Abstract
It is important that antibiotics prescriptions are based on antimicrobial susceptibility data to ensure effective treatment outcomes. The increasing availability of next-generation sequencing, bacterial whole genome sequencing (WGS) can facilitate a more reliable and faster alternative to traditional phenotyping for the detection and surveillance of AMR. This work proposes a machine learning approach that can predict the minimum inhibitory concentration (MIC) for a given antibiotic, here ciprofloxacin, on the basis of both genome-wide mutation profiles and profiles of acquired antimicrobial resistance genes. We analysed 704 Escherichia coli genomes combined with their respective MIC measurements for ciprofloxacin originating from different countries. The four most important predictors found by the model, mutations in gyrA residues Ser83 and Asp87, a mutation in parC residue Ser80 and presence of the qnrS1 gene, have been experimentally validated before. Using only these four predictors in a linear regression model, 65% and 93% of the test samples’ MIC were correctly predicted within a two- and a four-fold dilution range, respectively. The presented work does not treat machine learning as a black box model concept, but also identifies the genomic features that determine susceptibility. The recent progress in WGS technology in combination with machine learning analysis approaches indicates that in the near future WGS of bacteria might become cheaper and faster than a MIC measurement.
Similar content being viewed by others
Introduction
Antibiotics are an essential resource in the control of infectious diseases; they have been a major contributor to the decline of infection-associated mortality and morbidity in the twentieth century. However, the recent rise of antimicrobial resistance (AMR) threatens this situation1. Bacterial AMR is associated with a higher likelihood of therapeutic failure in case of infections. Accurate and fast prediction of AMR in bacteria is needed to select the optimal therapy.
With the increasing availability of next-generation sequencing, bacterial whole genome sequencing (WGS) is becoming a feasible alternative to traditional phenotyping for the detection and surveillance of AMR2,3,4. However, data analysis remains the weak point in this approach; fast and scalable methods are required to transform the ever-growing amount of genomic data into actionable clinical or epidemiological information5. Machine learning is a promising approach for this kind of data analysis.
AMR can be predicted in numerous ways. In addition to classic and highly standardized phenotypic testing of resistance, several methods of resistance prediction have been developed. Most novel methods use a genetic or genomic approach, although transcriptomic approaches have been investigated as well6,7,8. An important factor in the choice of the resistance prediction method is the microorganism under study. For example, the CRyPTIC consortium managed to predict resistance to four first-line drugs in Mycobacterium tuberculosis, using only known mutations extracted from WGS9. However, M. tuberculosis displays little-to-no horizontal gene transfer and low genomic evolution rate10, which makes it feasible to predict resistance only on the basis of known mutations11. For other bacteria, more advanced analysis methods such as machine learning need to be applied to allow for accurate prediction.
Machine learning has been applied to predict resistance from WGS data in several settings. To date, these methods have been restricted mostly to assign bacteria to binary categories, i.e. susceptible or non-susceptible8,12,13,14,15,16,17,18. Clinical breakpoints used to define susceptible and non-susceptible categories can change over time based on various protocols. Such binary categories do not allow following trends in subtle changes in susceptibility. Minimum inhibitory concentration (MIC) measures offer an adequate resolution to follow if susceptibility is changing in a population, which is useful for epidemiological purposes. Therefore, a resistance prediction method would preferably output a continuous estimate of resistance similar to MIC, instead of binary classification (S/R), as a number of studies already proposed19,20,21,22.
Additional issues should be considered when developing a reliable and useful prediction model. Genotypes are often geographically clustered23. This implies that if a prediction model is trained on data from one country, this model might not be generalizable to data from another country. Data from multiple countries are thus needed. A combinations of chromosomal mutations and acquired resistance genes may influence antimicrobial resistance together, it might be not enough to focus only on the point mutations or the acquired genes. Therefore, different data types need to be combined to obtain a biologically relevant set of input data. Lastly, while machine learning is able to analyse highly complex patterns of features, the model would preferably output generally understandable data. Among others k-mer profiles have been used to predict resistance20,21.
