Abstract
While carbon-based materials have spearheaded numerous breakthroughs in biomedicine, they also have procreated many logical concerns on their overall toxicity. Carbon dots (CDs) as a respectively new member have been extensively explored in nucleus directed delivery and bioimaging due to their intrinsic fluorescence properties coupled with their small size and surface properties. Although various in vitro/in vivo studies have shown that CDs are mostly biocompatible, sufficient information is lacking regarding genotoxicity of them and underlying mechanisms. This study aims to analyze the real-time cytotoxicity of super tiny CDs (2.05 ± 0.22 nm) on human breast cancer cells (MCF7) and human primary dermal fibroblast cell cultures (HDFa) by xCELLigence analysis system for further evaluating their genotoxicity and clastogenicity to evaluate the anti-tumor potential of CDs on breast adenocarcinoma. As combined with flow cytometry studies, comet assay and cytokinesis-block micronucleus assay suggest that the CDs can penetrate to the cell nuclei, interact with the genetic material, and explode DNA damage and G0/G1 phase arrest in cancer cells even at very low concentrations (0.025 ppm) which provide a strong foundation for the design of potentially promising CD-based functional nanomaterials for DNA-damage induced treatment in cancer therapy.
Similar content being viewed by others
Introduction
Functional nanomaterials that can better target cancer cells that will improve prevention and therapy could be accomplished by the combined efforts of nanotoxicologists, cancer biologists and nanobiomaterial scientists focused on toxicology and related cancer therapy. Engineering carbon-based nanomaterials and their applications are among the most dynamic fields in modern advanced materials science and engineering1,2,3. To date, various carbon nanomaterials such as carbon nanotubes4,5, fullerenes6, graphene7,8, graphene oxides9, carbon diamonds10,11, and carbon dots12,13 have been synthesized and reported by various researchers. Among these, CDs have become particularly interesting because of their unique physical and chemical properties such as thermal and electrical conductivity, high mechanical strength together with their unique optic and fluorescence features14,15,16. They have been used in bio-imaging, drug delivery, nucleus targeting, and labeling, photodynamic therapy, optoelectronics, solar cells, photocatalyst design, photodetectors, and many other biological and engineering fields17,18,19,20,21,22,23,24,25,26.
Several methods are available for the synthesis of CDs, namely, laser ablation15, electro-oxidation27 or oxidative acid treatment12; while nowadays, less laborsome and cheaper methods such as hydrothermal28 and thermal synthesis24,29,30, microwave-assisted hydrothermal synthesis31, ultrasound synthesis32, etc. are popularly exercised. CDs can be synthesized either from inorganic materials19,33 such as graphene34, carbon black35, and candle soot12 or biological carbon sources like food wastes, fruits, seeds and shells, plant extracts have been moving toward their application to biological sciences36,37,38.
Toxicity studies by various research groups have shown that CDs exhibit very low toxicity compared to heavy metal-based quantum dots when internalized by the cells26,39,40,41,42,43. In almost all of the cytotoxicity studies performed to date, CDs have been demonstrated to cause a negligible effect on cell viability at concentrations sufficient for drug delivery and bio-imaging. In the last 2 years, we have been focused on new types of CDs by tailoring the surface properties of them with the target of evaluating their potential in biological applications16,44,45. Very recently, utilizing different polymers as surface passivating agent and surface doping agent, we were able to show the immunostimulant and adjuvant-like effect of such CDs where no cytotoxic effect was observed44,45. CDs have been widely researched for nucleus-targeted delivery, nucleus labeling, photodynamic therapy, and optical monitoring of anticancer drugs46,47,48,49,50, however, so far there are few scientific reports in existence for addressing the genotoxic activities of CDs when they administrated on their own51,52,53,54,55,56. In a recent study, researchers showed the genotoxic responses of rat alveolar macrophages (NR8383) to amine-grafted graphene QDs which resulted in significant alterations in the expression of 2,898 genes after exposure for 24 h in which most of the down-regulated genes were reported the as they were responsive to “cell cycle”57. In a study on the use of graphene quantum dots (GQDs) as nucleus labeling of several cell lines (L929, HT-1080, MIA PaCa-2, HeLa, and MG-63 cells), researchers showed that internalization mechanism of the GQDs by healthy cells differ from tumor cells while GQDs showed to be entering into the nucleus regardless of the cell type. Moreover, the authors showed an altered number of L929 cells in the S phase as an indication of promoted cell proliferation in the presence of GQDs50. Yue et al.25 reported that ruthenium incorporated CDs with no apparent cytotoxicity have the concentration-dependent DNA photocleavage ability on HeLa cells upon the light irradiation (6.5 mW/cm2) showing the potential of CDs in imaging and photodynamic therapy (seeTable 2).
Nerium oleander (Oleander) is one of the most poisonous dwarf evergreen shrubs in the world58. Extracts from various parts of the plant show also anti-cancer59,60, anti-microbial61, anti-inflammatory62, anti-diabetic63, and neuroprotective activities62. Common ingredients of Oleander extract include polysaccharides containing rhamnose, galactose, arabinose, mannose, glucose, and galacturonic acid64,65,66,67. Other components and their concentrations may vary depending on the extraction method68. For instance, extracts of N. oleander leaves contain steroids, flavonoids, and terpenoids, etc.67. Our group lately presented the synthesis routes for Oleander based CDs using both thermal and microwave-based synthesis methodologies69,70 while we have shown that extract type (water or ethanol extraction) is one of the important parameters where the highest fluorescence and the lowest size was observed using water-based Oleander extract as a carbon source for CD synthesis.
The present study covers the effects of super tiny CDs on the cell viability of MCF7 tumor cells and normal HDFa cells together with the CD-induced differentiation in cell-cycle progression, genotoxicity, and clastogenicity on MCF7 cells. Our results suggest that CDs, alone or in combination with chemotherapeutics, may be exploited for the development of potentially promising functional nanomaterials for DNA-damage induced treatment in cancer therapy. They have the potential that could be extended to be used as new generation biolabeling and imaging agents as well. However, the possible influence of the cell cycle on cellular uptake of CDs and the mechanism of its effect on MCF7 cells needs further investigation.
Materials and methods
Plant material
Nerium oleander leaves collected from Esenler Region, Istanbul (41°01′37.70"N, 28°53′32.1"E) at 82 m May 2016 were booked in Izmir, Ege University Faculty of Pharmacy Herbarium (IZEF) with number 6056.
Chemicals
Ethanol and Polyethylene Glycol (PEG 10000N), Dulbecco's Modified Eagle's medium (DMEM), trypan blue solution, Dulbecco’s Phosphate Buffered Saline (DPBS), agarose with normal melting point and low melting point, dimethyl sulphoxide, ethidium bromide, Triton X-100, phosphate-buffered saline tablets, potassium chloride (KCl), Giemsa, ethylenediaminetetraacetic acid disodium salt dihydrate (Na2-EDTA) and cytochalasin B (Cyt-B), the positive control for the genotoxicity assays, ethyl methanesulphonate (EMS) (CAS no. 62-50-0, lot 1338043) were obtained from Sigma-Aldrich (Steinheim, Germany). Trypsin Buffer, Tyrosine Inhibitor Buffer, RNase Buffer, Propidium Iodide Stain Solutions were purchased from Becton Dickinson (BD). Sodium chloride and sodium hydroxide were purchased from Merck Chemicals (Darmstadt, Germany), whereas Chromosome medium B was purchased from Biochrom AG (Berlin, Germany). Frosted microscope slides were obtained from Menzel GmbH (Braunschweig, Germany. Human breast adenocarcinoma cell line (MCF7) and the human primary dermal fibroblast cell cultures (HDFa) were obtained from ATCC with number HTB-22 and PCS-201-12, respectively. Slides were visualized for Comet Assay by fluorescence microscopy using an Olympus BX51 System equipped with a video camera CCD-4230.
