Abstract
To screen the key immune genes in the development of cervical cancer, construct immune related gene pairs (IRGPs), and evaluate their influence on the prognosis of cervical cancer. Tumor Genome Atlas (TCGA) database and geo database were downloaded as training set and validation set respectively, and immune related gene data were downloaded from immport. IRGPs model is established by machine learning, and the model is analyzed and evaluated. Using the Uclcan to analyze the immune genes expression in cervical cancer, and to further explore the association with the expression level and the clinical stage and prognosis of cervical cancer. According to the analysis of training set, we identified 29 IRGPs as key gene pairs and constructed the model. The AUC value of the model was greater than 0.9, and the model group survival rate was conspicuous different (P < 0.001). The reliability of the model was confirmed in the validation group. Our IRGPs play an important role in the occurrence and development of cervical cancer, and can be used as a prognostic marker and potential new target of cervical cancer.
Similar content being viewed by others
Introduction
Cervical cancer is one of the four most common gynecological tumors1. Every year, at least 569,847 women in the world are diagnosed with cervical cancer, and more than 311,365 people are killed2. In recent years, the incidence rate of cervical cancer has decreased significantly through universal screening and health knowledge. However, the incidence rate of cervical cancer is still high in developing countries3. For women with low education in less developed areas, the coverage rate of cervical cancer screening is still very low4. Squamous cell carcinoma is the most common type of cervical cancer, accounting for 75% of cervical cancer cases, while adenocarcinoma only accounts for about 20%. In developing countries, 70% of cervical cancer patients have local infiltration or metastasis, which has led to the high mortality of cervical cancer in developing countries. Early cervical cancer is usually treated by radical hysterectomy. When there are risk factors such as lymph node metastasis and endometriosis that may lead to recurrence, they will be treated with chemotherapy5. The standard treatment for patients with locally advanced cervical cancer is conventional radiation therapy(CRT)6. The five-year survival rate of patients with locally advanced cervical cancer can be as high as 75–85% after surgical resection, radiotherapy, chemotherapy, CRT, and so on7. However, at present, all treatment methods are not effective for patients with paraaortic lymph node metastasis, and their three-year progression free survival time (PFS) and total survival time (OS) are 34% and 39%8,9, respectively. The five-year survival rate of cervical cancer patients with recurrence and metastasis was as low as 15%. Limited treatment is the main reason for this situation. Now, palliative chemotherapy is the most commonly used for patients with metastatic and recurrent cervical cancer10. The median survival time of patients with metastatic or recurrent cervical cancer treated with platinum/taxane chemotherapy and bevacizumab can be extended to 17 months11. However, these treatments are far from enough for most locally advanced and metastatic cervical cancer patients with positive lymph node metastasis.
In recent years, immunotherapy has been developed and increasingly used in cancer patients. For example, PD-L1 is overexpressed in a variety of tumor cells, including liver cancer cells and lung cancer cells, and plays an important role in regulating the immune response of tumor cells12,13,14,15. Currently, there are several clinical trials involving FDA-approved immunosuppressive checkpoint inhibitors, which attack tumor cells expressing PD-L1 by blocking the PD-L1/PD-1 signaling pathway, so as to improve the treatment and prognosis of patients. From the current situation, immunosuppressive therapy has achieved good results in many solid tumors16. The results of PD-1/PD-L1 inhibition in cervical cancer are also satisfactory. However, at present, immunoassay sites with a therapeutic effect are scarce, and the research on tumor immunotherapy is far from sufficient. In this study, we screened immune genes that are significantly related to the prognosis of cervical cancer, constructed an immune gene pair (IRGP) model based on these genes, and used it to verify the unique prognostic markers of cervical cancer.
Method
Data acquisition
Gene expression profile data of 255 patients with cervical squamous cell carcinoma were obtained from cancer and tumor gene map (TCGA https://www.tcga.org), and gene expression profile data set gse4400116 was obtained from gene expression compilation (GEO https://www.ncbi.nlm.nih.gov/geo/) database, including 300 samples of cervical cancer patients.
Acquisition of sample immune gene expression
2,498 immune related genes were downloaded from immport (https://www.immport.org/home), including antigen presenting cells, chemokines and their receptors, cytokines and their receptors, interferon, interleukin, etc. Using limma package in R (3.61), we compared the gene expression data of cervical cancer samples downloaded from TCGA database and geo database as training set and verification set with immune related genes, and extracted the expression amount of immune related genes in cervical cancer samples.