In this study, we focus on predicting a quantitative measure of ciprofloxacin resistance (MIC) for a geographically diverse population of E. coli using machine learning. We chose to study ciprofloxacin resistance in E. coli because of three reasons:
-
1.
Ciprofloxacin resistance in E. coli has been studied intensively
-
2.
Ciprofloxacin resistance in E. coli can be caused by a range of different chromosomal and plasmid-mediated mechanisms24
-
3.
Ciprofloxacin is commonly used in the treatment of E. coli infections across the globe.
In our selection of machine learning models, an important criterion was that high-scoring features could be extracted from the model. This would allow us to explore the reasoning behind each prediction and thus to interpret and understand the model. Also, if the trained model relies only on a few genomic features, when genetating predictions for new samples, it is enough to determine those few genomic attributes, WGS is not necessary.
Results
Our dataset consists of a phylogenetically diverse collection of E. coli strains, see Fig. 1. Strains in the test and train group are present throughout the whole phylogeny, although the groups are present predominantly in different parts of the phylogeny.
We trained a random forest model using genome-wide mutation profiles alongside the ResFinder-based profiles of acquired resistance genes. We ranked the predictors proposed by the model itself, see Supplementary Table S1. The model performed with high accuracy on the training set leave-one-country-out cross-validation using four predictors, see Supplementary Fig. S1. The addition of more features did not improve significantly the cross-validation results, therefore we kept only the first four, allowing for a simple and understandable model. We also trained a linear regression model on this restricted dataset.
Using these four predictors with the linear regression model, 264 out of the 266 test data samples were correctly classified at susceptible/non-susceptible level, 65% and 93% of the corresponding MIC values were correctly predicted within a two- and a four-fold dilution, see Table 1. All the four genetic features are experimentally proven to play an important role in ciprofloxacin resistance24.
These 4 predictors are the following:
-
gyrA mutation at amino acid #87
-
gyrA mutation at amino acid #83
-
parC mutation at amino acid #80
-
qnrS1 gene
All of the predictors above are binary (presence/absence) therefore there are \(2^4 = 16\) different possible prediction for any sample based on these features, see Supplementary Table S2. A linear regression model fitted on the log2 values of the MIC measurements could achieve similar performance as a more complex random forest model, see Fig. 2. Linear regression is preferred due to its simplistic nature. Having a random forest regressor with hundreds of decision trees and thousands of genomic features as predictors it is difficult to understand why the model made that particular prediction, leaving doubts of its clinical usefulness.
There are several previous works on predicting ciprofloxacin resistance for E. coli at susceptible/non-susceptible level. This makes a limited comparision possible between the presented method and the methods published in the literature. Limitation comes from the fact that the susceptible/non-susceptible outcome from the measurements are based on breakpoints which are not always reported. Also, some papers use disk diffusion test. Keeping these limitations in mind it is still worthy to compare our method to others. Pesesky et al.13 reported AUC of 0.9652–0.9786, while Hyun et al.18 reported 0.98 AUC. Moradigaravand et al.12 reported 0.97 precision and 0.81 recall while our linear model achieved 1.0 AUC, see Table 1 and 1.0 precision with 0.987 recall, see Supplementary Fig. S2.
Discussion
Here, we present an accurate method for predicting ciprofloxacin resistance for E. coli. With no built-in prior knowledge on which chromosomal mutations and which acquired resistance genes might be important, using a data-driven approach, we managed to create a machine learning model that was not only accurately predicting the susceptible/non-susceptible labels but also accurately predicting at MIC level. Additionally, the highlighted features of our approach could be narrowed down to four biologically understandable features, making the method simpler and therefore applicable to clinical microbiology practice.