Equipment
ELMA TI-H 5 model ultrasonic bath was used during the extraction process. The thermal synthesis was conducted using a Neuve muffle furnace. Characterization studies of CDs were performed on a Shimadzu UV-1800 UV–Vis spectrophotometer, Agilent Cary Eclipse fluorescence spectrophotometer, Malvern Zeta Sizer Nano ZS, and Perkin Elmer frontier FT-IR. X-ray photoelectron spectroscopy (XPS) screening was performed using the Specs-Flex XPS spectrometer (Al Kα 1,486.7 eV). Morphology of CDs was monitored by a JEOL JEM-1400 series 120 kV Transmission Electron Microscope (TEM) and the FEI Tecnai G2 F30 HR-TEM at 300 kV. Particle core radius was calculated by measuring at least 100 individual particles using Image J program. Cell-seeding calculations were carried out with the Cedex XS analyzer (Innovatis Inc.). xCELLigence system (ACEA Biosciences Inc.) was used as a real-time cell sorter. BD FACSAria III flow cytometer (BD Biosciences, US) and BD CELLQuest Pro software (BD Biosciences, US) were used for cell-cycle analysis. Fluorescence imaging was performed by a fluorescence microscope (Olympus BX51) equipped with a CCD-4230 video camera.
Preparing plant extracts
The fresh leaves of N. oleander were washed twice with distilled water and dried at incubator at 70 °C for 2 days. Dried leaves were then grinded to powders. The water extraction procedure was performed using ultra-pure water with the final concentration 12.5 g dry leaf/100 mL in ddH2O in an ultrasonic bath for 5 h at room temperature (RT). After extraction, clear extracts were obtained by centrifugation at 5,000 rpm for 15 min and stored at + 4 °C.
Synthesis of CD using N. oleander leaf extract
1 mL aqueous extract was dispersed 1 g PEG solution pre-prepared in 2 mL of ddH2O/ethanol solution (1:1 v/v). The mixture then placed in a muffle furnace and left for the caramelization process for 45 min at 300 °C. Resulting brown solid was cooled down to RT and dissolved in 6 mL ddH2O. CDs were separated by three cycles of centrifugation at 13,500 rpm for 20 min. The supernatants were collected and dried in a vacuum oven at 60 °C for 1–2 days.
Quantum yield of CDs
The quantum yield of the CD (diluted samples to obtain an absorbance value of less than 0.10) was determined using quinine sulfate in 0.1 M H2SO4 (quantum yield: 54%) as the standard sample following the procedure published previously71.
In vitro cytotoxicity of CDs on MCF7 and the HDFa cell lines by xCELLigence
The xCELLigence system was used according to the manufacturer's instructions. The cell index (CI) was obtained by measuring the change in the electrical impedance in the presence and absence of cells in the wells. MCF7 and HDFa cell lines were inoculated keeping the cell number as 1 × 104 cells/well and 3 × 104 cells/well, respectively, on 16-well plates of the xCELLigence system. Afterward, cell growth in each well of e-plates was monitored every 15 min and analyzed with RTCA Software 1.2. After ~ 18–24 h of cell transplantation, cells in the ‘logarithmic growth phase’ were exposed to varying concentrations of CDs dispersed in DMSO (0.0025 ppm, 0.025 ppm, 0.25 ppm, 2.5 ppm, and 50 ppm), and monitored real-time for 72 h. The cells growing in the growth media were used as control while 50 ppm aqueous of N. oleander extract was used to compare with CDs. Experiments were carried out at least quadruplicate. All experimental steps were conducted under dark conditions to prevent additional light-induced cellular damages.
In vitro genotoxicity of CDs on MCF7 cells by comet assay (SCGE)
Using the information obtained from the cytotoxicity studies, CD concentrations of 0.25, 2.5, and 50 ppm were selected for performing the alkaline comet assays. MCF7 cells in log-phase were plated in 96-well plates and incubated for 2 days. The negative control (Untreated cells that were grown in growth media, NC), and the positive control (Cells treated with 20 mM hydrogen peroxide) were inoculated in series. The comet assay was performed in alkaline conditions (pH > 13) as described previously72. Briefly, after cells were exposed to the CDs for 48 h and 72 h, the cells were collected and trypsinized. Centrifuged cells at 1,100 rpm for 5 min were counted and 1–3 × 104 cells were resuspended in 75 μL molten 0.5% low-melting-point agarose at 37 °C. The resuspended cells in agarose were put onto dry microscope slides pre-coated with 1% normal-melting agarose, and the agar solidified by keeping at RT for 10 min. The slides were thereafter immersed in cold lysing solution (2.5 M NaCl, 100 mM EDTA, 10 mM Tris, 1% Triton X-100) for at least 1 h at 4 °C. Afterward, they were transferred to an electrophoresis tank containing freshly-made electrophoresis buffer (1 mM EDTA, 300 mM NaOH; pH > 13), where they were kept for 20 min at room temperature to allow DNA unwinding. Electrophoresis was performed in the same buffer at RT for 15 min at 24 V and 300 mA (0.8 V/cm). The slides were then neutralized thrice with 0.4 M Tris buffer (pH 7.5), air-dried, and fixed in ethanol. All preparative steps were conducted under dark conditions to prevent additional DNA damages. Slides stained with ethidium bromide (0.1 mg/mL, 1:4) were analyzed under a fluorescence microscope equipped with a CCD-4230 video camera.
Slide scoring in comet assay
Comet images were analyzed following the method reported by Collins et al.73. The percentage of DNA in the comet tail from 100 cells per sample (duplicate, each with 50 cells/slide) was used as a measure of the amount of DNA damage. An intensity score from class 0 (undamaged) to class 4 (ultra-high damage) was assigned to each cell74. Observational blindness was employed, with the identity of the samples being withheld from the observer. Fifty cells per slide and two slides for each sample were examined to evaluate the DNA damage for each culture treated with CDs at different concentrations. The cells were classified by eye in the five categories based on the extent of DNA migration, undamaged (class 0), very little damage (class 1), moderate damage (class 2), high damage (class 3) ultrahigh damage (class 4). The ‘‘Arbitrary Unit (AU)’’ were used to express the extent of DNA damage and calculated using the following formula:
Ni = the number of scored cells in level i, i = the level of DNA damage (0, 1, 2, 3, 4).