Construction of immune related gene pairs (IRGPs)
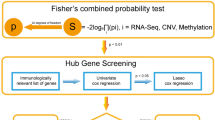
In the two groups of data processed in the previous step, the IRGP of the sample is calculated, and the relatively high change is selected according to the standard of media absolute deviation > 0.5. IRGP values are calculated by comparing gene expression levels in specific samples or profiles in pairs. The immune related genes are matched to compare the IRGPs. If the first IRG is larger than the second IRG, the output of the IRGP is 1; otherwise, the output is 0. If the ratio of IRGP score of 0 or 1 in training set and verification set is higher than 80%, then remove the IRGP and retain the remaining IRGP as candidate IRGP for prognosis prediction. The logistic rank test was used to screen the prognosis IRGP (FDR < 0.01). Cox risk regression analysis and glment in R (3.61) were used to perform tenfold cross validation to analyze the candidate IRGP and obtain the IRGP index. We constructed the best 29 gene pairs as immune gene pair model. We use ROC to calculate the optimal cutoff value of IRGP index, and use it as the basis to distinguish high and low risk groups.
IRGPs model validation
The single factor and multi factor Cox proportional risk analysis and survival analysis of TCGA and gse44001 cervical cancer samples were carried out with IRGPs model.
Infiltration of immune cells in cervical cancer samples.
In order to study the infiltration of immune cells in the high and low risk groups of cervical cancer, we used CIBERSORT17 to evaluate and predict the enrichment of immune cells in the samples. CIBERSORT is a tool for deconvolution of the expression matrix of immune cell subtypes based on the principle of linear support vector regression. RNA SEQ data can be used to estimate the infiltration of immune cells. CIBERSORT can analyze the relative abundance of 22 immune infiltrating cells in each sample, including NK cells, T cells, B cells and macrophages.
Functional enrichment analysis of GSEA, go and KEGG
Gene set enrichment analysis(GSEA) enrichment analysis was carried out for each gene related to immune prognosis using the fgsea package in R (3.61)18 .Cluster profiler19 was used to enrich Gene ontology(GO)function and KEGG pathway. Significant enrichment criteria: the absolute value of NES is greater than 1, the nomp value is less than 0.05, and the fdrq value is less than 0.25.
Expression of immune gene in cervical cancer
Ualcan 20 were used to analyze the expression of immune genes in cervical cancer.
Statistical analysis
Measured data were expressed as mean ± standard deviation (x ± s) and data were compared using t test. Kaplan Meier method was used for survival analysis. The receiver operating characteristic curve (ROC curve) and ROC analysis were completed by survivalROC(1.0.3). Single factor and multi factor analysis using Cox proportional risk regression model. P < 0.05 was statistically significant, P < 0. 01 as the difference has very significant statistical significance.
Ethical approval and consent to participate
This article does not contain any studies with patients or animals performed by any of the authors.
Results
Expression of immune related genes and construction of IRGPs in cervical cancer samples
We obtained gene expression data of 255 cervical cancer samples from TCGA database as training set, 300 cervical cancer samples from gse44001 as verification set, 2,498 immune related genes from immport, and 479 immune related genes from cervical cancer samples by comparing the two. Through these 479 immune related genes, we constructed 23,355 IRGPs. We remove more than 80% of the IRGP with a score of 0 or 1 from the training set and validation set, leaving 12,379 IRGP as candidates. Combine TCGA clinical data with training set data (Table 1). 73 prognosis related IRGPs were screened by lasso Cox proportional risk regression analysis. After 1,000 iterations, we selected 29 optimal IRGPs to build the immune prognosis model (Table 2).
IRGPs model validation
The immune prognosis model was applied to the training set, and the patients in each training set were scored. According to ROC curve analysis, the optimal cutoff value for patients to be divided into high and low risk groups is 1.124 (Fig. 1A). After evaluating the model, we found that AUC value of model 1, 3 and 5 years is 0.913 (Fig. 1B), 0.913 (Fig. 1C) and 0.912 (Fig. 1D). The results show that our immune prognosis gene has a high reliability for the model. The training set was divided into high-risk group (Fig. 2A) and high-risk group (Fig. 2B). The results showed that the overall survival rate (OS) of high-risk group was significantly lower than that of low-risk group. For TCGA training set data, single factor and multi factor Cox risk regression analysis showed that only IRGPs model showed significant prognostic effect in single factor Cox (Fig. 2C), while age and IRGPs could be significant independent prognostic factors in multi factor Cox (Fig. 2D). Applying this model to the validation set of gse44001 (Fig. 3A) (Table 3), survival analysis showed that the OS of patients in the high-risk group was significantly lower than that in the low-risk group (Fig. 3B). In the single factor and multi factor Cox analysis, IRGPs model and tumor size were significantly correlated with prognosis (Fig. 3C,D).