It was previously shown that accurate ciprofloxacin susceptible/non-susceptible binary prediction is possible for E. coli3,12,17. For some other bacteria-antibiotic combinations even MIC level predictions were performed19,20,21,22. This study goes beyond by not only predicting MIC level ciprofloxacin resistance for E. coli, but also highlighting the underlying reasoning behind the predictions. Furthermore, this study is one of very few that includes the presence or absence of genes located on mobile genetic elements (MGEs), in combination with chromosomal point mutations, in the machine learning algorithm. This is a crucial step since particularly in Gram-negative microorganisms such as E. coli, AMR is often encoded by genomic determinants located on MGE, or a combination of chromosomal and MGE encoded determinants, as is demonstrated in our study for ciprofloxacin. In addition, this study used data from different countries and regions thus ensuring potential variation in determinants that may contribute to ciprofloxacin resistance are represented in the data set.
Notably, a linear regression model based on only the four most important features of the random forest model performed nearly as well as the full model. These features comprise two gyrA mutations, one parC mutation and the presence of the qnrS1 gene. All features have been associated with ciprofloxacin resistance before24. In addition, the presence of a single determinant versus combinations of multiple of the four determinants predicted MIC ranges that were comparable to those observed experimentally and in clinical isolates24. For example, the single presence of the qnrS1 gene predicted a relatively low MIC but the combination of the qnrS1 gene with a single mutation in gyrA increased the predicted MIC substantially (Supplementary Table S2). Our results indicate that for prediction of ciprofloxacin susceptibility on the basis of whole-genome sequencing in E. coli, the analysis could be limited to only these four determinants.
It is worthy to note that the model was trained on all possible mutations. Also all acquired resistance genes were considered that appreared in the ResFinder gene database. Neither SNPs, nor resistance genes were pre-selected for ciprofloxacin during data preparation. Therefore the model could potentially discover novel, currently unknown mutation-based resistance encoding mechanisms which may be located in genes that are or are not yet known to contribute to resistance. For E. coli the ciprofloxacin resistance determinants that were predicted in our machine learning approach have been experimentally verified, but for other antibiotics, our approach could detect novel genomic variants associated with resistance.
Our study also has some limitations, which mostly pertain to the dataset. For strains with measured MICs in the range of 8–64 mg/L, our model performs worse than for strains with lower MICs. This is most likely due to the fact that the majority of resistant strains in our training data have an MIC of 32 mg/L, with only very few other resistant MICs. This hampers accurate prediction of MIC for more resistant E. coli. Additionally, our dataset is not yet diverse and complete enough to be applied on a wide scale. This is a common problem for many studies aiming to predict AMR from WGS data. Solving this would require continuous updating of databases and an adequate database structure, the latter we have addressed previously25.
In conclusion, we report a machine learning approach for a quantitative prediction of antibiotic resistance, which we applied for prediction of ciprofloxacin resistance in E. coli. In combination with continuous data base improvements, our approach could allow machine learning methods to enter routine clinical diagnostic and epidemiological practices to continuously improve predictions.
Methods
Data summary
In this study, 704 E. coli genomes combined with MIC measurement for ciprofloxacin were analysed25. Paired-end sequencing was performed on all isolates and the results were stored in FASTQ format. The isolates originated from five countries, Denmark, Italy, USA, UK, and Vietnam. The MIC distribution for these isolates is depicted in Table 2. Out of 704, 266 E. coli genomes had no country metadata available and were used as an independent test set. All data were deposited in the AMR Data Hub25 which consists of raw sequencing data, ciprofloxacin minimum inhibitory concentrations, and additional metadata such as the origin of the samples. All data is publicly available from the SRA EBI database with the following accession codes: PRJEB21131, PRJNA266657, PRJNA292901, PRJNA292904, PRJNA292902, PRJDB7087, PRJEB21880, PRJEB21997, PRJEB14086 and PRJEB16326.
Download and analysis scripts are available at https://github.com/patbaa/AMR_ciprofloxacin. iTOL phylogenetic tree is available at https://itol.embl.de/tree/14511722611491391569485969.