In vitro clastogenecity of CDs on MCF7 cells by cytokinesis-block micronucleus assay (CBMN)
The in vitro cytokinesis-block micronucleus assay (CBMN) was performed based on previously published procedures with minor modifications75. Cells were seeded at a density of 5 × 104 cells per well in a T25 flask and treated with CDs in dispersion. After the exposure period of 48 h and 72 h, cells were washed twice with PBS and re-incubated for 38 h in fresh medium containing 3 μg/mL Cytochalasin B. Further, cells were harvested and suspended in ice-cold KCl for 50 s at room temperature. The cells were then fixed in Carnoy’s solution (1:3 mixtures of acetic acid and methanol), and several drops of formaldehyde were added to preserve the cytoplasm. Immediately after centrifugation at 1,500 rpm (1,000g) for 10 min, cells were fixed again in Carnoy’s solution. Finally, the cells were dropped onto clean microscopic slides, air-dried, and stained with Giemsa. A total of 1,000 binucleated cells for each sample were examined microscopically for micronuclei as previously described76. The levels of chromosomal damage were reported as the fold induction of micronuclei compared with the untreated control. CBMN results were accepted only when (1) they were separated from the main nuclei, but included within the corresponding cytoplasm, (2) they had a chromatin material similar to that of the main nuclei, (3) they were coplanar to the main nuclei, (iv)they were 1/16th to 1/3rd of the mean diameter of the main nuclei. In the CBMN study, toxicity was evaluated by classifying cells according to the number of nuclei77.
Flow cytometry: cell cycle analysis of MCF7 cells treated with CDs
MCF7 breast cancer cells treated with CDs were analyzed by flow cytometry to determine the associated DNA index (DI) and to determine cell cycle phase distributions in these cells. 1 × 106 cell/mL cells were plated in 6-well cell culture plates and treated with CDs at three different concentrations (0.25, 2.5, and 50 ppm) and incubated for 48 h and 72 h at 37 °C, 5% CO2. The cells were detached with trypsin, centrifuged (400g, 5 min.) and washed with PBS. Cells were then collected by centrifuge and treated with 250 µL trypsin buffer and vortexed. The obtained solution was mixed with Tyrosine Inhibitor and RNase Buffer (200 µL) and the mixture was incubated for 10 min. Further, 200 μL cold (2–8 °C) Propidium Iodide (PI) Stain Solution was added into each tube and incubated for an additional 10 min at 4 °C in dark with rapid stirring. The data were analyzed by Flow Cytometry Analysis Software. Values were expressed as fractions of cells in cell cycle phases (the mean ± standard error). Each experiment was performed three times.
Statistical analysis
At least three independent experiments were carried out in triplicate for each evaluation. Data were expressed as the mean ± error (SE) and analyzed by repeated-measures ANOVA followed by the least significant difference post hoc test Bonferroni for the comet assay. An independent t test was used for the CBMN test. In all tests for comet assay and CBMN test differences were considered significant at p < 0.001 and p < 0.05 respectively. All the data analysis was carried out using software STATISTICA for comet assay and STATA MP/11 for the CBMN test.
Results and discussion
Characterization of physicochemical properties of CDs
Many studies have attempted to elucidate the mechanisms of nanoparticle toxicity and distinguish between their bulk counterparts78,79,80. It is well-known that the toxicity of nanoparticles (NPs) was highly dependent on their physicochemical characteristics81. Table 1 shows some physical properties of as-synthesized and purified CDs (Fig. 1). PEG was used as a surface passivation agent which mainly acts as surface functionalization precursors for developing highly tunable photoluminescence properties by stabilizing the dangling bonds and controlling the surface functional groups and surface states15,82,83. Use of N. Oleander as carbon source and PEG as passivating agent revealed super tiny CDs with a hydrodynamic size (Rh) of 2.05 ± 0.22 nm (Fig. 2a) and core radius of 1.79 ± 0.33 nm measured by DLS and TEM imaging, respectively. The HR-TEM image (inset in Fig. 1) shows that the CDs displayed a highly crystalline structure with a 0.21 nm lattice spacing that is attributed to the graphitic (sp2) carbon84. Surface ζ-potential of − 23.5 ± 6.21 mV which could be the sign of a surface with the higher density of oxygen-rich groups74,75.
Schematics showing the experimental procedure of CD preparation from N. Oleander aqueous extracts and TEM and HR-TEM (Inset) images of as-synthesized CDs displaying a highly crystalline structure with a 0.21 nm lattice spacing that is attributed to the graphitic (sp2) carbon. Inset: photograph of the CDs emitting green colored fluorescence under a UV beam of 365 nm.
CDs are exceptional carbon-based materials with superior optical properties that can be used in biomedical applications as they are trackable by fluorescence imaging. UV–Vis spectrum of CDs was presented in Fig. 2b. CDs showed a broad absorption peak located at around 245 nm which corresponds to a typical absorption of sp2 carbon network which was also supported by the HR-TEM imaging (Fig. 1)16,85,86. As can be seen in Fig. 2c, CDs showed the highest fluorescent emission at 466 nm upon excitation at 365 nm with the highest quantum yield of 7.12%. The fluorescence emission spectra of samples demonstrated an excitation-dependent feature originates from a combination of quantum confinement effects and the distribution of different emissive surface traps presented on the surface of CDs82,87.
FT-IR spectroscopy was carried out to characterize the surface properties of CDs. As depicted in Fig. 2d, CD showed a sharp peak around 3,600 cm−1 corresponds to free O–H groups present on the particle surface. The as-prepared CDs showed peaks belong to C–H stretches in methyl and methylene groups (2,800–3,000 cm−1)88. Peaks corresponding to C–O stretching located at 1,075–1,250 cm−1 might be associated with the partial oxidation of CD surfaces. Sharp peaks at 1,380 and 1,460 cm−1 were attributed to CH2 vibrations (Fig. 2d). Many oxygen-rich functional groups including C–O–C (1,150 cm−1), C–OH (1,250 cm−1) and C–OH stretching peak at 1,380 cm−1 could be indicative of a C–O–C asymmetric stretch or C–H bending arising from a methyl functional groups presenting on the CDs35,49,89.
Figure 3a depicts the XPS wide scan spectrum of the synthesized CDs. Two bands of the XPS survey spectrum at around 284.5 eV and 531.5 represented O1s and C1s, respectively, which indicates the atomic ratio of O/C is 32.8/67.2 as calculated from the survey spectrum. The high-resolution C1s XPS spectrum (Fig. 3b) was deconvoluted into three contributions at 283.4, 284.7, and 285.4 eV, which are associated with carbon in the states of sp2 C (C=C, C–C) and C–OR, respectively90,91,92. The deconvoluted O1s spectrum (Fig. 3c) had three components peaking at 529.4, 530.9, and 531.8 eV, which are due to the C=O, C–OH, and C–O–C groups, respectively91,92. XPS results support the UV–Vis spectrophotometry and FT-IR results that CDs are purely composed of C=C core and have a highly oxygenated and reactive surface.
In vitro cytotoxicity of CDs on MCF7 and HDFa cells
Although MTT assay is one of the most used methods for the assessment of in vitro cytotoxicity of materials, there are reports on the interference of carbon-based nanomaterials with the colorimetric assays93,94,95. The xCELLigence is a technology that gives the possibility to measure the cellular growth in real-time by measuring the net adhesion of cells on a custom-designed gold electrode following the changes in electrical impedance. Thus, it gives more pre-sized information on the long-term screening of cell viability and prevents the material-dye interaction-based false responses96 The cytotoxicity of the CDs were conducted on MCF7 and HDFa cells. The normalized cell index (CI) is indicative of the level of adhesion and therefore associated with the viability of the cells.