Infiltration of immune cells in cervical cancer samples
Most studies believe that the occurrence and development of tumor are closely related to immune cells, so it is an ideal method to study the infiltration of immune cells in tumor. We used CIBERSORT to analyze the infiltration of 22 kinds of immune cells in patients with high and low risk groups. Figure 4A shows the expression of immune cells in different risk groups. Macrophage M0 (Fig. 4B), activated mast cells (Fig. 4C) were significantly overexpressed in the high-risk group, while stationary dendritic cells (Fig. 4D), stationary mast cells (Fig. 4E), activated CD4T cells (Fig. 4F), and cd8t cells (Fig. 4G) were overexpressed in the low-risk group.
Functional enrichment analysis of GSEA, go and KEGG
We analyzed the function enrichment of IRGP in the model. The results of go analysis showed that IRGP in the model was mainly enriched in the binding of cytokines and their receptors, the binding of chemokines and their receptors, the binding of growth factors and their receptors, the binding of epidermal growth factor receptors, the binding of fibroblast growth factors and the activity of tyrosine kinase (Fig. 5A,B). KEGG results showed that these IRGP were involved in cytokine cytokine receptor interaction, chemokine signaling, tumor necrosis factor signaling, MAPK signaling, NF kappa B signaling, natural killer cell-mediated cytotoxicity, viral proteins and cytokines, and Th1 and Th2 cell differentiation (Fig. 5C,D). The results of GSEA (Fig. 6A) showed that these IRGP were significantly enriched in trace ribonucleoprotein complex (Fig. 6B), neurotransmitter transporter activity (Fig. 6C), endopeptidase activity (Fig. 6D), fibroblast growth factor receptor binding (Fig. 6E), hormone activity (Fig. 6F), fibroblast cell proliferation (Fig. 6G), and growth factor receptor binding (Fig. 6H).
Expression of immune gene in cervical cancer
We explored the expression of IRGP in cervical cancer using the ualcan model (Table 4). There were 6 low-level expression of IRGP in cervical cancer (Fig. 7) and 8 high-level expression of IRGP (Fig. 8). There were differences in the expression of 5 low expression IRGP and 8 high expression IRGP in different age groups (Fig. 9), and there were differences in the expression of 14 IRGP in different stages of cervical cancer (Fig. 10).
(A) Expression of CLCF1 in cervical cancer and normal tissues. (B) Expression of DLL4 in cervical cancer and normal tissues. (C) Expression of INHBA in cervical cancer and normal tissues (D) Expression of NOD1 in cervical cancer and normal tissues. (E) Expression of NRP1 in cervical cancer and normal tissues. (F) Expression of RBP7 in cervical cancer and normal tissues.
(A) Expression of CXCR3 in cervical cancer and normal tissues. (B) Expression of DUOX1 in cervical cancer and normal tissues. (C) Expression of FGFR3 in cervical cancer and normal tissues. (D) Expression of HLA-DQA2 in cervical cancer and normal tissues. (E) Expression of LTB4R2 in cervical cancer and normal tissues. (F) Expression of STC2 in cervical cancer and normal tissues. (G) Expression of TNFSF10 in cervical cancer and normal tissues. (H) Expression of CESC in cervical cancer and normal tissues.
(A) Expression of CLCF1 in cervical cancer and normal tissues. at different ages. (B) Expression of DLL4 in cervical cancer and normal tissues at different ages. (C) Expression of NOD1 in cervical cancer and normal tissues at different ages. (D) Expression of NRP1 in cervical cancer and normal tissues at different ages. (E) Expression of RBP7 in cervical cancer and normal tissues at different ages. (F) Expression of CXCR3 in cervical cancer and normal tissues at different ages. (G) Expression of DUOX1 in cervical cancer and normal tissues at different ages. (H) Expression of FGFR3 in cervical cancer and normal tissues at different ages. (I) Expression of HLA-DQA2 in cervical cancer and normal tissues at different ages. (J) Expression of LTB4R2 in cervical cancer and normal tissues at different ages. (K) Expression of STC2 in cervical cancer and normal tissues at different ages. (L) Expression of TNFSF10 in cervical cancer and normal tissues at different ages. (M) Expression of VAV3 in cervical cancer and normal tissues at different ages.
(A) Expression of CLCF1 in cervical cancer and normal tissues. at different stages. (B) Expression of CXCR3 in cervical cancer and normal tissues at different stages. (C) Expression of DLL4 in cervical cancer and normal tissues at different stages. (D) Expression of DUOX1 in cervical cancer and normal tissues at different stages. (E) Expression of FGFR3 in cervical cancer and normal tissues at different stages. (F) Expression of HLA-DQA2 in cervical cancer and normal tissues at different stages. (G) Expression of INHBA in cervical cancer and normal tissues at different stages. (H) Expression of LTB4R2 in cervical cancer and normal tissues at different stages. (I) Expression of NOD1 in cervical cancer and normal tissues at different stages. (J) Expression of NRP1 in cervical cancer and normal tissues at different stages. (K) Expression of RBP7 in cervical cancer and normal tissues at different stages. (L) Expression of STC2 in cervical cancer and normal tissues at different stages. (M) Expression of TNFSF10 in cervical cancer and normal tissues at different stages. (N) Expression of VAV3 in cervical cancer and normal tissues at different stages.
