mixture of poly-A primers and random hexamers. Subsequently, RT-PCR was performed for CIB1 (primer CIB1_9 and CIB1_10).
Western blot
Western blot was performed to confirm CIB1 knockout on protein level. Keratinocytes were grown in T75 cell culture flasks and lysed in radio-immunoprecipitation (RIPA) buffer supplemented with protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific) on ice; protein concentration was quantified using Pierce BCA protein assay (Thermo Fisher Scientific). 7.5 µg protein lysate per lane in Laemmli buffer were separated via SDS-PAGE on a 20% polyacrylamide gel and protein was transferred for 60 min at 350 mA onto nitrocellulose membrane. Polyclonal chicken anti-CIB1 antibody raised against amino acids 24–4321,22 was used at a dilution of 1:2,500. Monoclonal rabbit anti-vinculin antibody was used as loading control (Abcam, Cambridge, UK, ab129002, 1:10,000). Polyclonal goat HRP-conjugated antibodies to chicken IgY (Abcam, ab6877, 1:10,000) and rabbit IgG (Lifespan Biosciences, Seattle, WA, USA, LS-C60884, 1:3,000) were used as secondary antibodies. Signal was detected with SuperSignal West Pico Chemiluminescent Kit (Thermo Fisher Scientific) using a ChemiDoc SRX imager system (BioRad Laboratories, Hercules, CA, USA).
RNA-seq
Gene expression of the nine CIB1 knockout clones was compared to the nine mock transfected clones by RNA-seq. RNA quality of six samples was analyzed on a 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA) with the RNA 6000 Nano kit (Agilent Technologies) and all six samples had an RNA integrity number (RIN) between 9.6 and 1023. Nine replicates per group with a sequencing depth of 10 million reads per sample are considered powerful for this type of experiment24,25. Libraries were prepared using QuantSeq 3′ mRNA-Seq Library Prep Kit FWD for Illumina (Lexogen, Vienna, Austria) and sequenced on a HiSeq2500v4 instrument (Illumina) in standard 3′ single read rapid mode for 50 cycles. Library preparation and sequencing was performed by the NGS Core Facility of the Life & Brain Center of the University of Bonn. Quality control, mapping, and counting was performed by the integrated data analysis pipeline on Bluebee platform (Bluebee Holding, Rijswijk, Netherlands) using FASTQC (Babraham Institute, Cambridge, UK), STAR26, and HTSeq-count27. Filtering, normalization, and differential gene expression analysis was performed using edgeR28: Genes with fewer than 1 count per million in over 9 samples were not included in further analysis (removing 78.21% of the genes but only 0.27% of the counts). Normalization was done by calculating a scaling factor for each sample resulting in the lowest possible log-fold changes between samples using trimmed mean of M-values29. The scaling factors ranged from 0.953 to 1.072. Common, trended, and tag-wise dispersion was calculated using the quantile-adjusted conditional maximum likelihood method (qCML)30,31. Genes differentially expressed in the knockout clones compared to the mock transfected control clones were determined using an exact test30. Genes with a false discovery rate (FDR) < 0.05 were considered differentially expressed. To validate the results, all steps were repeated with CLC Genomics Workbench (Qiagen) according to vendor’s instructions. Sequence reads of the RNA-seq experiment can be accessed in the European Nucleotide Archive (ENA) under the accession number "PRJEB34355".
Validation of RNA-seq results by qRT-PCR
Expression of genes, that were differentially expressed with an FDR < 0.05 as calculated with either edgeR or CLC Genomics Workbench, and three housekeeping genes (GUSB, HPRT1, and TBP) was determined by qRT-PCR to validate the RNA-seq results. First, RNA samples were reverse transcribed using Verso cDNA synthesis kit (Thermo Fisher Scientific) according to manufacturer’s instructions. Each 20 µl reaction contained 10 µl Power SYBR Green PCR Master Mix (Thermo Fisher Scientific), 100 nM of each primer, and template cDNA corresponding to 12.5 ng RNA. For qPCR, cDNA was denatured at 95 °C for 10 min and amplified during 45 cycles using standard protocol in a 7500 Fast Real-Time PCR system (Thermo Fisher Scientific). Primer specificity was determined by melt curves. All reactions were performed in triplicates. Gene expression was normalized and quantified using qbase + (Biogazelle, Gent, Belgium). Confidence intervals were calculated using a Mann–Whitney U test.