All used files are listed at https://github.com/patbaa/AMR_ciprofloxacin/blob/master/meta.tsv with URLs provided. Isolates with accession codes and MIC measurements are also available at https://github.com/patbaa/AMR_ciprofloxacin/blob/master/supplementary_meta_table.csv.
Data preprocessing
Raw reads were mapped on the ATCC 25922 reference genome (https://www.ncbi.nlm.nih.gov/assembly/GCF_000743255.1) using BWA-MEM v0.7.1726 with default settings. Pileup files were generated with bcftools v1.927 with “–min-MQ 50” settings. Single-nucleotide polymorphisms (SNPs) and insertions-deletions (INDELs) were called using bcftools v1.9 with “–ploidity 1-m” flags. Further filtering was applied via bcftools v1.9 “\(\%\hbox {QUAL}>=50\) & \(\hbox {DP}>=20\)” flags. Bcftools output data was expressed as either a SNP (value: 1), an INDEL (value: 5) or no mutation (value: 0) per position in the reference genome. Exact numbers are irrelevant, as tree-based methods are not sensitive to the scale. The intention was to differentiate between reference alleles, SNPs and INDELs at a given position. Acquired resistance genes were identified using ResFinder v3.228 with a coverage threshold of 90% and an identity threshold of 90% using a database downloaded on 13th Apr. 2020. ResFinder was used with KMA v1.1.429. The ResFinder output data was expressed as presence (value: 1) or absence (value: 0) of resistance genes. The SNP/INDEL data and ResFinder data were subsequently merged which provided a matrix with more than 830,000 columns representing reference genome positions with at least one mutation and 175 columns representing detected resistance genes.
Phylogenetic tree generation
The merged variant call files were converted to a FASTA alignment using vcf2phylip v2.0, retaining positions that were called in at least 50% of isolates30. The invariant positions were removed from the alignment using snp-sites v2.4.031. The phylogeny was inferred using RAxML v8.2.9 in rapid bootstrap mode (-f a) with 100 bootstraps using a General Time Reversible model with Gamma rate heterogeneity including Lewis ascertainment bias correction (-m ASC_GTRGAMMA)32. The resulting phylogeny was visualized in iTOL33.
Metrics
We used the following metrics for the evaluation of the model:
\({\hbox{AUC-area under the receiver operating characteristics curve}}\)—we used the clinical breakpoint for ciprofloxacin, 1 mg/L, based on the Clinical & Laboratory Standards Institure guideline34 to encode whether samples are resistant or not.
\({{R}^{2}\hbox {score-coefficient of determination}}\)
where \(y_i\) is the measured value for sample i, \({\hat{y}}_i\) is the predicted value for sample i, \({\overline{y}}\) is the mean of the measured values.
\({{R }\hbox {-Pearson correlation coefficient}}\)
where cov is the covariance and \(\sigma \) is the standard deviation.
\({\hbox {ME-major error}}\)—when the sample is non-resistant by measurement, but it is predicted to be resistant. Non-resistant and resistant labels are derived from MIC via thresholding.
\({\hbox {VME-very major error}}\)—when the sample is resistant by measurement, but it is predicted to be non-resistant. Non-resistant and resistant labels are derived from MIC via thresholding.
\({\hbox {ACC-2-accuracy within two-fold dilution}}\)—the fraction of the samples with MIC properly predicted within a two-fold dilution. If the measured MIC is x, then the prediction is counted as properly predicted within a two-fold dilution if it falls to the [x/2;2x] interval. Dilution range is the natural scale for comparison of MIC predictions and measurements due to the logarithmic scale of the latter. As the MIC gives additional clinical information beyond the binary resistant/non-resistant outcome here we report both ACC-2 and ACC-4.
\({\hbox {ACC-4-accuracy within four-fold dilution}}\)—the fraction of the samples with MIC properly predicted within a four-fold dilution. If the measured MIC is x, then the prediction is counted as properly predicted within a four-fold dilution if it falls to the [x/4;4x] interval.