Figure 4a,b illustrates the normalized cell index of the MCF7 treated with varying concentrations of the CDs (0.0025–50 ppm) as compared to the PBS as the negative control. In all cases, cells were treated after 24 h following the seeding and the cell index was normalized at the point of treatment. The Oleander extract as control of the starting material resulted in cell death within a few hours after the treatment resulting in a sharp decrease in CI. After 24 h of treatment, CD at the highest concentration (50 ppm) revealed a significant decrease in cell growth as compared to the untreated controls. At lower concentrations, CDs did not show a similar growth trend as the untreated cells while no dose-dependent response was observed even after 72 h of treatment. These results agree well with experimental data taken from the literature that increased negative net charge and small size of CD increased the risk of cytotoxicity of them to tumor cells at high doses49,97. On the other hand, the story in the case of HDFa cells was different. As depicted in Fig. 4c, CDs did not show any cytotoxic effect at neither of the concentrations but also they significantly induced cell proliferation at CD concentrations of 0.25 ppm and below, while at 72 h of treatment with CDs even at the highest concentration resulted in increased cell proliferation which reveals that presence of CDs protected the cells from natural death. The CI value increased sixfold for the cells exposed to CDs at a concentration as low as 0.25 ppb as compared to the untreated cells. Oleandrin extract (50 ppm) was, as expected, showed to be cytotoxic for both of the cells regardless of the time of the exposure. Although the main reason is not clear yet, higher tolerability of non-cancerous cells, decreased or retarded cellular accumulation of CDs in healthy cells as compared to the cancerous cells have been highlighted in many recent reports to explain the enhancement of the cell proliferation of healthy cells after the CD exposure50,81,98,99,100. Yao et al.101 showed that CDs specifically interact with some cellular proteins of tumor cells and downregulate the level of some proteins and the activity of enzymes that are related to tumor cell invasion ability.
Time-dependent changes in cell index values and the average cell index values for (a), (b) MCF7 cells, and (c) HDFa cells at 48 h and 72 h after the treatment with varying concentrations of CDs as compared to the negative control (medium) and positive control (water extract of Oleandrin (50 ppm). Values represent mean ± SE, n = 3.
Evaluation of genotoxicity and clastogenicity of CDs
The genotoxicity and clastogenicity of CDs on MCF7 cells were investigated by comet assay and CBMN, respectively. Figure 5 represents AU values indicating CD-induced DNA damage at various levels. AU values of negative control were calculated to be 18.50 ± 4.20 and 33.75 ± 6.24 after 48 and 72 h respectively. In contrast, the level of DNA damage jumped to 337.75 ± 9.81 (48 h) and 292.5 ± 16.36 AU (72 h) in positive control where cells were treated with H2O2 (20 mM). Increased levels of DNA damages induced by carbon dots occurred with respect to the negative control (Fig. 5). Even at the lowest CD concentration (0.25 ppm), AU value was elevated more than twofold as compared to the untreated cells. The difference between selected concentration groups was significant (p < 0.05). CD-based oxidative DNA damage can be correlated with several factors that strongly define the extent of CD-induced DNA-damage, such as the small size of these nanoparticles together with the surface charge and surface functional groups of them which alters the interaction of CDs with the cell90,102,103,104,105,106. Comet tail formation attributed to the DNA breaks or failed DNA repair mechanisms induced by oxidative stress. As supported by FT-IR, XPS, and ʐ-potential results, CD surface bearing a high amount of oxygenated functional groups could lead to the production of ROSs, and as a consequence, the oxidative stress may lead the genomic instability97,107,108. A recent study by Zhou et al.90 also indicated that modulating the oxygenated groups, most effectively the number of ketonic carboxyl groups, ROS production by carbon dots can also be controlled, the highest the oxygenated groups on the surface the highest the ROS production capability.
(a) Representation of the different comet classes in the alkaline comet assay where MCF7 cells were visually scored into four classes according to the tail length: Type 0: undamaged, with no tail, Type 1: with a tail shorter than the diameter of the head (nucleus), Type 2: with the tail as long as 1–2 × the diameter of the head, and Type3: with a tail longer than 2 × of the diameter of the head, Type 4: ultra-high damaged, with a longer tail length than the head diameter. (b) Representative images of non-treated control MCF7 cells and cells treated with 2.5 ppm CDs after 48 h and 72 h. DNA damage in MCF7 cells presented as arbitrary units (AU) measured by the alkaline Comet assay after treatment of cells with CDs for (c) 48 h and (d) 72 h. H202 (20 mM) was used as a positive control. (**) There is a statistically significant difference between positive control and CD-treated cells (p < 0.001).
Clastogenic damage was also evaluated using CBMN for MCF7 cell lines (Fig. 6a,b) CDs contributed to a significant increase in DNA damage as compared to the negative control for all doses administered (0.25, 2.5 and 50 ppm) after their exposure for 48 h and 72 h (p < 0.01 and p < 0.05, respectively). A concentration-dependent enhancement of the level of clastogenic damage was observed at 48 h (Fig. 6a). After 72 h of exposure, the clastogenicity of CDs at 0.25 ppm was higher than CDs administrated at higher concentrations (Fig. 6b), and the micronucleus frequencies (MN%) decreased as compared to the lower concentrations after 48 h treatment. The difference between the MN value obtained for 0.25 ppm at two incubation times was significant (p < 0.05). The reduction overall in cellular damage at the 72nd hour can be explained by the induction of cellular damage repair mechanisms, which alters the protection of the tumor cell from further chemical attacks and reveal a subsequent recovery from existing ones at increased exposure time. The slight decrease in cell damage for higher CD concentrations can also be explained with the same phenomena. Decreased nanoparticle accumulation in tumor cells at increased exposure times have been also reported by many other researchers which might decrease the effect of CDs on tumor cells100. Another explanation of the occurring plateau might be the DNA damage which leads cells either to a necrotic death or irreversibly committing those to apoptotic cell death which eventually reduces the proportion of the highly damaged cells109.
Changes in micronucleus frequencies in MCF7 cell lines treated with different concentrations of CD depending on the exposure time (a) 48 h and (b) 72 h. The statistically significant difference as compared to the negative control was shown as *p < 0.05 and **p < 0.01. Values represent mean ± SE, n = 3.
