Abstract
Changes in insect herbivore performance under elevated atmosphere carbon dioxide concentrations e[CO2] are often driven by changes in the nutritional and defensive chemistry of their host plants. Studies addressing how the prolific pest cotton bollworm (Helicoverpa armigera) responds to e[CO2] show that performance usually declines, often associated with lower nutritional (e.g. nitrogen (N) concentrations) quality of host plants under e[CO2]. We investigated the impacts of e[CO2] on nutritional quality and anti-herbivore (jasmonate) defensive signalling in lucerne (Medicago sativa) when challenged by H. armigera. While foliar N decreased under e[CO2], other aspects of nutritional quality (soluble protein, amino acids, foliar C:N) were largely unaffected, potentially due to increased root nodulation under e[CO2]. In contrast, e[CO2] greatly reduced jasmonate signalling in M. sativa following H. armigera attack; jasmonic acid concentrations were ca. 56% lower in attacked plants grown under e[CO2]. Concurrent with this, relative growth rates of H. armigera were ca. 66% higher when feeding on e[CO2]-grown plants. In contrast with previous reports, which we meta-analytically summarise, we provide the first evidence that H. armigera performance can increase under e[CO2]. This may occur in plants, such as M. sativa, where e[CO2] has limited impacts on nutritional quality yet reduces jasmonate defence signalling.
Similar content being viewed by others
Introduction
With global populations expected to reach 11.2 billion by 2,100 there is an urgent need to ensure future food security, a challenge which is complicated by global climate change1. Invertebrate pests destroy enough food to feed a billion people a year2 which has, in part, fuelled interest in understanding which pests may become more problematic under predicted changes in the Earth’s climate3. In particular, unprecedented increases in atmospheric carbon dioxide (CO2) have the capacity to change plant chemistry which affect the susceptibility of crops to insect herbivores4,5,6.
The effects of elevated atmospheric CO2 concentration (e[CO2]) on plant nutritional and defensive chemistry, and their consequent effects on invertebrate herbivores, have received extensive attention7,8,9. In terms of nutritional quality, nitrogen availability is considered to be the limiting factor in insect herbivore diets10. Broadly speaking, e[CO2] causes foliar nitrogen (N) concentrations to decrease due to one or more processes, including dilution effects (i.e. relative to increased carbohydrate concentrations), reduced N uptake by roots, increased NH3 volatilization and reduced investment in the N-rich enzyme RUBISCO11,12,13,14. The impacts of e[CO2] on plant defences are less easily predicted, but some trends are emerging5. Plant defences against herbivorous arthropods are regulated by several phytohormonal pathways, including the jasmonic acid (JA), salicylic acid (SA) and ethylene signalling pathways15,16. Of these, the JA pathway is regarded as the master regulator of plant resistance to arthropod herbivores, especially chewing herbivores17. A growing number of studies, reviewed by Zavala, et al.5 and Ode, et al.6, suggest that e[CO2] downregulates constitutive and herbivore-induced activity of the JA signalling pathway.
Some pests, such as the cotton bollworm (Helicoverpa armigera Hübner), have now reached critical pest status because of their economic damage and rapid ability to invade new regions 18. In particular, H. armigera is a major global pest of diverse agricultural and horticultural crops causing upwards of US $ 7 billion annually in crop losses19. This has, in part, prompted research into which crops may be at risk from H. armigera under predicted atmospheric CO2 concentrations. To our knowledge, there are 10 published studies reporting the plant-mediated effects of e[CO2] on H. armigera across four plant families: Fabaceae20,21, Malvaceae22,23,24,25, Poaceae26,27,28 and Solanaceae29. Most of these studies reported that e[CO2] reduced many, but not all, H. armigera performance parameters20,21,22,23,24,25,26,27. Two studies, in contrast, reported that relative growth rates were unaffected by e[CO2]28,29. Where declines in performance have been reported this has largely been attributed to lower plant nutritional (i.e. nitrogen) quality under e[CO2]20,21,23,24,25,27.