Results
Validation of cell line
The human keratinocyte line NKc2115 was genetically analyzed to confirm its suitability for the presented project. Sanger sequencing of CIB1, TMC6, and TMC8 did not reveal loss of function variants and only frequent (MAF > 0.05) heterozygous SNPs (rs12449858, rs145016347, and rs7208422) were found. Normal expression level of the three genes was verified by RT-PCR. SNP array of gDNA revealed six numerical aberrations (Fig. S1): trisomy of chromosomes 5, 8q, 18, and 20; monosomy of chromosome 8p; and trisomy of chromosome 9 in a subpopulation of cells. Neither chromosomes 15 nor 17 containing the EV-causing genes showed numerical aberrations in SNP array. Therefore, the cells were suitable for genome editing and subsequent downstream analyses investigating the consequences of CIB1 depletion on gene expression.
Deletion activity of sgRNA
CRISPR/Cas9 was used with pairs of sgRNAs targeting both the 5′- and 3′- end of CIB1, to cause a deletion of the sequence between the target sites containing the complete gene. Four pairs of custom sgRNAs targeting both sides of the CIB1 gene were tested for their function in NKc21. Amplification of the entire CIB1 gene revealed a PCR product shortened by the intended length of ~ 3,500–3,700 bp depending on the pair used (Fig. S2). This experiment revealed three pairs of sgRNAs (1&3, 2&3, and 2&5) with sufficiently high activity for further use.
Generation of cell line model
In order to generate multiple CIB1 knockout cell lines, 73 clones were cultured from single NKc21 cells after genome editing. Successful CIB1 knockout was confirmed by PCR (Fig. 1A). In a first reaction (primers 1 & 8), the whole gene was amplified. A shortened amplicon, indicating the successful deletion of CIB1, was produced in 36 clones, 13 of these were heterozygously and 23 clones homozygously deleted (Fig. 1B). The second PCR (primers 5 & 6) targeted a region (exons 4–6) within CIB1, confirming a genome-wide deletion of CIB1 by absence of transcript in nine clones (Fig. 1C). Therefore, these nine clones were classified as complete CIB1 knockout clones and characterized in more detail. A PCR product spanning both sgRNA recognition sites was sequenced to locate the exact break points of the deletion. This revealed deletion of the complete CIB1 gene with break points near the sgRNA recognition sites (Table 2). The CIB1 protein was not detectable in any of these clones (Fig. 1D).
To obtain control cell lines with a similar genetic background, nine mock transfected and single cell sorted clones were generated in the same way as the knockout clones. CIB1 protein expression was confirmed in these clones by Western blot (Fig. 1D).
RNA-seq
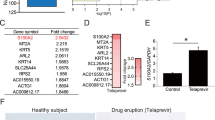
Transcriptome analyses by RNA-seq enabled the detection of differences in gene expression between the CIB1 ko clones and control clones. On average, 10.7 million reads were obtained per sample. After discarding reads that aligned to multiple regions, 7.1 million with a unique alignment in the human genome were retained (Fig. 2A). Per sample, 5.8 million reads were assigned to a transcript. MDS plot showed only small differences between knockout and control clones and the samples did not cluster by group (Fig. 2B). Differential gene expression analysis calculated by edgeR and CLC Genomic Workbench resulted in a total of six transcripts with an FDR < 0.05 (ABCA1, CIB1, FHOD1, FZD6, IL1RL1, and TNS2) (Table 3), four of them identical for both algorithms (ABCA1, CIB1, FHOD1, and TNS2) (Fig. 2C,D). While CIB1 expression was reduced more than 500-fold, the fold-changes of the other transcripts ranged between 0.178 and 2.07 (Fig. 2E).
