Abstract
Thermal stress response is an essential physiological trait that determines occurrence and temporal succession in nature, including response to climate change. We compared temperature-related demography in closely related heat-tolerant and heat-sensitive Brachionus rotifer species. We found significant differences in heat response, with the heat-sensitive species adopting a strategy of long survival and low population growth, while the heat-tolerant followed the opposite strategy. In both species, we examined the genetic basis of physiological variation by comparing gene expression across increasing temperatures. Comparative transcriptomic analyses identified shared and opposing responses to heat. Interestingly, expression of heat shock proteins (hsps) was strikingly different in the two species and mirrored differences in population growth rates, showing that hsp genes are likely a key component of a species’ adaptation to different temperatures. Temperature induction caused opposing patterns of expression in further functional categories including energy, carbohydrate and lipid metabolism, and in genes related to ribosomal proteins. In the heat-sensitive species, elevated temperatures caused up-regulation of genes related to meiosis induction and post-translational histone modifications. This work demonstrates the sweeping reorganizations of biological functions that accompany temperature adaptation in these two species and reveals potential molecular mechanisms that might be activated for adaptation to global warming.
Similar content being viewed by others
Introduction
On a global scale, a species’ occurrence is related to its tolerance of a particular range of environmental parameters such as temperature, salinity and precipitation. In aquatic ecosystems, temperature has a profound impact on an organism’s survival and performance, and can affect species abundance, spatio-temporal distribution, habitat colonization and species interactions1,2,3. There is large variation in the thermal tolerance among aquatic taxa. Many species can tolerate a broad range of temperatures, while others have specific and narrow temperature limits4,5. Importantly, this can impact temporal occurrence and temperature-dependent seasonal succession has been well documented among genetically similar species that might have evolved species-specific temperature specializations6,7,8,9. Therefore, knowledge of thermal boundaries is essential to understand how species have adapted to their environment and how they may respond to climate change.
Zooplankton represent an important component of aquatic ecosystems, as they transfer organic compounds and energy from primary producers (e.g. phytoplankton) to higher trophic levels10. Among zooplankton, monogonont rotifers are of particular interest because of their high, often cryptic, diversity, their frequent adaptation to specific environmental conditions, and high dispersal capability11,12. Species complexes formerly assumed to be ubiquitous generalists have been found to comprise cryptic species adapted to specific ecological conditions regarding temperature, habitat type, or salinity13,14,15. As their dispersal capabilities can be large, distribution and diversification seem less dependent on geographical barriers and historical factors, suggesting that ecological specialization is more likely to drive speciation13,14,15,16. As evidence of specialization, co-occurrence of differentially adapted, closely related species in a single locality is a common phenomenon in rotifers6,7,17. In these cases, morphologically similar species might have evolved different ecological specialties to reduce competition over resources in space or time15,18,19.
The best studied freshwater monogonont rotifer is the Brachionus calyciflorus species complex that has recently been resolved to four different species using integrative taxonomy: Brachionus calyciflorus sensu stricto (s.s.), Brachionus fernandoi, Brachionus dorcas, and Brachionus elevatus20,21. The species of this complex exhibit temporal succession, and their occurrences have been related to temperature in several studies7,8,17,22. More specifically, temperature constraints were shown to affect the temporal occurrence and abundance of B. calyciflorus cryptic species in different habitats in China7,17. Brachionus calyciflorus s.s. occurred mainly in summer at a maximum temperature of 32 °C7 while B. fernandoi mainly during winter and spring under a maximum temperature of 19 °C7. Both species generally exhibit a non-overlapping seasonal occurrence during summer and winter respectively, however, in November they co-occurred with B. fernandoi outnumbering B. calyciflorus s.s. in abundance7. Our previous comparative laboratory study on heat tolerance between different clones belonging to B. calyciflorus s.s. and B. fernandoi species has shown higher heat-tolerance of the former as compared to the latter23. In this study, acute heat-stress was imposed and the Critical Thermal Maximum (CTmax) was estimated as a proxy for survival. Heat resistance was species, but not clone, specific with values of CTmax varying 10 °C between the species23. Therefore, we confirmed that temperature tolerance likely plays a role in their temporal distribution23.