Similar responses may not hold true for all plant types, however. In their meta-analysis, Robinson, et al.4 showed that e[CO2] decreased N by just 10% in legumes, which was substantially less than reductions in N concentrations in non-leguminous plants (– 17%). This most likely stems from the fact that legumes form associations with N-fixing bacteria in their root nodules and e[CO2] often increases nitrogen fixation30,31. Any reductions in N concentrations of legumes arising under e[CO2] may therefore be less extensive than in non-leguminous plants4. e[CO2] may even lead to increases in some foliar amino acid concentrations where it substantially stimulates N-fixation32,33. Helicoverpa armigera attacks several legumes, including lucerne (alfalfa) (Medicago sativa L.), but to our knowledge no studies have yet addressed how e[CO2] affects H. armigera when feeding on this important forage legume.
The main objective of this study was to experimentally determine how e[CO2] affected M. sativa traits (biomass, root nodule abundance and density), nutritional chemistry (foliar carbon, nitrogen, protein and amino acid concentrations), defensive (JA) signalling and establish whether such changes were associated with changes in the growth rates of H. armigera. Our main hypothesis is that e[CO2] has negligible impacts on M. sativa nutritional status but depresses JA activity, which results in enhanced performance of H. armigera. Additionally, we conducted a simple a meta-analysis of previous studies to quantify overall effect of e[CO2] on the numerous H. armigera performance parameters reported to date.
Results
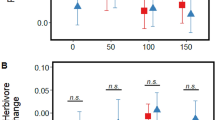
As detailed in the Methods, ambient CO2 (a[CO2]) and e[CO2] conditions were maintained at 400 and 640 ppm, respectively, and the experiment ran for eight weeks. Total plant biomass increased by an average of ca. 55% under e[CO2] (F1,4 = 11.30, P = 0.028), with this increase being reflected in the shoots (Fig. 1A). Root mass was unaffected by e[CO2] but root nodule abundance almost doubled under e[CO2] (Fig. 1B). In accordance with there being no change in root mass, but an increase in nodule numbers, there was an increase in nodule density on the roots of plants grown under e[CO2] compared to a[CO2] (58.9 and 41.0 nodules g-1 dry root mass, respectively) (F1,4 = 14.72, P = 0.019).
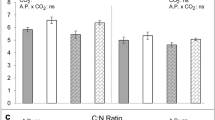
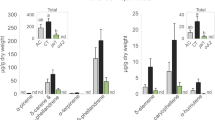
There was a significant decline (– 12%) in foliar concentrations of total nitrogen (Fig. 1C) under e[CO2], but no changes in carbon concentrations (F1,4 = 0.01, P = 0.929) resulting in a small but non-significant increase in foliar C:N ratio (Fig. 1D). Despite the decrease in nitrogen concentrations overall, soluble protein concentrations were very similar in plants grown under a[CO2] and e[CO2] conditions: 68.15 ± 2.65 and 66.45 ± 3.58 mg g−1, respectively (mean ± standard error, N = 12; F1,4 = 0.04, P = 0.846). Concentrations of total amino acids were also unaffected by CO2, (Fig. 2A; F1,4 = 0.01, P = 0.973). Both non-essential (Fig. 2B) and essential (Fig. 2C) amino acids were similarly unaffected by CO2 (F1,4 = 0.28, P = 0.624 and F1,4 = 3.41, P = 0.139, respectively), although there was a small decline in concentrations of tyrosine, arginine, leucine and phenylalanine under e[CO2] (Fig. 2C and Table S2).
Under a[CO2], herbivory by H. armigera triggered a sharp increase (ca. + 209%) in JA concentrations; this was not seen under e[CO2] and JA concentrations remained at similar levels as unchallenged plants grown under either CO2 regime (Fig. 3A). Concurrent with this, H. armigera developed significantly more rapidly (ca. 66% gain in mass per day) under e[CO2] compared to those feeding on plants grown at a[CO2] (Fig. 3B).
The meta-analytical comparison of the ten studies investigating the effects of e[CO2] on H. armigera performance (125 observations) quantitatively confirmed that there was an overall negative impact of e[CO2] on H. armigera performance and that no studies found positive effects (Fig. 4). This took into account all of the 125 performance parameters that were measured, including those that were unaffected or promoted by e[CO2]. The studies used a range (550–750 ppm) of CO2 concentrations for e[CO2] treatments with the average being 699.50 + 22.45 ppm (mean ± standard error). The e[CO2] level used in the current study was comparable with this (8.5% lower).