Differences in gene expression between the nine CIB1 knockouts and nine mock transfected clones were revealed using RNA-seq. (A) Workflow for analysis of RNA-seq data by STAR, HTSeq, and edgeR as well as CLC Genomics Workbench. The average number of remaining million reads after each calculation step are indicated. (B) MDS plot of all 18 samples by edgeR revealed small differences in gene expression caused by CIB1 knockout. (C) The Volcano plot shows difference in gene expression between CIB1 knockout clones and mock transfected clones. The gray box marks the area that is enlarged in (D). (D) Enlarged Volcano plot without CIB1 showing four differentially expressed genes above the threshold of FDR = 0.05 (green line). (E) Relative expression of genes differentially expressed as calculated by edgeR. Expression has been normalized to the average count per million of the nine control clones. Mock clones are depicted in blue and knock-out clones in red. Each data point is an independent clone.
Validation of RNA-seq results by qRT-PCR
RNA-seq results were verified by qRT-PCR using the same samples. After normalization with three housekeeping genes (GUSB, HPRT1, and TBP) relative expression of ABCA1, FHOD1, FZD6, IL1RL1, and TNS2 was quantified. qRT-PCR confirmed the fold-changes observed with RNA-seq. Differences in gene expression between CIB1 knockout clones and mock transfected clones were significant for ABCA1, FHOD1, TNS2, and IL1RL1 as calculated using the Mann–Whitney U test (Table 3).
Discussion
The presented study on a keratinocyte line with a complete CIB1 knockout showed a slight effect of CIB1 deficiency on expression of other genes. RNA-seq analysis of a high number of samples with an isogenic background revealed small changes in few genes. These identified low-level changes in single genes confirm the limited influence of CIB1 in keratinocytes. This is in accordance with expression data from keratinocytes of CIB1 deficient EV-patients, which showed small differences in the expression of only 40 genes7. No enrichment of a specific pathway was observed. It is to be expected that CIB1 and both TMC influence similar or even the same pathways as they form a complex and the stability of CIB1 is dependent on TMC8 at the least7. On the other hand, CIB1 deficiency has no effect on expression of TMC6 and TMC87, which is consistent in the presented data. TMC8 has been hypothesized to control β-HPV by promoting TNF-α induced apoptosis. Variants in TMC8 lead to NF-κB activation and pro-survival as response to TNF-α13. However, no indications for changed regulation of downstream targets of TNF-α or NF-κB have been identified in the presented or previous studies7, giving rise to the suspicion that CIB1/TMC6/TMC8 complex restricts β-HPV by a different mechanism. Fold-changes of the significantly differential expressed genes ABCA1, FZD6, IL1RL1, TNS2, and FHOD1 were small and these genes could only indirectly be connected to innate immunity to β-HPV (Fig. 3)22,32,33,34,35,36,37,38,39,40,41,42,43. Future studies might show whether these proteins play a relevant role in the pathomechanism of EV.
Known regulatory relationships between differentially expressed genes and their correlation to HPV. Network shows the links discussed in the literature between the differentially expressed genes and CIB1. Transcripts which were upregulated in the CIB1−/− clones in the presented study are depicted in green, downregulated transcripts in red.
EV patients with CIB1 deficiency have a clearly defined phenotype including the high susceptibility to β-HPV, development of plane warts, and the increased risk of developing NMSC. This suggests that the function of CIB1 in keratinocytes is the control of specific HPV, which would subsequently promote the development of NMSC7. No additional features shared by a significant subset of patients have been observed in many well documented patients6,44,45,46,47 with later identified homozygous CIB1 deficiency7. In contrast to the human phenotype, mice with Cib1 deficiency showed prolonged bleeding, impaired thrombus formation and hemostasis as well as male sterility48,49. The reason for the phenotypical variances between human and mice could be related to a different function of the protein in these species and has to be investigated in further studies.
In conclusion, CIB1 deficiency results in few changes in gene expression in the in vitro model developed in the presented study consistent with the clearly defined phenotype in patients7. These observations support the hypothesis of human CIB1 as a restriction factor for β-HPV and indicate that, in case of a CIB1 deficiency, other hypothetical functions of CIB1 can be compensated. The presented model can be used in future studies to investigate CIB1 interaction with β-HPV as well as influence on viral entry or proliferation.
Data availability
Sequence reads of the RNA-seq experiment can be accessed in the European Nucleotide Archive (ENA) under the accession number "PRJEB34355".