\({\hbox {MAFE-mean absolute fold error}}\)—The mean absolute difference between the log2 values of the prediction and the measurements.
Importance of the validation scheme
Proper validation is a key element in machine learning as most of the models have a large number of parameters which makes it easy for them to memorize the training dataset. In image recognition, popular convolutional neural networks can have more than 100 M parameters35. This number of parameters is orders of magnitudes larger than the number of pixels of a single image or even the number of the images in the whole usual benchmark data sets, such as ImageNet36. Having that many parameters it is possible to memorize the training data without generalizing any knowledge to the test data or for future use.
However, with having a proper validation scheme it is possible to fairly estimate the generalization power of a model. In many cases simply randomly splitting the samples into two groups to a test and a validation set is enough. If the data set is small, cross-validation can help, usually, K-fold cross-validation, where the data set is split into K set, each having the same size. Then, the model is trained on using data from \(K-1\) set and the predictions are made for the one set that was not used in the training process. Repeating the process, K times predictions can be generated for the whole data set in a way that the model did not see in training time any of the samples for which it is generating predictions. The weights of the model are reset between any two training.
K-fold cross-validation can produce too optimistic results if the samples are clustered. For example, when the data collection is biased, bacterial isolates from one country are predominantly resistant whilst isolates from other countries are predominantly susceptible to an antibiotic. In addition, genetic signatures are often clustered by country23. Due to such clustering, the model may predict the country of origin of the bacterial isolate, which may be correlated with the MIC, on both the training and the validation data sets, but it is not guaranteed that the same will happen in real-life usage later.
Leave-one-country-out validation
Here we propose a more strict and reliable validation method. Instead of randomly splitting the data into K different folds, we split the folds by country. Using this approach, the model is not rewarded if it only learns country-specific attributes. Leave-one-country-out validation was performed during the selection of the most important features in the data set, see Supplementary Table S1. The random forest model was fitted \(K=5\) times leaving out one country each time from the training data set. Then the feature importances were summed over each fold resulting in the final feature importance rankings.
It worth to look at Supplementary Table S1, which contains the feature importances calculated the way described above. For gyrA#87 we have fairly large values for all splits except, when the Vietnam data is left out from the training, suggesting that gyrA#87 mutant stains are mainly coming from Vietnam. The high feature importance of gyrA#83 for the split when Vietnam data is the test means that for the non-vietnamese data gyrA#83 mutation is the most descriptive.
Random forest model
For tabular data most often tree-like models perform the best. The random forest model is an ensemble of numerous (usually hundreds of) decision trees. In the training process, each tree is trained separately and each of them uses only a random fraction of the data, which ensures that the decision trees will not be identical. For a new sample, the prediction is the average of the prediction of the trees, or for classification the category that was predicted the most often by the individual trees. This ensemble technique ends up an accurate, robust, scalable model. The prediction error is usually large for each individual tree, but as long as the errors of the trees are uncorrelated, averaging their prediction lowers the final error.
Random forest regressor was trained with mean squared error criterion, min_samples_leaf = 1, min_samples_split = 2, and n_estimators = 200 for the feature selection. For the final evaluation mean squared error criterion, min_samples_leaf = 1, min_samples_split = 5, and n_estimators = 100 parameters were used. The random seed was fixed. Other parameters remained default. Scikit-learn v0.21.237 was used for fitting the model in Python 3.6.5.
Random forest feature importance
For decision trees the input variables, the features can be sorted by their importance. The importance can be defined in various ways; the used scikit-learn v0.21.237 implementation calculates the mean decrease impurity averaged over all the trees in the forest38,39. In this approach, the identification of the most important predictors becomes feasible even for cases when there are hundreds of thousands of features.
Model fitting
All models were fitted on the log2 values of the MIC, which is the natural scale for the MIC measurement. Later the predicted values were converted back to the MIC units.