Meta-analytical summary of previous studies addressing the plant-mediated impacts of e[CO2] on H. armigera performance. Effect size (95% confidence interval) shown with the number of observations reported in parentheses. Points to the left indicate significant decreases in performance under e[CO2] relative to H. armigera feeding under a[CO2]. Studies: Sharma et al. (2016)20, Khadar et al. (2014)21, Chen et al. (2005)23, (2007)22, Gang et al. (2007)25, Coll et al. (2008)24, Hall et al. (2020)28, Wu et al. (2006)27, Yin et al. (2010)26 and Guo et al. (2012)29.
Discussion
Our results show that e[CO2] accelerates the growth rates of H. armigera when feeding on M. sativa, to our knowledge the first example of H. armigera benefitting from e[CO2]. The meta-analytical summary confirmed quantitatively that, taken together, existing studies indicate that e[CO2] causes declines in H. armigera performance. We suggest that the increase in H. armigera growth rates in e[CO2] we observed was associated with the suppressed JA response, which potentially triggers downstream plant defences, and the relatively modest impacts of e[CO2] on host plant nutritional quality compared to previous studies.
While we did observe a significant decline in total N concentrations in e[CO2], we saw no changes in soluble protein or total amino acids and only marginal (non-significant) increases in foliar C:N. The decline in total N concentrations might, in part, be due to the lower concentrations of the essential (arginine, leucine and phenylalanine) and non-essential (tyrosine) under e[CO2]. This might not necessarily represent a decline in nutritional quality however, and may even reflect lower levels of plant defences such as flavonoids, which are key defences in M. sativa. Flavonoids are a part of the phenylpropanoid pathway and are ultimately derived from phenylalanine and tyrosine34. Moreover, arginine plays a critical role in nitric oxide metabolism, which regulates defence responses to biotic stress35. In addition, given that protein concentrations were unaffected by CO2, this suggests that decreases in these specific amino acids was not substantial enough to hinder protein synthesis. Collectively, this suggests that e[CO2] impacts on nutritional quality was limited.
The greater abundance of root nodules housing N-fixing bacteria under e[CO2] may have contributed to N levels and therefore limited the negative impacts of e[CO2] on nutritional quality. e[CO2] can increase N-fixation via several mechanisms, including larger numbers of N-fixing bacteria36, increased nodulation37 and enhanced nitrogenase activity38. This effect depends, however, on other factors such as nutrient availability39 and air temperature31. The two studies that used legumes to investigate the impacts of e[CO2] on H. armigera reported declines in total N (–18 to 25%)21 or protein content (–18 to 28%)20 in the foliage. In contrast, we observed more modest declines in total N (–12%) and almost no change in protein (–2.5%) or amino acid (+ 0.6%) concentrations. The effects of e[CO2] on root nodulation and amino acid concentrations were not reported in the other studies, but it would seem unlikely that N-fixation was being stimulated in these experiments. It seems reasonable to assume in the current study that the nutritional quality of the M. sativa did not appreciably decline under e[CO2] in the current study using the Lepidopteran H. armigera. It remains possible, however, that herbivores with different feeding guilds (e.g. aphids) and other M. sativa genotypes might have been affected differently.
While we did not quantify specific defensive metabolites, our results are consistent with the findings of Guo, et al.29 who also reported that e[CO2] depressed JA responses against H. armigera in tomato. Interestingly, this did not affect the relative growth rates of H. armigera. Guo, et al.29 speculated that this may have been due to lower N concentrations of plants grown under e[CO2], which we suggest did not occur to the same extent in the current study. The combination of depressed JA without substantive changes in nutritional quality under e[CO2] therefore most likely explains why H. armigera performed better on lucerne grown under e[CO2]. There is a small possibility that increased RGR was due to increased leaf temperature under e[CO2], though presumably this would have also occurred in studies reporting reduced performance under e[CO2] so this seems unlikely.