Discussion
Cervical cancer is one of the most common gynecological malignancies. HPV infection is considered to be the main cause of cervical cancer21,22 although the incidence rate of cervical cancer has been significantly decreased due to the development and promotion of HPV vaccine23. But incidence rate of cervical cancer is still high in developing countries and China's low income countries24. At present, for cervical cancer patients without invasion and lymphatic metastasis, the effect of surgery combined with radiotherapy and chemotherapy is better. If metastasis and infiltration occur, the treatment effect of cervical cancer patients will become very unsatisfactory. In recent years, immunotherapy has performed well in a variety of cancers including cervical cancer25,26,27. Blocking PD-L1 / PD-1 signaling pathway to attack tumor cells expressing PD-L1 is the current mainstream method28. Although the anticancer activity of PD-1 and PD-L1 inhibitors is exciting, such immunotherapy is not effective for all patients, and a meta-analysis shows that patients who receive PD-1 / PD-L1 inhibitors have a higher risk of rash, thyroid dysfunction, pruritus, pneumonia and colitis29,30,31. Therefore, it is of great significance for the detection and treatment of cervical cancer to predict and find more biomarkers that may be related to immune prognosis.
At present, most of the prognostic genes need to be standardized to reduce the errors caused by sequencing platform and samples. In this study, the scores of IRGPs constructed by us are calculated from the gene expression data of the same sample, which can not only ignore the impact of different platforms, but also do not need to standardize and scale the data. This method has been used in many studies, including cancer molecular classification, with high reliability32,33.
In this study, we screened 29 pairs of IRGP to construct the immune prognosis model related to the overall survival rate of cervical cancer patients. The AUC values of the model in 1, 3 and 5 years were all greater than 0.9. According to these 29 pairs of IRGP, they were divided into high-risk group and low-risk group. In TCGA training group and GSE44001 verification group, the OS of high-risk group was significantly lower than that of low-risk group (P < 0.01). These 29 pairs of IRGP have a good effect on sample discrimination. We found that macrophage Mo and activated mast cells were significantly over expressed in high-risk group by immunocyte infiltration analysis of samples. The existing research shows that mast cells and macrophages play an important role in cervical cancer, which can promote the development of cervical cancer by promoting lymphangiogenesis and angiogenesis34,35,36. However, in the low-risk group, the expression of static dendritic cells, static mast cells, activated CD4T cells and cd8t cells is high. Although the effect of CD4T cells on cervical cancer has not been agreed, the cd8t cells are closely related to the better prognosis of cervical cancer patients37,38,39, there is evidence that dendritic cells will decrease in patients with high HPV infection, which indicates that high expression of dendritic cells is beneficial to resist cervical cancer40, which is consistent with our results. The enrichment analysis of go and GSEA showed that these immune genes were mainly involved in the binding of cytokines and their receptors, the binding of chemokines and their receptors, the binding of growth factors and their receptors, the binding of epidermal growth factor receptors, the activity of metalloendopeptidase, the binding of fibroblast growth factors and their receptors, hormone activity, fibroblast proliferation, and the binding process of growth factor receptors.As we all know, cytokines and chemokines are the key factors in the immune response. for example: In cervical cancer, IL-10 can interfere with the differentiation of dendritic cells and thus play a strong immunosuppressive effect,TGF—β 1 can inhibit T cell proliferation and attenuate immune response41. Research shows that growth factors and epidermal growth factors are closely related to the growth of cervical cancer and the survival rate of cervical cancer patients. High expression of growth factors and epidermal growth factors often predict poor prognosis42,43,44. Growth of fibroblasts can stimulate angiogenesis at the early stage of tumor The proliferation and invasion of cancer cells and the remodeling of extracellular matrix promote the growth of cervical cancer45,46. KEGG results showed that these immune genes were mainly enriched in chemokine signaling pathway, tumor necrosis factor signaling pathway, MAPK signaling pathway, NF kappa B signaling pathway, natural killer cell-mediated cytotoxicity, viral protein and cytokine, and Th1 and Th2 cell differentiation. Th1 and Th2 may be involved in the pathogenesis and growth of cervical cancer. Th1 may be the target of predicting chemotherapy response of advanced cervical cancer47,48,49,50, while other pathways are classical signal pathways related to cancer. Immune cytokines play an important role in cervical lesions. Torres et al. Found that IL-10 is highly expressed in the cervix of women with persistent HPV, which may be related to the persistence of HPV and the promotion of disease progression. Further research by their team showed that copy individuals of IL-4, IL-6, IL-10 and TGFB1 were significantly associated with cervical cancer, and could be used as biomarkers for susceptibility to the disease51,52.These 29 pairs of IRGP have 47 different immune genes, most of which are cytokines, antimicrobial agents and natural killer cells, which are involved in various stimulation reactions and play a key role. In cervical cancer, HPV can inhibit the apoptosis of cervical cancer cells by down regulating NOD153. In our sample, we also found that the expression of NOD1 in tumor tissue is low and there are differences in different ages and stages (Figs. 7D, 9C, 10I). Sang Yeon Cho et al. Found that duox1 is highly expressed in cervical squamous cell carcinoma and can play a good prognostic role by increasing the amount of innate immune cells54. The analysis also showed that DUOX1 is highly expressed in tumor tissues and related to age and grade (Figs. 8B, 9G, 10D). Stc2 can promote the proliferation of cervical cancer cells and increase the resistance to cisplatin55, while high expression of DDL4 is usually associated with low pelvic lymph node metastasis and survival rate of cervical cancer56. Therefore, we believe that the IRGP constructed in this study plays an important role in the development and prognosis of cervical cancer.