Influence of CDs on the cell cycle of MCF7 cells
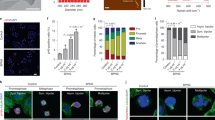
Considering the genotoxicity and clastogenicity induced by CDs, further analysis of the effects of CDs on the cell-cycle phases and apoptosis of MCF7 cells were performed. Cell proliferation is dependent on the cell-cycle progression, in which cells pass through the G0/G1 phase to the S phase and the G2/M phase. As depicted in Fig. 7a, after 48 h of treatment, 57.5%, 34.5% and 8.0% of untreated control cells were in the G0/G1, S and G2/M phases, respectively. Cells treated with CDs at even 0.25 ppm resulted in a decrease in the number of cells in the S phase (p < 0.05), and consequently a slight increase in the number of cells in the G0/G1 phases as compared to the control group. At increased CD concentrations, it has also been shown that induced cell cycle arrest in the G0/G1 phase is happening in a concentration-dependent manner, and the data were statistically significant as compared to the control group (p < 0.001). The effect of CDs on the cell cycle was shown to be time-dependent. As depicted in Fig. 6a,b, at 72 h, the changes in the cell cycle phases differentiated dramatically; 89.5%, 90.5% and 94.2% of MCF7 cells were in the G0/G1 phase respectively for cells treated with 0.25, 2.5 and 50 ppm CD (p < 0.005 for different concentrations of CDs), while only 76.6% of the control population was in the G0/G1 phase (p < 0.001). The significant differences (p < 0.001) in cell cycles for changing CD concentrations showing that prolonged delay in the G0/G1 phase is induced by CDs even at the lowest concentrations (0.25 ppm) which resulted in slower cell growth and delayed the entrance to S phase (Fig. 7a). As compared to untreated cells, CD (2.5 ppm) treated cells in S phase decreased by around 35% and 82% after incubation for 48 h and 72 h, respectively (Fig. 7b). Together with the genotoxicity and clastogenic analysis, the significant arrest of the cell population in G0/G1 phase, and the dramatic decrease in the population of cells in the S and G2/M phases support that CDs could trigger MCF7 cell apoptosis by inducing anomalies at G0/G1 phase by DNA damage and mutagenic stimulate (Fig. 7c). A comparison of the present study and previous literature is provided in Table 2. Most of the listed studies showed that negatively charged CDs with a size lower than 10 nm which can freely enter or interact with the nucleus while there are a couple of examples reporting nucleus targeting by larger sized CDs47,92,100, relatively few of them addressing the CD-induced anomalies in cell-cycle49,108,110,111. Our results suggest that CDs could have the potential as both drug carriers interacting with the cell-nucleus and therapeutic agents against tumor cells (Scheme 1). However, more studies should be done to handle this enormous therapeutic potential taking account the safety manners.
Statistical analysis of G0/G1, S, and G2/M populations in MCF cells (NC) and cells treated with (a) varying concentrations of CDs (0.25–50 ppm) for 48 h and 72 h. (b) Comparison of cell cycle phase profile of NC and CD (0.25 ppm) treated cells in different cellular phases. (c) Schematic representation of the possible effect of CDs in the cell cycle progression.
Conclusions
Although benign biocompatibility is attributed to carbon, systematic, and reliable biosafety assessment of carbon-based nanomaterials still needs to be conducted. Carbon dots serve as a useful and usable platform for a wide range of biological and biomedical applications including bio-imaging and nucleus targeted delivery/imaging. Herein, a systematic toxicity analysis of CDs produced from the extract of N. oleander via thermal synthesis method was conducted. In contradiction to many of the previous studies, we concluded that CDs have concentration and time-dependent cytotoxic potentials (at 50 ppm) over MCF7 cells while they comforted the proliferation of healthy HDFa cells even at highest CD concentrations. CDs caused severe DNA damage evident by the formation of COMET tail, micronuclei in MCF7 cells even at concentrations as low as 0.25 ppm. The interference of CDs with cell-cycle progression resulted in cell arrest in G0/G1 phases which showed that they can interact with genetic material and could trigger MCF7 cell apoptosis. Inducing Oxidative Stress Responses and interference with the cell cycle machinery could be due to the structure and surface properties of CDs while other mechanisms could also be involved. Although further studies are warranted to investigate the mutagenicity or carcinogenicity potency of CD in mammalian cells, this work shows evidence that CDs with super tiny size and high amount of oxygen on the surface can specifically affect the cellular function of tumor cells, and thus they have the potential to be used alone as an anti-cancer therapeutic material that can selectively target cancer cells by inducing series of DNA damage.
References
Cha, C., Shin, S. R., Annabi, N., Dokmeci, M. R. & Khademhosseini, A. Carbon-based nanomaterials: multifunctional materials for biomedical engineering. ACS Nano 7, 2891–2897 (2013).
Chawla, J. & Kumar, A. Ranking carbon-based nanomaterials using cytotoxicity to minimize public health risks. Int J Environ Eng Manag 4, 301–308 (2013).
Chen, C. & Haifang, W. Biomedical applications and toxicology of carbon nanomaterials. Nanomaterials https://doi.org/10.1002/9783527692866 (2016).
Visalli, G. et al. Toxicological assessment of multi-walled carbon nanotubes on A549 human lung epithelial cells. Toxicol. In Vitro 29, 352–362 (2015).
Kumar, S., Mehdipour, H. & Ostrikov, K. K. Plasma-enabled graded nanotube biosensing arrays on a Si nanodevice platform: catalyst-free integration and in situ detection of nucleation events. Adv. Mater. 25, 69–74 (2013).
Georgakilas, V., Perman, J. A., Tucek, J. & Zboril, R. Broad family of carbon nanoallotropes: classification, chemistry, and applications of fullerenes, carbon dots, nanotubes, graphene, nanodiamonds, and combined superstructures. Chem. Rev. 115, 4744–4822 (2015).
Ke, Q. & Wang, J. Graphene-based materials for supercapacitor electrodes—a review. J. Mater. 2, 37–54 (2016).
Lee, J.-K. et al. The seeded growth of graphene. Sci. Rep. 4, 5682 (2015).
Han, J. W. & Kim, J. Reduced graphene oxide—silver nanoparticle nanocomposite: a potential anticancer nanotherapy. Int. J. Nanomed. 10, 6257–6276 (2015).
Adach, K., Fijalkowski, M. & Skolimowski, J. Antioxidant effect of hydroxylated diamond nanoparticles measured in soybean oil. Fuller. Nanotub. Carbon Nanostruct. 23, 1024–1032 (2015).
Krueger, A. New carbon materials: biological applications of functionalized nanodiamond materials. Chem. A Eur. J. 14, 1382–1390 (2008).
Liu, H., Ye, T. & Mao, C. Fluorescent carbon nanoparticles derived from candle soot. Angew. Chemie Int. Ed. 46, 6473–6475 (2007).
Wu, F., Su, H., Wang, K., Wong, W.-K. & Zhu, X. Facile synthesis of N-rich carbon quantum dots from porphyrins as efficient probes for bioimaging and biosensing in living cells. Int. J. Nanomed. 12, 7375–7391 (2017).
Wang, J. et al. Carbon nanodots featuring efficient FRET for two-photon photodynamic cancer therapy with a low fs laser power density. Biomaterials 35, 9372–9381 (2014).
Sun, Y. P. et al. Quantum-sized carbon dots for bright and colorful photoluminescence. J. Am. Chem. Soc. 128, 7756–7757 (2006).
Alas, M. O. & Genc, R. An investigation into the role of macromolecules of different polarity as passivating agent on the physical, chemical and structural properties of fluorescent carbon nanodots. J. Nanoparticle Res. 19, 185 (2017).
Tao, H. et al. In vivo NIR fluorescence imaging, biodistribution, and toxicology of photoluminescent carbon dots produced from carbon nanotubes and graphite. Small 8, 281–290 (2012).
Zhang, X. et al. Color-switchable electroluminescence of carbon dot light-emitting diodes. ACS Nano 7, 11234–11241 (2013).