The exact mechanism by which e[CO2] affects JA in other systems remains unclear5. One possibility is that JA patterns operate via circadian regulation, peaking during the day when photosynthesis rates are highest and intercellular CO2 is lowest40. Intracellular CO2 is lowest during the night which is linked to lower JA concentrations, so higher CO2 may more generally result in lower JA concentrations5. Moreover, e[CO2] has been linked to enhanced SA concentrations, which often has an antagonistic relationship with the JA pathway41.
In reporting these findings, we aim to highlight that the effects of e[CO2] on this important insect herbivore cannot be universally assumed to be negative or neutral even though this has consistently been the case in previous studies. In particular, compromised defence signalling (potentially leading to lower downstream production of defences) in e[CO2] relative to a[CO2], together with relatively modest declines in nutritional quality may explain enhanced H. armigera performance in some instances.
Methods
Chambers and environmental conditions
The study was conducted in six naturally lit glasshouse chambers (3 × 5 × 3 m; width × length × height) with UV transparent plexiglass (6 mm thick) walls and roof. Air temperature was regulated at 25 °C (± 0.25 °C). Humidity was controlled at 60% (± 6%). Three of the chambers were maintained at ambient CO2 (aCO2; 400 µmol mol−1) and three chambers at eCO2 (640 µmol mol−1). The elevated concentration reflects low-mid predictions for 2,100 depending on scenario42. Environmental conditions were monitored continuously throughout the experiment and temperature readings were verified with portable temperature loggers.
Experimental procedure
Lucerne (M. sativa cv. Sequel) was grown from seed (Seedmark, Adelaide, Australia) in 72 pots (70 mm diameter) filled with 700 g of native soil mix (Turtle Nursery and Landscape Supplies; see Table S1 for compositional details). These were distributed evenly between the six chambers, such that 36 plants were grown under either a[CO2] or e[CO2] (12 plants per chamber). Plants were watered with ca. 70 mL of tap water three times a week. After seven weeks, herbivores were applied to 12 plants grown under a[CO2] or e[CO2] (i.e. four plants in each of the six chambers). Specifically, H. armigera eggs (supplied by CSIRO, Narrabri) were individually hatched and reared on an artificial diet 43 at 20ºC under a 15:9 h photoperiod (L:D) for 7 days. One larva was weighed and applied to the base of the plants. White mesh (organza) bags (125 × 170 mm) were applied tightly around the rim of all pots (including herbivore free plants) to retain herbivores on their allocated plants. Seven days later, the herbivores were removed and reweighed before removing plants from the soil with root washing in water. Relative growth rates (RGR) were calculated [(change in mass / initial mass) / days]. The number of active (pink) root nodules was recorded for all of the herbivore-free plants. Roots and shoots from these plants were then snap frozen, freeze dried, ground and weighed prior to chemical analysis. All of the chemical analysis was conducted on ground tissue from herbivore-free plants using a sub-sample of the collective foliar material from each plant.
Chemical analysis
Twelve plant samples, selected at random across chambers, for both CO2 treatments were used to determine total carbon, nitrogen and soluble protein concentrations in ground foliage. Carbon and nitrogen were quantified (using ca. 6 mg material) with an elemental combustion analyser (FLASH EA 112 Series CHN analyser, Thermo-Finnigan, Waltham, MA, USA). For soluble protein analysis, modified from Jones, et al.44, 1 mL of 0.1 M NaOH was added to ca. 23 mg of material and homogenised at 25 °C for 30 min. The mixture was then centrifuged at 12,000 rpm for 5 min. The supernatant was removed and added to a clean microtube. A 1:4 dilution of each extract was made and dilutions were measured in technical triplicate on a on CLARIOstar High Performance Monochromator multimode microplate reader (BMG labtech, Offenburg, Germany) using the Bradford assay modified for a 96-well plate44,45. Protein concentrations were calculated using a standard curve of bovine serum albumin.