There are also some deficiencies in our research. Although we select data samples from two databases for analysis, and use more advanced methods to reduce the errors caused by platforms, samples, etc., this is still a retrospective analysis. If we can carry out a prospective study or obtain clinical samples and evaluate them with Western blot or immunohistochemistry, it will be more convincing.
Conclusion
We constructed an immune gene pair model which is closely related to the prognosis of cervical cancer patients. The model contains 29 IRGP and 47 immune-related genes. The biological functions of these 47 immune-related genes are closely related to the occurrence and development of cervical cancer. Therefore, we think that these IRGPs may be the target of predicting or diagnosing cervical cancer, and suggest that immunotherapy can improve the prognosis of cervical cancer patients by regulating these IRGPs.
Data availability
All data are available. Please contact us to access if it is needed.
References
Stewart Bernard, W. et al. Cancer prevention as part of precision medicine: “plenty to be done”. Carcinogenesis 37(1), 2–9. https://doi.org/10.1093/carcin/bgv166 (2016).
Bray, F. et al. Global cancer statistics 2018: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. https://doi.org/10.3322/caac.21492 (2018).
Passos Camila, M., Sales Jacqueline, B., Maia Emanuella, G., Caldeira Thaís, C M., Rodrigues Roberta, D., Figueiredo N., Claro Rafael M. (2020). Trends in access to female cancer screening in Brazil, 2007–16. J Public Health (Oxf), https://doi.org/10.1093/pubmed/fdaa028
Asrabuddhe, V. V., Parham, G. P., Mwanahamuntu, M. H. & Vermund, S. H. Cervical cancer prevention in low- and middle-income countries: feasible, affordable, essential. Cancer Prevent. Res. 5, 11–17. https://doi.org/10.1158/1940-6207.CAPR-11-0540 (2012).
Koh Wui-Jin., Abu-Rustum Nadeem R., Bean Sarah., Bradley Kristin., Campos Susana M., Cho Kathleen R., Chon Hye Sook., Chu Christina., Clark Rachel., Cohn David., Crispens Marta Ann., Damast Shari., Dorigo Oliver., Eifel Patricia J., Fisher Christine M., Frederick Peter., Gaffney David K., Han Ernest., Huh Warner K., Lurain John R., Mariani Andrea., Mutch David., Nagel Christa., Nekhlyudov Larissa., Fader Amanda Nickles., Remmenga Steven W., Reynolds R Kevin., Tillmanns Todd., Ueda Stefanie., Wyse Emily., Yashar Catheryn M., McMillian Nicole R., Scavone Jillian L.(2019). Cervical Cancer, Version 3.2019, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc Netw., 17(1), 64–84. https://doi.org/10.6004/jnccn.2019.0001
Chen, J. et al. Nanotechnology in the management of cervical cancer. Rev. Med. Virol. 25(Suppl. 1), 72–83. https://doi.org/10.1002/rmv.1825 (2015).
Varia, M. A. et al. Cervical carcinoma metastatic to para-aortic nodes: extended field radiation therapy with concomitant 5-fluorouracil and cisplatin chemotherapy: a Gynecologic Oncology Group study. Int. J. Radiat. Oncol. Biol. Phys. 42(5), 1015–23. https://doi.org/10.1016/s0360-3016(98)00267-3 (1998).