Understanding the ability of species to adapt to environmental change necessitates knowledge of the genomic basis of the relevant adaptations. The rapid development of high-throughput sequencing technologies and whole transcriptome profiling (RNA-seq) has enabled a deeper investigation of adaptive and functional variation in model and non-model species24. Regulation of gene expression is an essential mechanism underlying physiological robustness as well as phenotypic plasticity25. Since selection acts on the sequence itself (DNA), but also on expression, transcriptome data are particularly useful in revealing the genetic basis of adaptation, i.e., genes contributing to fitness by either structural (non-synonymous substitutions) or expression differences. In recently diverged snake species for example, significant expression differentiation was identified with little and non-coding sequence variation across populations, demonstrating that expression differentiation was the exclusive genetic basis of polygenic adaptation26. By profiling transcriptional changes induced by temperature, it is possible to identify the gene regions or pathways that are likely to be targets of temperature-driven selection27,28.
Rotifers have been used as model organisms to understand complex processes such as the evolution of sex, aging, and stress responses to toxic compounds29,30,31,32,33. However, transcription studies on temperature response have often focused mainly on genes involved in generation of oxidative stress and genes encoding for heat shock proteins (hsps)34,35,36,37. Heat shock proteins are divided into several groups (families) of different molecular weights (kDa): e.g. hsp90, hsp70, hsp60, hsp40, and small proteins. hsp70s, in combination with other proteins, play a vital role in stress tolerance and survival under adverse conditions. They reduce accumulation of peptide aggregates and promote the correct folding of newly synthesized proteins38. hsp90s also play a major role in stress tolerance, mainly by removing incorrectly folded proteins. Furthermore, they regulate the activity of other proteins (e.g., kinases) and stabilize the cytoskeleton39. Induction of hsp genes is an evolutionary old and conserved mechanism, and is described from prokaryotes to higher eukaryotes40. However, the specific genes involved and the conditions of induction vary among taxa41. In rotifers, particularly Brachionus species, members of the hsp70 and hsp40 families increase heat shock survival, suggesting that there may be coordination among heat shock proteins in which hsp40 works synergistically to regulate hsp70’s activity (as shown in B. manjavacas37).
To investigate the marked variation in thermal tolerance between two closely related species in the former B. calyciflorus species complex, we compared life-history demography and gene expression under mild to high temperature conditions. We used life-table experiments to examine survival, fecundity, and population growth rate differences between the two species. We collected transcriptomic data (RNA-seq) to examine the genetic basis of physiological response and its difference between the two species. The mechanisms we identify are important to understand how physiology determines species’ temporal distribution, and how this might be affected by different scenarios of climate change.
Materials and methods
Rotifer culture and life table experimental conditions
In a previous laboratory study, we quantified acute heat response of different clones belonging to B. calyciflorus s.s. (10 clones) and B. fernandoi (5 clones)23. Thermal tolerance was estimated as a bi-dimensional phenotypic trait affected by both the intensity and duration of the heat by measuring time to incapacitation which is considered a proxy for survival42. Cross-species differences were revealed in the maximum temperature (CTmax) that the species were able to tolerate23. For the purpose of this study we selected two asexually reproducing clones with known differences in the maximum temperature they can sustain (CTmax) under acute heat exposure23. One clone represented the heat-tolerant B. calyciflorus s.s. (clone IGB; CTmax = 43.18 °C) and the other one the heat-sensitive B. fernandoi (clone A10; CTmax = 38.49 °C)23. Both clones originate from Northern Germany and were reared under laboratory conditions for more than 10 years. Species classification was previously confirmed by amplifying a portion of the ITS1 genetic marker23. Stock cultures were maintained in WC medium43 at 20 °C under a 16:8 light:dark photoperiod. A food combination of two algae species, Monoraphidium minutum (Culture collection Göttingen, strain SAG-243-1) and Cryptomonas sp. (Culture collection Göttingen, strain SAG-26-80), was provided weekly.