Fourteen plant samples, selected at random across chambers, for both CO2 treatments were used for amino acid analysis. Soluble amino acids were extracted from ca. 75 mg of foliar tissue with 525 μl 80% methanol, simultaneously heated and vortexed at 50 °C/850 rpm respectively. Samples were centrifuged and the supernatant filtered through 0.22 μm pore size nylon membrane. Underivatised amino acids were separated by reverse-phase high-performance liquid chromatography (HPLC) using an Agilent 1260 Infinity HPLC system equipped with an Agilent Poroshell 120 EC-C18 column (4.6 × 150 mm, 2.7 µm). Using a flow rate of 0.6 mL/min and an injection volume of 7 µl, analyte peaks were detected with a Corona charged aerosol detector (CAD; Corona CAD veo; Thermo Fisher Scientific Inc.) and eluted using two mobile phases (Solvent A: 0.4% heptafluorobutyric acid and 0.02% trifluoroacetic acid (TFA) in distilled water, Solvent B: 0.1% TFA in acetonitrile, modified from Furota, et al.46. Amino acid standards (0, 0.125 and 2 µmol 1–1) containing 16 amino acids were used to calibrate the analysis; arginine, histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine and valine are essential (i.e. unable to be synthesized by insects de novo) and seven are non-essential; alanine, asparagine, aspartic acid, glutamic acid, glutamine, proline and tyrosine.
The procedures used for phytohormone analysis have previously been described in Hall et al. (2020)28. Jasmonic acid (JA) was analysed in six samples, selected at random, for combinations of CO2 and herbivore treatments (24 in total). Samples were extracted using the Bligh-Dyer method47 to remove interfering compounds. Ground leaf material (ca. 50 mg) was mixed with 500 µL of 70% methanol and 100 ppb of deuterated JA (d5-JA) as internal standard. Samples were then mixed for 30 min at 4 ºC in a rotator mixer, after which 180 µl of chloroform was added and samples vortexed for 30 s. The last step was repeated and then 200 µl of water was added, samples were then centrifuged for 10 min at 6,000 rpm at room temperature. The water/methanol solution was pipetted to a clean 2 ml Eppendorf tube and filtered using a 0.22 µm PTFE syringe filter. The extracts were analysed using an Acuity Ultra Performance Liquid Chromatography (UPLC) coupled to a Xevo triple quadrupole mass spectrometer (Waters Corporation, Milford, USA). Five microliters from each sample were injected into a 2.1 mm × 50 mm × 1.7 µm, C18 reverse phase column. The mobile phase consisting of water (A) and acetonitrile (B) both containing 0.1% (v/v) formic acid was passed through the column at a constant flow rate of 0.6 mL min−1 over a linear gradient (A%, t min): 80% A at 0 min; 50% A at 2 min; 0% A and 2.1 min. JA was detected by Electrospray ionization tandem mass spectrometry (ESI–MS/MS) in negative ion mode. Identification of JA was determined based on the fragmentation pattern of an authentic JA standard. JA was quantified using a calibration curve of the JA standard which was adjusted for sample recovery based on the concentration of the internal standard. Final JA concentrations were standardised by dry weight of the sample. The internal standard, d5-JA, was purchased from CDN Isotopes (Quebec, Canada). HPLC grade methanol, chloroform, and JA were purchased from Sigma-Aldrich (MO, USA).
Statistical analysis
One-way ANOVAs with CO2 regime as the fixed factor were used to analyse most plant responses (biomass, nodule number and density, C, N, protein and amino acid concentrations) and herbivore RGR. For foliar JA concentrations a two-way ANOVA (CO2 × herbivory) was used with Fisher’s LSD test applied to determine differences between specific treatments. To avoid pseudoreplication, chamber was included as a block term with three chambers replicating each CO2 regime. Log transformations were applied for nodule number and nodule density to meet assumptions of normality and heteroscedasticity. Logit transformations were applied to C concentrations, foliar C:N ratio, some amino acids (see Table S2) for the same reasons. Satisfactory transformation was not possible in one instance and a Kruskal–Wallis test was applied (see Table S2 for details). Analysis was conducted using Genstat (version 18, VSN International, Hemel Hempstead, UK).