Randall Leslie, M., Monk Bradley, J., Darcy Kathleen, M., Tian, C., Burger Robert, A., Liao, S.-Y., Peters William, A., Stock Richard, J., Fruehauf John, P. (2009). Markers of angiogenesis in high-risk, early-stage cervical cancer: a Gynecologic Oncology Group study. Gynecol. Oncol., 112(3), 583-9. https://doi.org/10.1016/j.ygyno.2008.11.013
Boussios, S. et al. Management of patients with recurrent/advanced cervical cancer beyond first line platinum regimens: where do we stand? A literature review. Crit. Rev. Oncol. Hematol. 108, 164–174. https://doi.org/10.1016/j.critrevonc.2016.11.006 (2016).
Tewari, K. S. et al. Bevacizumab for advanced cervical cancer: final overall survival and adverse event analysis of a randomised, controlled, open-label, phase 3 trial (gynecologic oncology group 240). Lancet 390, 1654–1663. https://doi.org/10.1016/S0140-6736(17)31607-0 (2017).
Minion, L. E. & Tewari, K. S. Cervical cancer - state of the science: from angiogenesis blockade to checkpoint inhibition. Gynecol. Oncol. 148, 609–621. https://doi.org/10.1016/j.ygyno.2018.01.009 (2018).
Facchinetti, F. et al. Moving immune checkpoint blockade in thoracic tumors beyond NSCLC. J Thorac Oncol. 11, 1819–1836 (2016).
Lastwika, K. J. et al. Control of PD-L1 expression by oncogenic activation of the AKT/mTOR pathway in non-small cell lung cancer. Cancer Res. 76, 227–238 (2016).
Brahmer, J. R. et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 366(26), 2455–2465 (2012).
Constantinidou, A., Alifieris, C. & Trafalis, D. T. Targeting programmed cell death -1 (PD-1) and ligand (PD-L1): a new era in cancer active immunotherapy. Pharmacol. Ther. 18, 30173–30176. https://doi.org/10.1016/j.pharmthera.2018.09.008 (2018).
Gettinger, S. et al. Five-year follow-up of nivolumab in previously treated advanced non-small-cell lung cancer: results from the CA209-003 study. J. Clin. Oncol. 36, 1675–1684. https://doi.org/10.1200/JCO.2017.77.0412 (2018).
Yoo-Young, L. et al. Genetic profiling to predict recurrence of early cervical cancer. Gynecol. Oncol. 131(3), 650–654. https://doi.org/10.1016/j.ygyno.2013.10.003 (2013).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 12(5), 453–457. https://doi.org/10.1038/nmeth.3337 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 102(43), 15545–15550. https://doi.org/10.1073/pnas.0506580102 (2005).
Guang chuang, Y., Li-Gen, W., Yanyan, H. & Qing-Yu, H. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5), 284–287 (2012).
Smith, R. A. et al. Cancer screening in the United States, 2015: a review of current American cancer society guidelines and current issues in cancer screening. CA Cancer J Clin 65(1), 30–54. https://doi.org/10.3322/caac.21261 (2015).
Brianti, P., De Flammineis, E., Mercuri, S.R. Review of HPV-related diseases and cancers. New Microbiol. 40, 80–85 (2017).
Wang, X., Huang, X. & Zhang, Y. Involvement of human papillomaviruses in cervical cancer. Front Microbiol 9, 2896. https://doi.org/10.3389/fmicb.2018.02896 (2018).
Braaten, K.P., Laufer, M.R., Human papillomavirus (HPV), HPV-related disease, and the HPV vaccine. Rev Obstet Gynecol. (2008) 1, 2–10. https://medreviews.com/journal/reviews-in-obstetrics-gynecology.
Zhang, L., Zhao, Y., Tu, Q., Xue, X., Zhu, X., Zhao, K.-N. (2020). The role of programmed cell death ligand-1/ programmed cell death-1 (PD-L1/PD-1) in HPV-induced cervical cancer and potential for their use in blockade therapy. Curr. Med. Chem., https://doi.org/10.2174/0929867327666200128105459.
Shibata, T., Lieblong, B.J., Sasagawa, T., Nakagawa, M. (2019). The promise of combining cancer vaccine and checkpoint blockade for treating HPV-related cancer. Cancer Treat. Rev., 78(undefined), 8–16. https://doi.org/10.1016/j.ctrv.2019.07.001
Zhou, C., Tuong, Z. K. & Frazer, Ian H. Papillomavirus immune evasion strategies target the infected cell and the local immune system. Front Oncol 9, 682. https://doi.org/10.3389/fonc.2019.00682 (2019).
Liu, Y. et al. PD-1/PD-L1 inhibitors in cervical cancer. Front Pharmacol 10, 65. https://doi.org/10.3389/fphar.2019.00065 (2019).