Before starting the life table experiments, cultures were exposed to a period of gradual acclimatization by increasing the temperature 2 °C every 2 days until reaching the experimental temperature. Because of this, the acclimation period varied among cultures, with the longest adaptation period (2 weeks) for the highest temperature (32 °C). After reaching the experimental temperature, we maintained the rotifer cultures for one week (at least two generations) before starting the experiment to reduce potential maternal effects. Food was supplied ad libitum daily. For B. calyciflorus s.s., experiments were conducted at four temperatures (20 °C, 23 °C, 26 °C, 32 °C), while for B. fernandoi three temperature assays (20 °C, 23 °C, 26 °C) were used. We tried several times to acclimate B. fernandoi to 32 °C, but high mortality always led to culture collapse before the initiation of the experiment.
During the experiments single females bearing a subitaneous, asexual egg were isolated from the culture, placed in a microtitre well, and inspected thoroughly for hatched neonates. Life-table experiments were started by introducing one neonate into a new well to 1 ml of algal suspension composed of Monoraphidium minutum (5 × 105 cells/ml) and Cryptomonas sp. (2 × 104 cells/ml), to avoid food limitation. For each temperature and species, at least 24 individuals were recorded. Survival and reproduction were recorded every 12 h and any newly hatched neonates were removed. Every 24 h the maternal individuals were transferred into a new well with fresh food suspension. The experiment was conducted in the dark and continued until all individuals of each cohort died44,45.
Computations and statistical analysis
The variables of average life span (survival), age-specific survival (lx) and age-specific fecundity (mx) were estimated for each species and temperature assay (x was defined as the age interval in days, lx as the proportion of surviving individuals at the beginning of the age interval and mx as the number of offspring produced per female alive from the start until the end of any age interval46. The intrinsic rate of population increase (r) was also estimated from these data using Lotka’s equation (Eq. 1)47:
Kaplan–Meier survival curves were calculated to compare survival across species and temperatures. Differences between survival functions were analyzed pairwise using a log-rank test. To further estimate the effect of species, temperature, and their interaction on lifespan and fecundity, we used generalized linear models (GLMs). We selected the best-fitting model based on the AIC criterion48. To compare fecundity among the species and temperature treatments we used the non-parametric Kruskal–Wallis (K–W) one-way analysis of variance. For pairwise comparisons the pairwise Wilcox test was used with a Bonferroni correction. Non parametric tests were used for comparisons, as all of the assumptions to perform a parametric Analysis of Variance (ANOVA; normal distribution of the data, normal distribution of the residuals, and homoscedasticity) were violated. To compare the intrinsic rate of increase (r), the 95% confidence interval was estimated via bootstrapping with 199 iterations49 using a custom n R script. All statistical analyses were performed using R 3.4.150.
Sample cultivation, collection, and RNA isolation
Samples for RNA-seq were first cultivated as batch cultures in 1 l glass bottles containing WC medium at 20 °C under 16:8 light:dark photoperiod. The same two algae combination was provided ad libitum every 2 days for 2 weeks before the RNA-seq experiment to allow for substantial population growth. For each species and temperature, we sub-sampled the initial stock into new flasks to create four replicates of 200 ml each, containing approximately 1,000 individuals. We placed the flasks into water-baths adjusted to the experimental temperature and heat-exposed the rotifers for 4 h. Experimental temperatures were selected according to the life-table results in order to represent control (20 °C; temperature in which both clones were acclimated), mild heat treatment (23 °C for B. fernandoi; 26 °C for B. calyciflorus s.s.; intermediate temperature between control and high heat exposure), and high heat treatment (26 °C for B. fernandoi and 32 °C for B. calyciflorus s.s.; temperature in which the population growth rate starts to decline) (Fig. 1). In addition, preliminary experiments during which temperature was increased 2 °C every 4 days showed that 32 °C and 26 °C represent the threshold temperature above which population growth rate became negative, for B calyciflorus s.s. and B. fernandoi, respectively. For specimen collection we filtered each replicate (200 ml) through a 30 μm sieve, re-suspended what remained on the filter in WC medium, and centrifuged it at 2,000×g for 10 min to pellet phytoplankton and other debris, before transferring the rotifers (remaining in the supernatant) into 1 ml of TRIzol LS and storing them at − 80 °C until RNA extraction. For RNA extraction, we used four replicates from each temperature and species with the exception of B. fernandoi at mild heat, where two replicates were used. Each replicate, comprised approximately of 1,000 individuals, was reared in independent flasks of 200 ml.