Meta-analysis
Original research papers were identified via searches on Web of Science and BIOSIS on 8 April 2020 using the search terms ‘carbon dioxide’ AND ‘Helicoverpa armigera’ together with ‘CO2’ AND ‘Helicoverpa armigera’. After removal of duplicates, 54 records were identified for initial screening (Fig. S1). Examination of titles and/or abstracts resulted in 36 studies being excluded on the grounds that they were not relevant or did not contain data (e.g. reviews). The remaining 18 full-text articles were assessed for eligibility, resulting in the removal of eight studies for specific non-compliance issues (listed in Fig. S1). Performance were parameters classified as abundance, feeding efficiency, growth/development, mortality/survival and reproduction were used. Numerical data were extracted from graphical figures using DigitizeIt (v2.3.3; Bormisoft, Braunschweig, Germany). For performance parameters where higher numerical values indicated poorer herbivore performance (e.g. mortality), a negative sign was applied to the value. We used responses measured on plants that excluded other treatments (e.g. transgenic Bt or silicon supplementation) and selected one performance parameter in the few cases where responses were essentially duplicates (e.g. relative growth rate and mean relative growth rate).
Meta-analysis were conducted using the package metafor48 in the R statistical platform. The effect size (Hedges’ d) was calculated for each pair of performance responses (i.e. at a[CO2] and e[CO2]). Where more than one e[CO2] level was applied, both levels were included as separate entries. This measure of effect size compares two means using a pooled standard deviation and bias correction and reflects the number of standard deviations by which the means differ49. Positive values arise when herbivores performed better on plants grown in e[CO2] compared to control plants (a[CO2] plants) whereas negative values indicate the opposite (i.e. they perform worse on e[CO2] grown plants).
Data availability
All meta-analysis and empirical data are posted on the figshare repository https://doi.org/10.6084/m9.figshare.12480068.
Change history
07 December 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Gregory, P. J., Johnson, S. N., Newton, A. C. & Ingram, J. S. I. Integrating pests and pathogens into the climate change/food security debate. J. Exp. Bot. 60, 2827–2838 (2009).
Birch, A. N. E., Begg, G. S. & Squire, G. R. How agro-ecological research helps to address food security issues under new IPM and pesticide reduction policies for global crop production systems. J. Exp. Bot. 62, 3251–3261 (2011).
Johnson, S. N. & Jones, T. H. Global Climate Change and Terrestrial Invertebrates (John Wiley & Son Ltd., New York, 2017).
Robinson, E. A., Ryan, G. D. & Newman, J. A. A meta-analytical review of the effects of elevated CO2 on plant-arthropod interactions highlights the importance of interacting environmental and biological variables. New Phytol. 194, 321–336 (2012).
Zavala, J. A., Nabity, P. D. & DeLucia, E. H. An emerging understanding of mechanisms governing insect herbivory under elevated CO2. Annu. Rev. Entomol. 58, 79–97 (2013).
Ode, P. J., Johnson, S. N. & Moore, B. D. Atmospheric change and induced plant secondary metabolites—Are we reshaping the building blocks of multi-trophic interactions? Curr. Opin. Ins. Sci. 5, 57–65 (2014).
DeLucia, E. H., Nabity, P. D., Zavala, J. A. & Berenbaum, M. R. Climage change: Resetting plant–insect interactions. Plant Physiol. 160, 1677–1685 (2012).
Facey, S. L., Ellsworth, D. S., Staley, J. T., Wright, D. J. & Johnson, S. N. Upsetting the order: How climate and atmospheric change affects herbivore–enemy interactions. Curr. Opin. Insect Sci. 5, 66–74 (2014).
Newman, J. A., Anand, M., Henry, H. A. L., Hunt, S. & Gedalof, Z. Climate Change Biology (CABI, 2011).
Mattson, W. J. Herbivory in relation to plant nitrogen content. Annu. Rev. Ecol. Syst. 11, 119–161 (1980).
Drake, B. G., Gonzalez-Meler, M. A. & Long, S. P. More efficient plants: A consequence of rising atmospheric CO2? Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 609–639 (1997).
Stiling, P. & Cornelissen, T. How does elevated carbon dioxide (CO2) affect plant–herbivore interactions? A field experiment and meta-analysis of CO2-mediated changes on plant chemistry and herbivore performance. Glob. Change Biol. 13, 1823–1842 (2007).
Pang, J. et al. A new explanation of the N concentration decrease in tissues of rice (Oryza sativa L.) exposed to elevated atmospheric pCO2. Environ. Exp. Bot. 57, 98–105 (2006).
Taub, D. R. & Wang, X. Z. Why are nitrogen concentrations in plant tissues lower under elevated CO2? A critical examination of the hypotheses. J. Integr. Plant Biol. 50, 1365–1374 (2008).