Sul, J. et al. FDA approval summary: Pembrolizumab for the treatment of patients with metastatic non-small cell lung cancer whose tumors express programmed death-ligand. Oncologist. 21, 643–650. https://doi.org/10.1634/theoncologist.2015-0498 (2016).
Garon, E. B. et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 372, 2018–2028. https://doi.org/10.1056/NEJMoa1501824 (2015).
Nishijima, T. F., Shachar, S. S., Nyrop, K. A. & Muss, H. B. Safety and tolerability of PD-1/PD-L1 inhibitors compared with chemotherapy in patients with advanced cancer: a meta-analysis. Oncologist. 22, 470–479. https://doi.org/10.1634/theoncologist.2016-0419 (2017).
Li, B., Cui, Y., Diehn, M. & Li, R. Development and validation of an individualized immune prognostic signature in early-stage nonsquamous non-small cell lung cancer. JAMA Oncol.. 3(11), 1529–1537 (2017).
Peng, P. L. et al. Identification of a novel gene pairs signature in the prognosis of gastric cancer. Cancer Med. 7(2), 344–350 (2018).
Dolores, U.-B., Marta, C.-M., Emilio, C., Margarita, G.-R. & de Esparza, A.-R. The role of macrophages and mast cells in lymphangiogenesis and angiogenesis in cervical carcinogenesis. Exp. Mol. Pathol. 89(2), 190–196. https://doi.org/10.1016/j.yexmp.2010.06.002 (2010).
Wang, Q., Steger, A., Mahner, S., Jeschke, U. & Heidegger, H. The formation and therapeutic update of tumor-associated macrophages in cervical cancer. Int J Mol Sci https://doi.org/10.3390/ijms20133310 (2019).
Xiao-Jing, C. et al. Hypoxia-induced ZEB1 promotes cervical cancer progression via CCL8-dependent tumour-associated macrophage recruitment. Cell Death Dis 10(7), 508. https://doi.org/10.1038/s41419-019-1748-1 (2019).
Punt, S., van Vliet Marjolein, E., Spaans Vivian, M., de Kroon Cornelis D., Fleuren Gert, J., Gorter, A., Jordanova Ekaterina, S.(2015). FoxP3(+) and IL-17(+) cells are correlated with improved prognosis in cervical adenocarcinoma. Cancer Immunol. Immunother., 64(6), 745–53. https://doi.org/10.1007/s00262-015-1678-4
Punt, S., Houwing-Duistermaat Jeanine, J., Schulkens Iris, A., Thijssen Victor, L., Osse Elisabeth, M., de Kroon Cornelis, D., Griffioen Arjan, W., Fleuren Gert, J., Gorter A., Jordanova Ekaterina S.(2015). Correlations between immune response and vascularization qRT-PCR gene expression clusters in squamous cervical cancer. Mol. Cancer, 14, 71. https://doi.org/10.1186/s12943-015-0350-0
Carus A., Ladekarl M., Hager H., Nedergaard B S., Donskov F.(2013). Tumour-associated CD66b+ neutrophil count is an independent prognostic factor for recurrence in localised cervical cancer. Br. J. Cancer, 108(10), 2116–22. https://doi.org/10.1038/bjc.2013.167
Zhihong, W., Yuhua, H., Ni, X. & Qiling, Ma. Elevated expression of secreted autocrine growth factor progranulin increases cervical cancer growth. Cell Biochem. Biophys. 71(1), 189–193. https://doi.org/10.1007/s12013-014-0183-2 (2015).
Jimenez-Flores, R. et al. High-risk human papilloma virus infection decreases the frequency of dendritic Langerhans’ cells in the human female genital tract. Immunology 117(2), 220–228. https://doi.org/10.1111/j.1365-2567.2005.02282.x (2006).
Torres-Poveda, K. et al. Role of IL-10 and TGF-β1 in local immunosuppression in HPV-associated cervical neoplasia. World J Clin Oncol. 5(4), 753–763. https://doi.org/10.5306/wjco.v5.i4.753 (2014).
Soonthornthum, T., Arias-Pulido, H., Joste, N., Lomo, L., Muller, C., Rutledge, T., Verschraegen, C. (2011). Epidermal growth factor receptor as a biomarker for cervical cancer. Ann. Oncol., 22(10), 2166–78. https://doi.org/10.1093/annonc/mdq723
Wei-Jie, T. et al. Prognostic impact of epidermal growth factor receptor overexpression in patients with cervical cancer: a meta-analysis. PLoS ONE 11(7), e0158787. https://doi.org/10.1371/journal.pone.0158787 (2016).
Neta, E., Morgan, T., Peter, O., Tuttleton, A. S. & Douglas, H. Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-kappaB-dependent manner. Cancer Cell 17(2), 135–147. https://doi.org/10.1016/j.ccr.2009.12.041 (2010).