(Figure made in Inkscape v.1.0; https://inkscape.org/).
Experimental design for transcriptomic responses to heat exposure between heat-tolerant and the heat-sensitive Brachionus species. Temperature regimes were chosen to represent control (20 °C), mild (26/23 °C) and high (32/26 °C) heat treatment for each species.
Samples in TRIzol LS, after having been homogenized using a Tissue Lyzer (4 min, 50 Hz), were incubated overnight at room temperature. A total of 500 μl of chloroform was added to each sample and samples were centrifuged for 15 min at 4 °C to facilitate phase separation51. The colorless, upper aqueous phase was transferred into an RNeasy Mini Kit column (Qiagen, Germany) and proceeded to RNA precipitation according to the manufacturer’s instructions. Total RNA concentration was estimated using a NanoDrop 1000 spectrophotometer (ThermoFischer Scientific, Germany). Quality of total RNA was examined using Agilent Bioanalyzer 2100 (Agilent Technologies, USA).
For transcriptomic library preparation, enrichment of mRNA from total RNA (3 μg) was performed with poly (A) capture using NEXTflex Poly (A) Beads. Strand-specific libraries were constructed using NEXTflex Rapid Directional RNA-Seq Kit (Bioo Scientific, USA) according to manufacturer’s instructions. Final elution was performed in 16 μl of elution buffer and a PCR amplification of 14 cycles was performed51. Libraries were quantified using Qubit dsDNA HS Assay Kit (Invitrogen, Germany) and quality control was performed using Agilent Bioanalyzer 2100 (Agilent Technologies, USA).
Transcriptome sequencing, assembly and annotation
Libraries were sequenced as 150 bp paired-end (PE) reads using an Illumina HiSeq 4000 sequencing system, performed by Novogene (Hong Kong, China). Raw data have been deposited in the NCBI Short Read Archive under the accessions numbers (SRA: SRR10426055-76). Adapter sequences were trimmed and low quality reads were filtered using a 5 bp sliding window with a mean quality threshold of 20 and minimum read length of 36 bp using Trimmomatic v0.3652. Read quality (before and after quality-filtering) was assessed using FastQC v0.11.553.
Processed reads were assembled de novo with Trinity v.2.5.154. Two separate transcriptome assemblies were created for B. calyciflorus s.s. and B. fernandoi, using all reads generated for each respective species. For the assembly and further read quantification we did not utilize the recently produced B. calyciflorus s.s. genome as this would introduce a bias, since the B. fernandoi reads would not map as well to the reference as those from B. calyciflorus s.s.. We, however, used the genome assembly to filter out contaminating algae sequences. Since each replicate contained rotifer individuals and algae from the culture medium, removing contaminants (i.e. reads belonging to algae) was essential before any further analysis. To filter out contamination, we used a custom perl script which, using the blastn algorithm (ncbi-blast-2.6.055), assigns all contigs either to a local algae database (Monoraphidium minutum, Chlamydomonas reinhardtii and, Cryptomonas sp) or to the respective B. calyciflorus s.s. genome assembly56. Transcripts were only assigned as of rotifer origin when the top hit was to the B. calyciflorus s.s. genome and the bit-score gain over matches to the next species was > 100. The same custom script was further used to remove ribosomal RNA reads by performing a blastn search to a local database consisting of 18S and 28S sequences of Brachionus species downloaded from NCBI.