References
de Jong, S. J. et al. Epidermodysplasia verruciformis: Inborn errors of immunity to human beta-papillomaviruses. Front. Microbiol. 9, 1222 (2018).
Orth, G. Epidermodysplasia verruciformis: A model for understanding the oncogenicity of human papillomaviruses. Ciba Found. Symp. 120, 157–174 (1986).
Orth, G. et al. Characterization of two types of human papillomaviruses in lesions of epidermodysplasia verruciformis. Proc. Natl. Acad. Sci. USA 75, 1537–1541 (1978).
Lewandowsky, F. & Lutz, W. Ein Fall einer bisher nicht beschriebenen Hauterkrankung (Epidermodysplasia verruciformis). Arch. Dermatol. Syphilol. 141, 193–203 (1922).
Majewski, S. & Jablonska, S. Skin autografts in epidermodysplasia verruciformis: Human papillomavirus-associated cutaneous changes need over 20 years for malignant conversion. Cancer Res. 57, 4214–4216 (1997).
Imahorn, E. et al. Novel TMC8 splice site mutation in epidermodysplasia verruciformis and review of HPV infections in patients with the disease. J. Eur. Acad. Dermatol. Venereol. 31, 1722–1726 (2017).
de Jong, S. J. et al. The human CIB1-EVER1-EVER2 complex governs keratinocyte-intrinsic immunity to β-papillomaviruses. J. Exp. Med. 215, 2289–2310 (2018).
Youssefian, L. et al. Epidermodysplasia verruciformis: Genetic heterogeneity and novel EVER1 and EVER2 mutations revealed by genome-wide analysis. J. Investig. Dermatol. https://doi.org/10.1016/j.jid.2018.07.010 (2018).
Landini, M. M. et al. Lack of EVER2 protein in two epidermodysplasia verruciformis patients with skin cancer presenting previously unreported homozygous genetic deletions in the EVER2 gene. J. Investig. Dermatol. 132, 1305–1308 (2011).
Leisner, T. M., Freeman, T. C., Black, J. L. & Parise, L. V. CIB1: A small protein with big ambitions. FASEB J. 30, 2640–2650 (2016).
Lazarczyk, M. et al. Regulation of cellular zinc balance as a potential mechanism of EVER-mediated protection against pathogenesis by cutaneous oncogenic human papillomaviruses. J. Exp. Med. 205, 35–42 (2008).
Lazarczyk, M., Cassonnet, P., Pons, C., Jacob, Y. & Favre, M. The EVER proteins as a natural barrier against papillomaviruses: A new insight into the pathogenesis of human papillomavirus infections. Microbiol. Mol. Biol. Rev. 73, 348–370 (2009).
Gaud, G., Guillemot, D., Jacob, Y., Favre, M. & Vuillier, F. EVER2 protein binds TRADD to promote TNF-α-induced apoptosis. Cell Death Dis. 4, e499–e499 (2013).
Vuillier, F. et al. Loss of the HPV-infection resistance EVER2 protein impairs NF-κB signaling pathways in keratinocytes. PLoS ONE 9, e89479 (2014).
Chamcheu, J. C. et al. Immortalized keratinocytes derived from patients with epidermolytic ichthyosis reproduce the disease phenotype: A useful in vitro model for testing new treatments. Br. J. Dermatol. 164, 263–272 (2011).
Todaro, G. J. & Green, H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17, 299–313 (1963).
Ramoz, N. et al. Mutations in two adjacent novel genes are associated with epidermodysplasia verruciformis. Nat. Genet. 32, 579–581 (2002).
Aushev, M., Koller, U., Mussolino, C., Cathomen, T. & Reichelt, J. Traceless targeting and isolation of gene-edited immortalized keratinocytes from epidermolysis bullosa simplex patients. Mol. Ther. Methods Clin. Dev. 6, 112–123 (2017).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Leisner, T. M., Liu, M., Jaffer, Z. M., Chernoff, J. & Parise, L. V. Essential role of CIB1 in regulating PAK1 activation and cell migration. J. Cell Biol. 170, 465–476 (2005).