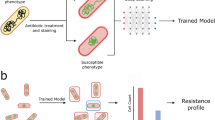
Study pipeline
The pipeline of this study is shown in Fig. 3. First, the raw reads were converted to a numerical table indicating mutations and plasmid related resistant genes. In the second step, a random forest model is fitted on the train data via leave-one-country cross-validation. Features importances were averaged over each fold. Then the highest-ranking features were kept which significantly reduced the dimensionality of the data. Using this low dimensional training data a random forest model and a linear regression was fitted. For fitting the models always the log2 MIC values were used as a natural scale for the MIC measurements.
Workflow of the study. First, a random forest model was fitted to the training data with leave-one-country-out validation. Feature importances of the fitted models are averaged over all the folds and the four best features are kept. Then the random forest model and a linear regression model were fitted on all the training samples using only the four best features. And model performances are tested using the independent test dataset.
At the last step, the performance of the models was evaluated on the unseen test data using the same restricted feature set.
References
Lederberg, J. Infectious history. Science 288, 287–293 (2000).
Otto, M. Next-generation sequencing to monitor the spread of antimicrobial resistance. Genome Med. 9, 68 (2017).
Stoesser, N. et al. Predicting antimicrobial susceptibilities for Escherichia coli and klebsiella pneumoniae isolates using whole genomic sequence data. J. Antimicrob. Chemother. 68, 2234–2244 (2013).
Su, M., Satola, S. W. & Read, T. D. Genome-based prediction of bacterial antibiotic resistance. J. Clin. Microbiol. 57, e01405-18 (2019).
Köser, C. U., Ellington, M. J. & Peacock, S. J. Whole-genome sequencing to control antimicrobial resistance. Trends Genet. 30, 401–407 (2014).
Barczak, A. K. et al. Rna signatures allow rapid identification of pathogens and antibiotic susceptibilities. Proc. Natl. Acad. Sci. 109, 6217–6222 (2012).
Khaledi, A. et al. Transcriptome profiling of antimicrobial resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 60, 4722–4733 (2016).
Khaledi, A. et al. Fighting antimicrobial resistance in Pseudomonas aeruginosa with machine learning-enabled molecular diagnostics. bioRxivarXiv:643676 (2019).
Consortium, C. & the 100, . G. P. Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N. Engl. J. Med. 379, 1403–1415 (2018).
Duchêne, S. et al. Genome-scale rates of evolutionary change in bacteria. Microb. Genom. 2 (2016).
Veyrier, F., Pletzer, D., Turenne, C. & Behr, M. A. Phylogenetic detection of horizontal gene transfer during the step-wise genesis of mycobacterium tuberculosis. BMC Evolut. Biol. 9, 196 (2009).
Moradigaravand, D. et al. Prediction of antibiotic resistance in Escherichia coli from large-scale pan-genome data. PLoS Comput. Biol. 14, e1006258 (2018).
Pesesky, M. W. et al. Evaluation of machine learning and rules-based approaches for predicting antimicrobial resistance profiles in gram-negative bacilli from whole genome sequence data. Front. Microbiol. 7, 1887 (2016).
Davis, J. J. et al. Antimicrobial resistance prediction in patric and rast. Sci. Rep. 6, 27930 (2016).
Yang, Y. et al. Machine learning for classifying tuberculosis drug-resistance from dna sequencing data. Bioinformatics 34, 1666–1671 (2017).
Kouchaki, S. et al. Application of machine learning techniques to tuberculosis drug resistance analysis. Bioinformatics 35, 2276–2282 (2018).
Her, H.-L. & Wu, Y.-W. A pan-genome-based machine learning approach for predicting antimicrobial resistance activities of the Escherichia coli strains. Bioinformatics 34, i89–i95 (2018).
Hyun, J. C., Kavvas, E. S., Monk, J. M. & Palsson, B. O. Machine learning with random subspace ensembles identifies antimicrobial resistance determinants from pan-genomes of three pathogens. PLoS Comput. Biol. 16, e1007608 (2020).
Eyre, D. W. et al. WGS to predict antibiotic mics for neisseria gonorrhoeae. J. Antimicrob. Chemother. 72, 1937–1947 (2017).