Identification of differentially expressed genes and pathways
Gene-level quantification estimates produced by RSEM57 were imported into R/Bioconductor with the tximport package. The tximport package produces count matrices from gene-level quantification files with effective gene length taken into account58. To detect differential gene expression, we passed the estimated count matrices from tximport to DESeq259 and analyzed the two species separately. To build the model for the differential expression analysis we removed low count genes (< 10) and genes that were not present in at least 2 replicates. To examine intra-species temperature specific expression pattern in B. calyciflorus s.s., we performed pairwise contrasts within the model in the following combinations: 20 vs. 26 °C (control vs. mild heat), 20 vs. 32 °C (control vs. high heat), and 26 vs. 32 °C (mild vs. high heat) (Fig. 1). For B. fernandoi similarly we performed pairwise comparisons in the following combinations: 20 vs. 23 °C (control vs. mild heat), 20 vs. 26 °C (control vs. high heat), and 23 vs. 26 °C (mild vs. high heat) (Fig. 1). We used a false discovery rate (FDR) threshold of 0.05 to correct for multiple testing. All differentially expressed genes (DEGs) were annotated against the NCBI non-redundant (nr) database using blastx (e-value cutoff 1e−10). We assessed overall temperature-dependent patterns of expression by plotting a two-dimensional principal component analysis (PCA) of log-transformed counts for each species separately. Heatmaps representing differential expressed genes were constructed by using the Heatmapper program60.
We further categorized differential expression at the gene-pathway level using the online Kyoto Encyclopedia of Genes and Genomes (KEGG) automatic server for KEGG pathway analysis61,62,63, which clusters genes based on their association in biochemical pathways. To estimate whole KEGG pathway expression, genes belonging to the same KEGG pathway were clustered together and a differential pathway expression analysis was performed. Pathways were considered differentially expressed at a false discovery rate (FDR) below 0.05. Analyses and visualization was performed using the “gage”64, “clusterProfiler”65, “pathview”66, and “ggplot2”67 R packages. Heatmaps representing differentially expressed KEGG pathways were constructed in R50.
To capture similar or contrasting patterns of expression among the two species, orthologous genes were identified using OrthoFinder68. From this, we compared expression of orthologous genes between B. fernandoi and B. calyciflorous using Clust69. In Clust, gene clusters (groups) are identified that are consistently co-expressed (well-correlated) in both shared and contrasting patterns between species. Within this, we chose patterns that were biologically meaningful for further analysis. These groups/clusters were checked for functional enrichment in any KEGG pathway, using a Fisher Exact Test and correcting for false positives (FDR = 0.05).
Results
Life history responses to elevated temperatures
Survival, fecundity and the resulting population growth rate of both species were strongly affected by temperature. For both species, survival was significantly reduced from control to mild and from mild to high heat (Fig. 2a, all p-values available in Supplementary Table 1). Cross-species survival comparisons revealed significant differences at 20 °C (p < 0.001) and 23 °C (p = 0.008), with B. fernandoi surviving longer than B. calyciflorus s.s (Fig. 2a, Supplementary Table 1). Fecundity analysis with the K–W test revealed that both B. calyciflorus s.s. and B. fernandoi had a significant lower fecundity at the highest imposed heat (B. calyciflorus s.s.: 20 °C vs. 32 °C, p < 0.001; 23 °C vs. 32 °C, p = 0.001; 26 °C vs. 32 °C, p = 0.002; B. fernandoi: 20 °C vs. 26 °C, p < 0.001; 23 °C vs. 26 °C, p = 0.004, Supplementary Fig. 1A, Supplementary Table 2). Across the two species, significant differences were observed at 26 °C, in which the heat-tolerant B. calyciflorus s.s. had higher fecundity than the heat-sensitive B. fernandoi (Bcal26 vs. Bfer26, p < 0.001, Supplementary Fig. 1A, Supplementary Table 2). Generalized linear models revealed that both survival and fecundity were significantly dependent on temperature (GLM: survival, p < 0.001; fecundity, p < 0.001), species (GLM: survival, p < 0.001; fecundity, p = 0.006), and their interaction (GLM: survival, p < 0.001; fecundity, p = 0.001) (Supplementary Figs. 1B, 1C).