Naik, U. P., Patel, P. M. & Parise, L. V. Identification of a novel calcium-binding protein that interacts with the integrin alphaIIb cytoplasmic domain. J. Biol. Chem. 272, 4651–4654 (1997).
Schroeder, A. et al. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7, 3 (2006).
Conesa, A. et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 13 (2016).
Liu, Y., Zhou, J. & White, K. P. RNA-seq differential expression studies: More sequence or more replication?. Bioinformatics 30, 301–304 (2014).
Dobin, A. et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010).
Robinson, M. D. & Smyth, G. K. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics 23, 2881–2887 (2007).
Robinson, M. D. & Smyth, G. K. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9, 321–332 (2008).
Das, R., Mahabeleshwar, G. H. & Kundu, G. C. Osteopontin induces AP-1-mediated secretion of urokinase-type plasminogen activator through c-Src-dependent epidermal growth factor receptor transactivation in breast cancer cells. J. Biol. Chem. 279, 11051–11064 (2004).
Kyo, S., Klumpp, D. J., Inoue, M., Kanaya, T. & Laimins, L. A. Expression of AP1 during cellular differentiation determines human papillomavirus E6/E7 expression in stratified epithelial cells. J. Gen. Virol. 78(Pt 2), 401–411 (1997).
Peto, M., Tolle-Ersu, I., Kreysch, H. G. & Klock, G. Epidermal growth factor induction of human papillomavirus type 16 E6/E7 MRNA in tumor cells involves two AP-1 binding sites in the viral enhancer. J. Gen. Virol. 76(Pt 8), 1945–1958 (1995).
Calderwood, D. A. et al. Integrin beta cytoplasmic domain interactions with phosphotyrosine-binding domains: A structural prototype for diversity in integrin signaling. Proc. Natl. Acad. Sci. USA 100, 2272–2277 (2003).
Goldschmidt, M. E., McLeod, K. J. & Taylor, W. R. Integrin-mediated mechanotransduction in vascular smooth muscle cells: Frequency and force response characteristics. Circ. Res. 88, 674–680 (2001).
Zhou, X., Yin, Z., Guo, X., Hajjar, D. P. & Han, J. Inhibition of ERK1/2 and activation of liver X receptor synergistically induce macrophage ABCA1 expression and cholesterol efflux. J. Biol. Chem. 285, 6316–6326 (2010).
Gerdes, M. J. et al. Activator protein-1 activity regulates epithelial tumor cell identity. Cancer Res. 66, 7578–7588 (2006).
Leisner, T. M., Moran, C., Holly, S. P. & Parise, L. V. CIB1 prevents nuclear GAPDH accumulation and non-apoptotic tumor cell death via AKT and ERK signaling. Oncogene 32, 4017–4027 (2013).
Tanaka, T., Narazaki, M. & Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 6, a016295 (2014).
Kulhan, N. G. et al. Could interleukin-33 and its suppressor of tumorigenicity 2 (ST2) receptor have a role in cervical human papillomavirus (HPV) infections?. Gynecol. Endocrinol. https://doi.org/10.1080/09513590.2019.1590699 (2019).
Jin, M., Komine, M., Tsuda, H., Oshio, T. & Ohtsuki, M. Interleukin-33 is expressed in the lesional epidermis in herpes virus infection but not in verruca vulgaris. J. Dermatol. 45, 855–857 (2018).
Oyoshi, M. K., Venturelli, N. & Geha, R. S. Thymic stromal lymphopoietin and IL-33 promote skin inflammation and vaccinia virus replication in a mouse model of atopic dermatitis. J. Allergy Clin. Immunol. 138, 283–286 (2016).
Arnold, A. W. et al. Homozygosity for the c.917A→T (P.N306l) polymorphism in the EVER2/TMC8 gene of two sisters with epidermodysplasia verruciformis Lewandowsky-Lutz originally described by Wilhelm Lutz. Dermatology 222, 81–86 (2011).
Kienzler, J. L. et al. Epidermodysplasie verruciforme. Données ultrastructurales, virologiques et photobiologiques; à propos d’une observation. Ann. Dermatol. Venereol. 106, 549–563 (1979).
Lutz, W. A propos de l’epidermodysplasie verruciforme. Dermatologica 92, 30–43 (1946).