Paulo, S., Palomares, E. & Martinez-Ferrero, E. Graphene and carbon quantum dot-based materials in photovoltaic devices: from synthesis to applications. Nanomaterials 6, 157 (2016).
De, B., Voit, B. & Karak, N. Carbon dot reduced Cu2O nanohybrid/hyperbranched epoxy nanocomposite: mechanical, thermal and photocatalytic activity. RSC Adv. 4, 58453–58459 (2014).
Guo, D.-Y., Shan, C.-X., Qu, S.-N. & Shen, D.-Z. Highly sensitive ultraviolet photodetectors fabricated from ZnO quantum dots/carbon nanodots hybrid films. Sci. Rep. 4, 7469 (2014).
Yang, C.-H. et al. Correction: Yang, C.-H., et al. Immobilization of Brassica oleracea Chlorophyllase 1 (BoCLH1) and Candida rugosa Lipase (CRL) in magnetic alginate beads: an enzymatic evaluation in the corresponding proteins. Molecules 2014, 19, 11800–11815. Molecules 20, 7325–8 (2015).
Alas, M. O., Güngör, A., Genc, R. & Erdem, E. Feeling the power: robust supercapacitor from nanostructured conductive polymer fostered with Mn+2 and carbon dots. Nanoscale Accepted A, 1–15 (2019).
Genc, R. et al. High-capacitance hybrid supercapacitor based on multi-colored fluorescent carbon-dots. Sci. Rep. 7, 11222 (2017).
Yue, L. et al. Red-emissive ruthenium-containing carbon dots for bioimaging and photodynamic cancer therapy. ACS Appl. Nano Mater. 3, 869–876 (2020).
Wu, F. et al. Porphyrin-implanted carbon nanodots for photoacoustic imaging and in vivo breast cancer ablation. ACS Appl. Bio Mater. 1, 110–117 (2018).
Sun, X. et al. Microwave-assisted ultrafast and facile synthesis of fluorescent carbon nanoparticles from a single precursor: preparation, characterization and their application for the highly selective detection of explosive picric acid. J. Mater. Chem. A 4, 4161–4171 (2016).
Kuo, T. R. et al. One-pot green hydrothermal synthesis of fluorescent nitrogen-doped carbon nanodots for in vivo bioimaging. Anal. Bioanal. Chem. 408, 77–82 (2016).
Chen, M., Wang, W. & Wu, X. One-pot green synthesis of water-soluble carbon nanodots with multicolor photoluminescence from polyethylene glycol. J. Mater. Chem. B 2, 3937 (2014).
Nanoparticles, F. C. et al. An investigation into the role of macromolecules of different polarity as passivating agent on the physical, chemical and structural properties of fluorescent carbon nanodots. J. Nanopart. Res. 19(5), 185 (2017).
Zhu, H. et al. Microwave synthesis of fluorescent carbon nanoparticles with electrochemiluminescence properties. Chem. Commun. https://doi.org/10.1039/b907612c (2009).
Gao, H., Sapelkin, A. V., Titirici, M. M. & Sukhorukov, G. B. In situ synthesis of fluorescent carbon dots/polyelectrolyte nanocomposite microcapsules with reduced permeability and ultrasound sensitivity. ACS Nano 10, 9608–9615 (2016).
Baker, S. N. N. & Baker, G. A. A. Luminescent carbon nanodots: Emergent nanolights. Angew. Chemie Int. Ed. 49, 6726–6744 (2010).
Zheng, X. T., Ananthanarayanan, A., Luo, K. Q. & Chen, P. Glowing graphene quantum dots and carbon dots: properties, syntheses, and biological applications. Small 11, 1620–1636 (2015).
Wang, Z. et al. Photoluminescent carbon quantum dot grafted silica nanoparticles directly synthesized from rice husk biomass. J. Mater. Chem. B 5, 4679–4689 (2017).
Sahu, S., Behera, B., Maiti, K. & Mohapatra, S. Simple one-step synthesis of highly luminescent carbon dots from orange juice: application as excellent bio-imaging agents. Chem. Commun. 48, 8835–8837. https://doi.org/10.1039/c2cc33796g (2012).
Mehtaa, V. N., Jha, S., Singhalc, R. K. & Kailasaa, S. K. Preparation of multicolor emitting carbon dots for HeLa cell imaging. New J. Chem. 38, 6152–6160 (2014).
Gupta, A. et al. Paper strip based and live cell ultrasensitive lead sensor using carbon dots synthesized from biological media. Sens. Actuators B Chem. 232, 107–114 (2016).
Shi, L. et al. Eco-friendly synthesis of nitrogen-doped carbon nanodots from wool for multicolor cell imaging, patterning, and biosensing. Sens. Actuators B Chem. 235, 316–324 (2016).
Hsu, P.-C., Chen, P.-C., Ou, C.-M., Chang, H.-Y. & Chang, H.-T. Extremely high inhibition activity of photoluminescent carbon nanodots toward cancer cells. J. Mater. Chem. B 1, 1774–1781 (2013).
Ko, H. Y. et al. In vivo imaging of tumour bearing near infrared fluorescence-emitting carbon nanodots derived from tire soot. Chem. Commun. 49, 10290–10292 (2013).
Ray, S. C., Saha, A., Jana, N. R. & Sarkar, R. Fluorescent carbon nanoparticles: synthesis, characterization, and bioimaging application. J. Phys. Chem. C 113, 18546–18551 (2009).
Wu, F. et al. Near-infrared emissive lanthanide hybridized carbon quantum dots for bioimaging applications. J. Mater. Chem. B 4, 6366–6372 (2016).
Ayaz, F., Alaş, M. Ö, Oğuz, M. & Genç, R. Aluminum doped carbon nanodots as potent adjuvants on the mammalian macrophages. Mol. Biol. Rep. 46, 2405–2415 (2019).
Ayaz, F., Alas, M. O. & Genc, R. Differential immunomodulatory effect of carbon dots influenced by the type of surface passivation agent. Inflammation https://doi.org/10.1007/s10753-019-01165-0 (2019).
Kuo, C.-W., Chueh, D.-Y., Singh, N., Chien, F.-C. & Chen, P. Targeted nuclear delivery using peptide-coated quantum dots. Bioconjug. Chem. 22, 1073–1080 (2011).
Jung, Y. K., Shin, E. & Kim, B.-S. Cell nucleus-targeting zwitterionic carbon dots. Sci. Rep. 5, 18807 (2015).
Yang, L. et al. Doxorubicin conjugated functionalizable carbon dots for nucleus targeted delivery and enhanced therapeutic efficacy. Nanoscale 8, 6801–6809 (2016).
Havrdova, M. et al. Toxicity of carbon dots—effect of surface functionalization on the cell viability, reactive oxygen species generation and cell cycle. Carbon 99, 238–248 (2016).
Kumawat, M. K., Thakur, M., Gurung, R. B. & Srivastava, R. Graphene quantum dots for cell proliferation, nucleus imaging, and photoluminescent sensing applications. Sci. Rep. 7, 15858 (2017).
Bayat, A., Masoum, S. & Hosseini, E. S. Natural plant precursor for the facile and eco-friendly synthesis of carbon nanodots with multifunctional aspects. J. Mol. Liq. 281, 134–140 (2019).