(Figure produced by using R 3.4.150).
Kaplan–Meier survival curves (a), and population growth rate/r (b) for the heat-tolerant B. calyciflorus s.s. (Bcal) and the heat-sensitive B. fernandoi (Bfer) under different temperature conditions. Solid lines denote responses of B. fernandoi to different temperatures (20 °C, 23 °C, 26 °C), while dashed lines represent responses of B. calyciflorus s.s (20 °C, 23 °C, 26 °C, 32 °C). Boxes denote the 95% confidence intervals estimated via bootstrap with 199 iterations.
The intrinsic rate of population increase (r) was above zero for both species, indicating a positive growth rate at all tested temperatures. However, r was always higher for the heat-tolerant B. calyciflorus s.s. than for the heat-sensitive B. fernandoi (Fig. 2b). For B. calycifloryus s.s, r increased with the increase of temperature until its maximum at 26 °C. In contrast, r remained constant in B. fernandoi from 20 °C to 23 °C, and decreased at 26 °C.
Comparative transcriptomics and cross-temperature differential gene expression
Sequencing of the B. calyciflorus s.s. and B. fernandoi transcriptomes generated 670,242,336 and 358,527,278 quality-filtered PE reads, respectively, with approximately equal numbers of reads among libraries (Supplementary Table 3). Information about the de novo assemblies, assembled unigenes, and KEGG KO term assignment can be found in Table 1.
We first examined the expression data for temperature-dependent responses in each species by a two-dimensional plot of the first two principal components of a PCA. This analysis showed temperature-dependent separation of replicates for the heat-tolerant, B. calyciflorus s.s.. This separation was less clear for the heat-sensitive species, in which replicates from 20 °C and 23 °C clustered together, while all samples treated at 26 °C formed a separate cluster (Supplementary Fig. 2). Overall, we found a greater number of differentially expressed genes (DEGs) in the heat-tolerant species than the heat-sensitive. In both species we captured the largest number of DEGs in pairwise comparison control vs. high heat (Fig. 3A,B). Further, in the heat-tolerant species, we captured a greater number of genes differentially expressed between control and mild heat (1,268) than between mild and high heat (337). In contrast, in the heat-sensitive species we found the opposite pattern, i.e., 40 DEGs between control and mild heat and 532 DEGs between mild and high heat (Fig. 3A,B).

(Figure made in Inkscape v.1.0; https://inkscape.org/).
Venn diagrams showing the number of differentially expressed genes in pairwise temperature comparisons as determine by the DESeq2 analyses (FDR = 0.05) for the heat-tolerant B. calyciflorus s.s. (a) and the heat-sensitive B. fernandoi (b). Blue color represents control treatment (C), orange color represents mild heat treatment (M), and red color represents high heat treatment (H). Up- and down- regulation is defined from the perspective of the higher temperature over the lower one.
We focused on pairwise comparison of mild vs. high heat in both species as we had evidence that there is a strong effect of temperature on population growth rate in this transition. In B. calyciflorus s.s., among the 337 genes differentially expressed between mild (26 °C) and high (32 °C) heat, were genes encoding for RNA polymerases (upregulated with heat), histone proteins (up- or downregulated) and N-acetyltranferases (upregulated with heat; Supplementary Fig. 3A). In heat-sensitive B. fernandoi, among the 532 differentially expressed genes between mild (23 °C) and high (26 °C) heat, we found genes encoding for several histone (H3 and H4) methyltranferase proteins (all up-regulated with heat; Supplementary Fig. 3B).
We further focused on genes that are differentially expressed in all the three pairwise comparisons. These genes are considered the most responsive to temperature, exhibiting either up- or down-regulation along with temperature increase. In the heat tolerant B calyciflorus s.s., there were 23 genes differentially expressed in all pairwise comparisons. Among these, genes encoding for ribosomal proteins and glutathione S-transferase were up-regulated at lower temperatures, while proteases related genes were up-regulated at higher temperatures (Supplementary Fig. 4A). In the heat-sensitive B. fernandoi, there were two genes that were differentially expressed in all pairwise comparisons. These genes encode for E3 ubiquitin-ligase and a mediator of RNA polymerase and were both up-regulated at higher temperatures (Supplementary Fig. 4B).