Nguyen, M. et al. Developing an in silico minimum inhibitory concentration panel test for Klebsiella pneumoniae. Sci. Rep. 8, 421 (2018).
Nguyen, M. et al. Using machine learning to predict antimicrobial mics and associated genomic features for nontyphoidal salmonella. J. Clin. Microbiol. 57, e01260-18 (2019).
Li, Y. et al. Penicillin-binding protein transpeptidase signatures for tracking and predicting \(\beta \)-lactam resistance levels in streptococcus pneumoniae. MBio 7, e00756-16 (2016).
Novembre, J. et al. Genes mirror geography within europe. Nature 456, 98 (2008).
van der Putten, B. C. et al. Quantifying the contribution of four resistance mechanisms to ciprofloxacin mic in Escherichia coli: A systematic review. J. Antimicrob. Chemother. 74, 298–310 (2018).
Matamoros, S. et al. Accelerating surveillance and research of antimicrobial resistance-an online repository for sharing of antimicrobial susceptibility data associated with whole genome sequences. bioRxiv arXiv:532267 (2019).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with bwa-mem. arXiv preprint arXiv:1303.3997 (2013).
Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011).
Zankari, E. et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644 (2012).
Clausen, P. T., Aarestrup, F. M. & Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 19, 307 (2018).
Ortiz, E. M. vcf2phylip v2.0: convert a VCF matrix into several matrix formats for phylogenetic analysis. (version v2.0). Zenodo (2019) https://doi.org/10.5281/zenodo.2540861.
Page, A. J. et al. SNP-sites: rapid efficient extraction of SNPS from multi-fasta alignments. Microb. Genom. 2 (2016).
Stamatakis, A. Raxml version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Letunic, I. & Bork, P. Interactive tree of life (itol) v4: Recent updates and new developments. Nucleic Acids Res. 47(W1), W256–W259 (2019).
CLSI. Fluoroquinolone Breakpoints for Enterobacteriaceae and Pseudomonas aeruginosa, 1st edn (Clinical and Laboratory Standards Institute, Wayne, 2019).
Simonyan, K. & Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv preprintarXiv:1409.1556 (2014).
Deng, J. et al. Imagenet: A large-scale hierarchical image database. In 2009 IEEE Conference on Computer Vision and Pattern Recognition 248–255 (IEEE, 2009).
Pedregosa, F. et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Louppe, G., Wehenkel, L., Sutera, A. & Geurts, P. Understanding variable importances in forests of randomized trees. Advances in neural information processing systems 431–439 (2013).
Breiman, L. Classification and Regression Trees (Routledge, Abingdon, 2017).
Acknowledgements
This study was supported by the COMPARE Consortium, which has received funding from the European Union’s Horizon 2020 research and innovation programme under Grant agreement No. 643476. I.C. acknowledges support from National Research, Development and Innovation Fund of Hungary, Project no. FIEK_16-1-2016-0005
Author information
Authors and Affiliations
Consortia
Contributions
All authors contributed to the conceptualization of the study. B.A.P., B.C.L.P. and C.S. wrote the manuscript, S.M., B.C.L.P., R.S.H., O.L., C.S. collected the data from various public data sources. B.A.P., E.G., D.A.A. performed machine learning modeling. The COMPARE ML-AMR group contributed data and technical help. All authors contributed to the study with insightful discussion. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pataki, B.Á., Matamoros, S., van der Putten, B.C.L. et al. Understanding and predicting ciprofloxacin minimum inhibitory concentration in Escherichia coli with machine learning. Sci Rep 10, 15026 (2020). https://doi.org/10.1038/s41598-020-71693-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-71693-5
This article is cited by
-
Interpretable machine learning-based decision support for prediction of antibiotic resistance for complicated urinary tract infections
npj Antimicrobials and Resistance (2023)
-
Feasibility of predicting allele specific expression from DNA sequencing using machine learning
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.