Rueda, L. A. & Rodriguez, G. Verrugas humanas por virus papova. Correlacion clinica, histologica y ultraestructural. Med. Cutanea Ibero Lat. Am. 2, 113–136 (1976).
Naik, M. U. et al. CIB1 deficiency results in impaired thrombosis: The potential role of CIB1 in outside-in signaling through integrin alpha IIb beta 3. J. Thromb. Haemost. 7, 1906–1914 (2009).
Yuan, W. et al. CIB1 is an endogenous inhibitor of agonist-induced integrin alphaIIbbeta3 activation. J. Cell Biol. 172, 169–175 (2006).
Aochi, S. et al. A novel homozygous mutation of the EVER1/TMC6 gene in a Japanese patient with epidermodysplasia verruciformis. Br. J. Dermatol. 157, 1265–1266 (2007).
Tate, G., Suzuki, T., Kishimoto, K. & Mitsuya, T. Novel mutations of EVER1/TMC6 gene in a Japanese patient with epidermodysplasia verruciformis. J. Hum. Genet. 49, 223–225 (2004).
Sunohara, M. et al. Dye laser photodynamic therapy for Bowen’s disease in a patient with epidermodysplasia verruciformis. Osaka City Med. J. 58, 77–82 (2012).
Zuo, Y.-G., Ma, D., Zhang, Y., Qiao, J. & Wang, B. Identification of a novel mutation and a genetic polymorphism of EVER1 gene in two families with epidermodysplasia verruciformis. J. Dermatol. Sci. 44, 153–159 (2006).
Gober, M. D. et al. Novel homozygous frameshift mutation of EVER1 gene in an epidermodysplasia verruciformis patient. J. Investig Dermatol. 127, 817–820 (2007).
Rady, P. L. et al. Novel homozygous nonsense TMC8 mutation detected in patients with epidermodysplasia verruciformis from a Brazilian family. Br. J. Dermatol. 157, 831–833 (2007).
Heuser, S. et al. The levels of epithelial anchor proteins β-catenin and ZO-1 are altered by E7 of HPV5 and HPV8. J. Gen. Virol. https://doi.org/10.1099/jgv.0.000363 (2015).
Berthelot, C. et al. Treatment of a patient with epidermodysplasia verruciformis carrying a novel EVER2 mutation with imiquimod. J. Am. Acad. Dermatol. 56, 882–886 (2007).
Sun, X. K., Chen, J. F. & Xu, A. E. A homozygous nonsense mutation in the EVER2 gene leads to epidermodysplasia verruciformis. Clin. Exp. Dermatol. 30, 573–574 (2005).
Miyauchi, T. et al. Genetic analysis of a novel splice-site mutation in TMC8 reveals the in vivo importance of the transmembrane channel-like domain of TMC8. Br. J. Dermatol. 175, 803–806 (2016).
Mizuno, Y. et al. Merkel cell polyomavirus-positive Merkel cell carcinoma in a patient with epidermodysplasia verruciformis. Acta. Dermatol. Venereol. 95, 98–99 (2015).
Acknowledgements
We thank the University of Basel, the Basel University Hospital, and the Department of Biomedicine (DBM) for institutional support, the DBM FACS core facility for assistance with single cell sorting, the DBM bioinformatics core facility for advice for data analysis, and the members of the DBM genome editing club for discussions. Further, we thank Hedwig Wariwoda (Basel, Switzerland) for assistance with cell culture maintenance, Francis Jacob (Basel, Switzerland) for technical help with genome editing, and Hans Törmä (Uppsala, Sweden) for providing the keratinocyte line NKc21.
Funding
This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
All authors were involved in conception and/or design of the work. E.I., M.A., S.H., P.H., and B.B. performed data collection. E.I. and S.H. performed data analysis. E.I., S.C., J.R., P.H.I., and B.B. performed data interpretation. P.H.I. and B.B. supervised the project. E.I. wrote the draft of the article. All authors critically revised the article and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Imahorn, E., Aushev, M., Herms, S. et al. Gene expression is stable in a complete CIB1 knockout keratinocyte model. Sci Rep 10, 14952 (2020). https://doi.org/10.1038/s41598-020-71889-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-71889-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.