Howe, G. A. & Jander, G. Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41–66 (2008).
Wu, J. Q. & Baldwin, I. T. New insights into plant responses to the attack from insect herbivores. Annu. Rev. Genet. 44, 1–24 (2010).
Erb, M., Meldau, S. & Howe, G. A. Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 17, 250–259 (2012).
Anderson, C. J. et al. Hybridization and gene flow in the mega-pest lineage of moth, Helicoverpa. Proc. Natl. Acad. Sci. U.S.A. 115, 5034–5039 (2018).
Jones, C. M., Parry, H., Tay, W. T., Reynolds, D. R. & Chapman, J. W. Movement ecology of pest Helicoverpa: Implications for ongoing spread. Annu. Rev. Entomol. 64, 277–295 (2019).
Sharma, H. C. et al. Elevated CO2 influences host plant defense response in chickpea against Helicoverpa armigera. Arthropod-Plant Interact. 10, 171–181 (2016).
Khadar, B. A., Prabhuraj, A., Rao, M. S., Sreenivas, A. G. & Naganagoud, A. Influence of elevated CO2 associated with chickpea on growth performance of gram caterpillar, Helicoverpa armigera (Hüb.). Appl. Ecol. Environ. Res. 12, 345–353 (2014).
Chen, F., Wu, G., Parajulee, M. N. & Ge, F. Long-term impacts of elevated carbon dioxide and transgenic Bt cotton on performance and feeding of three generations of cotton bollworm. Entomol. Exp. Appl. 124, 27–35 (2007).
Chen, F. J., Wu, G., Ge, F., Parajulee, M. N. & Shrestha, R. B. Effects of elevated CO2 and transgenic Bt cotton on plant chemistry, performance, and feeding of an insect herbivore, the cotton bollworm. Entomol. Exp. Appl. 115, 341–350 (2005).
Coll, M. & Hughes, L. Effects of elevated CO2 on an insect omnivore: A test for nutritional effects mediated by host plants and prey. Agric. Ecosyst. Environ. 123, 271–279 (2008).
Gang, W., Chen, F. J., Sun, Y. C. & Feng, G. Response of successive three generations of cotton bollworm, Helicoverpa armigera (Hübner), fed on cotton bolls under elevated CO2. J. Environ. Sci. 19, 1318–1325 (2007).
Yin, J., Sun, Y. C., Wu, G. & Ge, F. Effects of elevated CO2 associated with maize on multiple generations of the cotton bollworm, Helicoverpa armigera. Entomol. Exp. Appl. 136, 12–20 (2010).
Wu, G., Chen, F. J. & Ge, F. Response of multiple generations of cotton bollworm Helicoverpa armigera Hübner, feeding on spring wheat, to elevated CO2. J. Appl. Entomol. 130, 2–9 (2006).
Hall, C. R., Mikhael, M., Hartley, S. E. & Johnson, S. N. Elevated atmospheric CO2 suppresses jasmonate and silicon-based defences without affecting herbivores. Funct. Ecol. 34, 993–1002 (2020).
Guo, H. J. et al. Elevated CO2 reduces the resistance and tolerance of tomato plants to Helicoverpa armigera by suppressing the JA signaling pathway. PloS One 7, e41426, https://doi.org/10.1371/journal.pone.0041426 (2012).
Soussana, J. F. & Hartwig, U. A. The effects of elevated CO2 on symbiotic N2 fixation: A link between the carbon and nitrogen cycles in grassland ecosystems. Plant Soil 187, 321–332 (1996).
Johnson, S. N., Gherlenda, A. N., Frew, A. & Ryalls, J. M. W. The importance of testing multiple environmental factors in legume-insect research: Replication, reviewers and rebuttal. Front. Plant Sci. 7, 489, https://doi.org/10.3389/fpls.2016.00489 (2016).
Guo, H. et al. Pea aphid promotes amino acid metabolism both in Medicago truncatula and bacteriocytes to favor aphid population growth under elevated CO2. Global Change Biol. 19, 3210–3223 (2013).
Johnson, S. N., Ryalls, J. M. W. & Karley, A. J. Global climate change and crop resistance to aphids: contrasting responses of lucerne genotypes to elevated atmospheric carbon dioxide. Ann. Appl. Biol. 165, 62–72 (2014).