Chatzimitakos, T. G., Kasouni, A. I., Troganis, A. N. & Stalikas, C. D. Carbonization of human fingernails: toward the sustainable production of multifunctional nitrogen and sulfur codoped carbon nanodots with highly luminescent probing and cell proliferative/migration properties. ACS Appl. Mater. Interfaces 10, 16024–16032 (2018).
Cailotto, S. et al. Carbon dots from sugars and ascorbic acid: role of the precursors on morphology, properties, toxicity, and drug uptake. ACS Med. Chem. Lett. 9, 832–837 (2018).
Li, D. et al. Fluorescent carbon dots derived from Maillard reaction products: their properties, biodistribution, cytotoxicity, and antioxidant activity. J. Agric. Food Chem. 66, 1569–1575 (2018).
Gomez, I. J., Arnaiz, B., Cacioppo, M., Arcudi, F. & Prato, M. Nitrogen-doped carbon nanodots for bioimaging and delivery of paclitaxel. J. Mater. Chem. B 6, 5540–5548 (2018).
Zhang, X., Lu, J., Zhou, X., Guo, C. & Wang, C. Rapid microwave synthesis of N-doped carbon nanodots with high fluorescence brightness for cell imaging and sensitive detection of iron (III). Opt. Mater. 64, 1–8 (2017).
Xu, L., Zhao, J. & Wang, Z. Genotoxic response and damage recovery of macrophages to graphene quantum dots. Sci. Total Environ. 664, 536–545 (2019).
Khan, I., Kant, C., Sanwaria, A. & Meena, L. Acute cardiac toxicity of Nerium oleander/indicum poisoning (kaner) poisoning. Heart Views 11, 115–116 (2010).
Slingerland, M., Cerella, C., Guchelaar, H. J., Diederich, M. & Gelderblom, H. Cardiac glycosides in cancer therapy: from preclinical investigations towards clinical trials. Invest. New Drugs https://doi.org/10.1007/s10637-013-9984-1 (2013).
Cerella, C., Dicato, M. & Diederich, M. Assembling the puzzle of anti-cancer mechanisms triggered by cardiac glycosides. Mitochondrion https://doi.org/10.1016/j.mito.2012.06.003 (2013).
El Sawi, N. M., Geweely, N. S., Qusti, S., Mohamed, M. & Kamel, A. Cytotoxicity and antimicrobial activity of Nerium oleander extracts. J. Appl. Anim. Res. 37, 25–31 (2010).
Benson, K. F., Newman, R. A. & Jensen, G. S. Antioxidant, anti-inflammatory, anti-apoptotic, and skin regenerative properties of an Aloe vera-based extract of Nerium oleander leaves (nae-8(®)). Clin. Cosmet. Investig. Dermatol. 8, 239–248 (2015).
Mwafy, S. N. & Yassin, M. M. Antidiabetic activity evaluation of glimepiride and Nerium oleander extract on insulin, glucose levels and some liver enzymes activities in experimental diabetic rat model. Pak. J. Biol. Sci. 14, 984–990 (2011).
Panosyan, A. & Al-Mukarish, S. Nerium oleander extract. (2006). USA Patent Pub No: US2006/0188585 A1
Carbik, I., Başer, K. H. C., Özel, H. Z., Ergun, B. & Wagner, H. Immunologically active polysaccharides from the aqueous extract of Nerium oleander. Planta Med. 56, 668 (1990).
Dey, P. & Chaudhuri, T. K. Comparative phytochemical profiling and effects of Nerium oleander extracts on the activities of murine peritoneal macrophages. Arch. Biol. Sci. 68, 515–531 (2016).
Sinha, S. N. & Biswas, K. A concise review on Nerium oleander L.—an important medicinal plant. Trop. Plant Res. 3, 408–412 (2016).
Chaudhary, K. K., Prasad, D., Sandhu, B. & Chaudhary, K. Preliminary pharmacognostic and phytochemical studies on Nerium oleander Linn. (White cultivar). J. Pharmacogn. Phytochem. 4, 185–188 (2015).
Simsek, S., Ozge Alas, M., Ozbek, B. & Genc, R. Evaluation of the physical properties of fluorescent carbon nanodots synthesized using Nerium oleander extracts by microwave-assisted synthesis methods. J. Mater. Res. Technol. 8, 2721–2731 (2019).
Simsek, S., Alas, M. O., Ozbek, B. & Genc, R. Fluorescent carbon dots from Nerium oleander: effects of physical conditions and the extract types. J. Fluoresc. Accepted A, 1–14 (2019).
Wang, Q. et al. Fluorescent carbon dots as an efficient siRNA nanocarrier for its interference therapy in gastric cancer cells. J. Nanobiotechnol. 12, 58 (2014).
Nygren, J., Suhonen, S., Norppa, H. & Linnainmaa, K. DNA damage in bronchial epithelial and mesothelial cells with and without associated crocidolite asbestos fibers. Environ. Mol. Mutagen. 44, 477–482 (2004).
Collins, A. R., Ai-guo, M. & Duthie, S. J. The kinetics of repair of oxidative DNA damage (strand breaks and oxidised pyrimidines) in human cells. Mutat. Res. Repair 336, 69–77 (1995).
Sun, L. W. et al. Toxic effects of aminophenols on aquatic life using the zebrafish embryo test and the comet assay. Bull. Environ. Contam. Toxicol. 73, 628–634 (2004).
Wu, X., Lu, W. & Mersch-Sundermann, V. Benzo(a)pyrene induced micronucleus formation was modulated by persistent organic pollutants (POPs) in metabolically competent human HepG2 cells. Toxicol. Lett. 144, 143–150 (2003).
Fenech, M. Cytokinesis-block micronucleus cytome assay. Nat. Protoc. 2, 1084–1104 (2007).
Fenech, M. & Morley, A. A. Measurement of micronuclei in lymphocytes. Mutat. Res. Mutagen. Relat. Subj. 147, 29–36 (1985).
Ema, M., Hougaard, K. S., Kishimoto, A. & Honda, K. Reproductive and developmental toxicity of carbon-based nanomaterials: a literature review. Nanotoxicology 10, 391–412 (2016).
Sargent, L. M., Reynolds, S. H. & Castranova, V. Potential pulmonary effects of engineered carbon nanotubes: in vitro genotoxic effects. Nanotoxicology 4, 396–408 (2010).
Ali-boucetta, H. & Kostarelos, K. Pharmacology of carbon nanotubes: toxicokinetics, excretion and tissue accumulation. Adv. Drug Deliv. Rev. 65, 2111–2119 (2013).
Lu, F. et al. Hydroxyl functionalized carbon dots with strong radical scavenging ability promote cell proliferation. Mater. Res. Express 6, 65030 (2019).
Li, X., Zhang, S., Kulinich, S. A., Liu, Y. & Zeng, H. Engineering surface states of carbon dots to achieve controllable luminescence for solid-luminescent composites and sensitive Be2+ detection. Sci. Rep. 4, 4976 (2015).
Li, L. & Dong, T. Photoluminescence tuning in carbon dots: surface passivation or/and functionalization, heteroatom doping. J. Mater. Chem. C 6, 7944–7970 (2018).