Fullár, A., Dudás, J., Oláh, L., Hollósi, P., Papp, Z., Sobel, G., Karászi, K., Paku, S., Baghy, K., Kovalszky, I. (2015). Remodeling of extracellular matrix by normal and tumor-associated fibroblasts promotes cervical cancer progression. BMC Cancer, 15, 256. https://doi.org/10.1186/s12885-015-1272-3
Zhang, W., Tian, X., Mumtahana, F., Jiao, J., Zhang, T., Croce Kimiko D., Ma, D., Kong, B., Cui B. (2015). The existence of Th22, pure Th17 and Th1 cells in CIN and Cervical Cancer along with their frequency variation in different stages of cervical cancer. BMC Cancer, 15, 717. https://doi.org/10.1186/s12885-015-1767-y
Wenguang, Ma., Kun, W., Jongqiang, Du., Junqi, L. & Ge, L. Multi-dose parecoxib provides an immunoprotective effect by balancing T helper 1 (Th1), Th2, Th17 and regulatory T cytokines following laparoscopy in patients with cervical cancer. Mol. Med. Rep. 11(4), 2999–3008. https://doi.org/10.3892/mmr.2014.3003 (2015).
Alpana, S., Medha, R., Abhigyan, S. & Manoj, S. Cytokines (TH1 and TH2) in patients with advanced cervical cancer undergoing neoadjuvant chemoradiation: correlation with treatment response. Int. J. Gynecol. Cancer 19(7), 1269–1275. https://doi.org/10.1111/IGC.0b013e3181a8efcc (2009).
Sreenivas, A. et al. Functional tumor infiltrating TH1 and TH2 effectors in large early-stage cervical cancer are suppressed by regulatory T cells. Int. J. Gynecol. Cancer 22(7), 1130–1137. https://doi.org/10.1097/IGC.0b013e318262aa53 (2012).
Torres-Poveda, K, Burguete-García, A.I., Cruz, M., et al. The SNP at -592 of human IL-10 gene is associated with serum IL-10 levels and increased risk for human papillomavirus cervical lesion development. Infect Agent Cancer. 7(1):32. https://doi.org/10.1186/1750-9378-7-32
Torres-Poveda, K., Burguete-García, A.I., Bahena-Román, M., et al. Risk allelic load in Th2 and Th3 cytokines genes as biomarker of susceptibility to HPV-16 positive cervical cancer: a case control study. BMC Cancer. 2016;16, 330. https://doi.org/10.1186/s12885-016-2364-4
Liu, X., Ma, H., Fei, L., Jiang, M., Xia, M., Bai, L., Pi, X., Chen, S., Yu, L. (2020). HPV-mediated down-regulation of NOD1 inhibits apoptosis in cervical cancer. Infect. Agents Cancer, 15, 6. https://doi.org/10.1186/s13027-020-0272-3
Yeon, C. S. et al. Dual oxidase 1 and NADPH oxidase 2 exert favorable effects in cervical cancer patients by activating immune response. BMC Cancer 19(1), 1078. https://doi.org/10.1186/s12885-019-6202-3 (2019).
Yuxia, W., Ying, G., Hairong, C., Guichun, Y. & Wenhua, T. Stanniocalcin 2 promotes cell proliferation and cisplatin resistance in cervical cancer. Biochem. Biophys. Res. Commun. 466(3), 362–368. https://doi.org/10.1016/j.bbrc.2015.09.029 (2015).
Shanshan, Y. et al. DLL4 as a predictor of pelvic lymph node metastasis and a novel prognostic biomarker in patients with early-stage cervical cancer. Tumour Biol. 37(4), 5063–5074. https://doi.org/10.1007/s13277-015-4312-3 (2016).
Acknowledgements
HAN NIE, Fanqin Bu and Jiasheng Xu are the co-first authors of this article; HAN NIE is the 1st of the first authors; Fanqin Bu is the 2nd of the first authors and Jiasheng Xu is the 3rd of the first authors. The study did not accept any funding.
Author information
Authors and Affiliations
Contributions
H.N. research design and drafting the manuscript. F.B. helping to revision the manuscript. J.X. literature search; T.L. revision of the manuscript and writing guidance. J.H. review and revision of the manuscript and writing guidance.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nie, H., Bu, F., Xu, J. et al. 29 immune-related genes pairs signature predict the prognosis of cervical cancer patients. Sci Rep 10, 14152 (2020). https://doi.org/10.1038/s41598-020-70500-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-70500-5
This article is cited by
-
A promising natural killer cell-based model and a nomogram for the prognostic prediction of clear-cell renal cell carcinoma
European Journal of Medical Research (2024)
-
A risk prediction model mediated by genes of APOD/APOC1/SQLE associates with prognosis in cervical cancer
BMC Women's Health (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.