Deng, Y. & Lu, S. Biosynthesis and regulation of phenylpropanoids in plants. Crit. Rev. Plant Sci. 36, 257–290 (2017).
Winter, G., Todd, C. D., Trovato, M., Forlani, G. & Funck, D. Physiological implications of arginine metabolism in plants. Front. Plant Sci. 6, 534, https://doi.org/10.3389/fpls.2015.00534 (2015).
Schortemeyer, M., Hartwig, U. A., Hendrey, G. R. & Sadowsky, M. J. Microbial community changes in the rhizospheres of white clover and perennial ryegrass exposed to Free Air Carbon dioxide Enrichment (FACE). Soil Biol. Biochem. 28, 1717–1724 (1996).
Ryle, G. J. A. & Powell, C. E. The influence of elevated CO2 and temperature on biomass production of continuously defoliated white clover. Plant Cell Environ. 15, 593–599 (1992).
Norby, R. J. Nodulation and nitrogenase activity in nitrogen-fixing woody plants stimulated by CO2 enrichment of the atmosphere. Physiol. Plantarum 71, 77–82 (1987).
Edwards, E. J., McCaffery, S. & Evans, J. R. Phosphorus availability and elevated CO2 affect biological nitrogen fixation and nutrient fluxes in a clover-dominated sward. New Phytol. 169, 157–167 (2006).
Goodspeed, D., Chehab, E. W., Min-Venditti, A., Braam, J. & Covington, M. F. Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci. U.S.A. 109, 4674–4677 (2012).
Thaler, J. S., Humphrey, P. T. & Whiteman, N. K. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17, 260–270 (2012).
IPCC. Climate Change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change (IPCC, 2014).
Teakle, R. E. & Jensen, J. M. in Handbook of Insect Rearing, Vol. 2 (eds R. Singh & R.F. Moore) 312–322 (Elsevier, London, 1985).
Jones, C. G., Hare, J. D. & Compton, S. J. Measuring plant protein with the Bradford assay. 1. Evaluation and standard method. J. Chem. Ecol. 15, 979–992 (1989).
Bradford, M. M. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Furota, S., Ogawa, N. O., Takano, Y., Yoshimura, T. & Ohkouchi, N. Quantitative analysis of underivatized amino acids in the sub- to several-nanomolar range by ion-pair HPLC using a corona-charged aerosol detector (HPLC-CAD). J. Chromatogr. B 1095, 191–197 (2018).
Bligh, E. G. & Dyer, W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959).
Viechtbauer, W. Conducting meta-analyses in R with the metafor Package. J. Stat. Softw. 36, 1–48 (2010).
Hedges, L. V. & Olkin, I. Statistical Methods for Meta-Analysis (Academic Press, New York, 1985).
Acknowledgements
The Invertebrate Biology class of 2017 at Western Sydney University are thanked for collecting the data with the assistance of technical officers (Michael Duncan and Sharleen Knox) and demonstrators (Rhiannon Rowe, Barbara May, Scott Bevins, Caitlyn Drayton-Taylor, Giles Ross, Jon Finch and Jess Ezveld). Meena Mikhael is thanked for assistance with the phytohormone analysis.
Funding
This study was funded by Australian Research Council (FT17 0100342).
Author information
Authors and Affiliations
Contributions
S.N.J. designed the study and oversaw data collection. J.M.W. and C.R.H. conducted the chemical analysis. S.N.J. undertook analysis and prepared the manuscript with J.M.W. and C.R.H.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Johnson, S.N., Waterman, J.M. & Hall, C.R. Increased insect herbivore performance under elevated CO2 is associated with lower plant defence signalling and minimal declines in nutritional quality. Sci Rep 10, 14553 (2020). https://doi.org/10.1038/s41598-020-70823-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-70823-3
This article is cited by
-
Hyposidra talaca (Geometridae: Lepidoptera) outbreak in tea gardens: management strategies and future prospects
Journal of Plant Diseases and Protection (2024)
-
Benefits of silicon-enhanced root nodulation in a model legume are contingent upon rhizobial efficacy
Plant and Soil (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.