Song, P. et al. The chemical redox modulated switch-on fluorescence of carbon dots for probing alkaline phosphatase and its application in an immunoassay. RSC Adv. 8, 162–169 (2018).
Jaiswal, A., Ghosh, S. S. & Chattopadhyay, A. One step synthesis of C-dots by microwave mediated caramelization of poly(ethylene glycol). Chem. Commun. 48, 407 (2012).
Zhao, S., Li, C., Huang, H., Liu, Y. & Kang, Z. Carbon nanodots modified cobalt phosphate as efficient electrocatalyst for water oxidation. J. Mater. 1, 236–244 (2015).
Gan, Z., Xu, H. & Hao, Y. Mechanism for excitation-dependent photoluminescence from graphene quantum dots and other graphene oxide derivates: consensus, debates and challenges. Nanoscale 8, 7794–7807 (2016).
Liu, X., Pang, J., Xu, F. & Zhang, X. Simple approach to synthesize amino-functionalized carbon dots by carbonization of chitosan. Sci. Rep. 6, 31100 (2016).
Niu, J. et al. Facile synthesis and optical properties of nitrogen-doped carbon dots. New J. Chem. 38, 1522 (2014).
Sarkar, S. et al. Graphene quantum dots from graphite by liquid exfoliation showing excitation-independent emission, fluorescence upconversion and delayed fluorescence. Phys. Chem. Chem. Phys. 18, 21278–21287 (2016).
Siddique, A. B., Pramanick, A. K., Chatterjee, S. & Ray, M. Amorphous carbon dots and their remarkable ability to detect 2,4,6-trinitrophenol. Sci. Rep. 8, 9770 (2018).
Liu, H. et al. Blue and cyan fluorescent carbon dots: one-pot synthesis, selective cell imaging and their antiviral activity. RSC Adv. 7, 28016–28023 (2017).
Ong, K. J. et al. Widespread nanoparticle-assay interference: implications for nanotoxicity testing. PLoS ONE 9, e90650 (2014).
Holder, A. L., Goth-Goldstein, R., Lucas, D. & Koshland, C. P. Particle-induced artifacts in the MTT and LDH viability assays. Chem. Res. Toxicol. 25, 1885–1892 (2012).
Kroll, A., Pillukat, M. H., Hahn, D. & Schnekenburger, J. Interference of engineered nanoparticles with in vitro toxicity assays. Arch. Toxicol. 86, 1123–1136 (2012).
Kho, D. et al. Application of xCELLigence RTCA biosensor technology for revealing the profile and window of drug responsiveness in real time. Biosensors 5, 199–222 (2015).
Zhou, Y., Sun, H., Wang, F., Ren, J. & Qu, X. How functional groups influence the ROS generation and cytotoxicity of graphene quantum dots. Chem. Commun. 53, 10588–10591 (2017).
Hamd-Ghadareh, S., Salimi, A., Parsa, S. & Fathi, F. Simultaneous biosensing of CA125 and CA15-3 tumor markers and imaging of OVCAR-3 and MCF-7 cells lines via bi-color FRET phenomenon using dual blue-green luminescent carbon dots with single excitation wavelength. Int. J. Biol. Macromol. 118, 617–628 (2018).
Lin, X. J. et al. Multifunctional fluorescent carbon dots inhibit the invasiveness of lung cancer cells. New J. Chem. 42, 15311–15314 (2018).
Hill, S. A. et al. Selective photothermal killing of cancer cells using LED-activated nucleus targeting fluorescent carbon dots. Nanoscale Adv. 1, 2840–2846 (2019).
Yao, H. et al. Synthesis of ginsenoside Re-based carbon dots applied for bioimaging and effective inhibition of cancer cells. Int. J. Nanomed. 13, 6249–6264 (2018).
Ema, M. et al. Reproductive and developmental toxicity of carbon-based nanomaterials: a literature review. Nanotoxicology 10, 391–412 (2016).
Panzarini, E. Biocompatibility of carbon nanoparticles in HeLa cells is dictated by synthesis and sterilization procedures. Nanosci. Nanometrol. 2, 1 (2016).
Mesari, T. et al. High surface adsorption properties of carbon-based nanomaterials are responsible for mortality, swimming inhibition, and biochemical responses in Artemia salina larvae. Aquat. Toxicol. 163, 121–129 (2015).
Qiu, Y. et al. Antioxidant chemistry of graphene-based materials and its role in oxidation protection technology. Nanoscale 6, 11744–11755 (2014).
Tabish, T. A., Zhang, S. & Winyard, P. G. Developing the next generation of graphene-based platforms for cancer therapeutics: the potential role of reactive oxygen species. Redox Biol. 15, 34–40 (2018).
Wang, D., Zhu, L., Chen, J.-F. & Dai, L. Can graphene quantum dots cause DNA damage in cells?. Nanoscale 7, 9894–9901 (2015).
Periasamy, V. S., Athinarayanan, J., Alfawaz, M. A. & Alshatwi, A. A. Carbon nanoparticle induced cytotoxicity in human mesenchymal stem cells through upregulation of TNF3, NFKBIA and BCL2L1 genes. Chemosphere 144, 275–284 (2016).
Borges, H. L., Linden, R. & Wang, J. Y. J. DNA damage-induced cell death: lessons from the central nervous system. Cell Res. 18, 17–26 (2008).
Kalytchuk, S. et al. Carbon dot nanothermometry: intracellular photoluminescence lifetime thermal sensing. ACS Nano 11, 1432–1442 (2017).
Li, C.-L. et al. Carbon dots prepared from ginger exhibiting efficient inhibition of human hepatocellular carcinoma cells. J. Mater. Chem. B 2, 4564–4571 (2014).
Zhang, Y. et al. The synthesis and functional study of multicolor nitrogen-doped carbon dots for live cell nuclear imaging. Molecules 25, 306 (2020).
Acknowledgements
Sinem Şimşek gratefully acknowledges TUBITAK (The Scientific and Technological Research Council of Turkey) for the graduate scholarship.
Author information
Authors and Affiliations
Contributions
The manuscript was written through contributions of R.G., and D.B. and S.S. R.G. planned and designed C-Dot studies, S.S. performed the experiments on C-Dots. R.G., B.Ö and S.S. collected and analyzed the data. R.G planned and designed the studies on in vitro cytotoxicity of CDs. D.Y. and S.S carried out in vitro experiments and analyzed the data with R.G. D.B planned and design genotoxicity and clastogenicity studies. D.Y and A.A.Ş performed the genotoxicity and clastogenicity experiments, collected and analyzed the data. All authors take full responsibility for the content of the paper and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Şimşek, S., Şüküroğlu, A.A., Yetkin, D. et al. DNA-damage and cell cycle arrest initiated anti-cancer potency of super tiny carbon dots on MCF7 cell line. Sci Rep 10, 13880 (2020). https://doi.org/10.1038/s41598-020-70796-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-70796-3
This article is cited by
-
Triphenylphosphonium conjugated gold nanotriangles impact Pi3K/AKT pathway in breast cancer cells: a photodynamic therapy approach
Scientific Reports (2023)
-
Carbon nanodots as sensitive and selective nanomaterials in pharmaceutical analysis
Journal of Materials Science (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.