Cross-temperature differentially expressed genes in heat shock response
We examined patterns of expression in hsp genes to evaluate their specific contribution to the heat shock response (HSR) in both species. In both species, we found a significant down-regulation of heat shock protein genes at temperatures where population growth rate was maximized (Figs. 1B, 4A,B). In general, genes encoding for hsp had higher expression at the lowest temperature treatment (20 °C) for the heat-tolerant, B. calyciflorus s.s. and at the highest temperature for the heat-sensitive, B. fernandoi. Genes encoding for hsp27 and hsp70 followed this pattern. Genes encoding for hsp10, hsp40, and hsp60 were differentially expressed only in B. calyciflorus s.s. where they followed the same pattern. Genes encoding for hsp90 beta were up-regulated under the highest imposed temperature regime in both species. In B. calyciflorus s.s., genes encoding for hsp20 were also up-regulated under the highest imposed temperature (32 °C), following the induction pattern of hsp90 beta (Fig. 4A).

(Figure produced by the Heatmapper program60).
Heat map using normalized counts to show patterns of expression in hsp genes for the heat-tolerant B. calyciflorus s.s. (a), and the heat-sensitive B. fernandoi (b). Blue color (C) represents control treatment, orange color (M) represents mild heat treatment, and red color (H) represents high heat treatment. The symbols indicate the pairwise comparison in which differential gene expression (DE) was significant (p < 0.05). The normalized counts (relative expression normalized with DESeq2) were used for the heat maps and the color key represents a spectrum of lowest gene expression (blue) to highest gene expression (red).
Cross-temperature differential KEGG pathway expression
To capture shared and contrasting patterns at the level of gene-pathways, we examined whole KEGG pathway expression in two pairwise comparisons, control vs. mild heat and mild vs. high heat, as both of these comparisons represent a stepwise temperature induction. Overall, mild heat resulted in down-regulation of pathways related to genetic information processing in heat-tolerant, B. calyciflorus s.s., and up-regulation of metabolism related pathways in the heat-sensitive, B. fernandoi. More specifically, pathways “ribosome”, “proteasome”, and “oxidative phosphorylation” were down-regulated under mild heat exposure for B. calyciflorus s.s. and up-regulated for B. fernandoi (Fig. 5A, Supplementary Figs. 5, 6). In general, high heat caused up-regulation of pathways related mainly to metabolism such as carbohydrate metabolism and lipid metabolism and down-regulation of pathways related to signal transduction for the heat-tolerant B. calyciflorus s.s. (Fig. 5B, Supplementary Fig. 7). The opposite was observed for the heat-sensitive, B fernandoi, in which pathways related to signal transduction were up-regulated, while pathways related to carbohydrate and lipid metabolism were down-regulation under high heat. Genes involved in meiosis pathway were also up-regulated under high heat, while genes involved in “ribosome” pathway were down-regulated for this species (Fig. 5B, Supplementary Fig. 8).

(Figure made in Inkscape v.1.0; https://inkscape.org/).
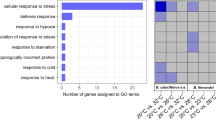
Distribution of differential expressed (DE) KEGG pathways (FDR = 0.05) in main and secondary KEGG biological categories in control vs. mild heat exposure (a), and in mild vs. high heat exposure (b). Left side represent changes captured for the heat tolerant, B. calyciflorus s.s. while right side depicts changes in the heat-sensitive B. fernandoi.
Cross-species co-expression among groups of genes
To capture similar or contrasting patterns of expression between the two species, we searched for clusters of orthologous genes exhibiting co-expression. Analysis with Clust revealed the presence of 8 such gene groups/clusters (Supplementary Fig. 9). We have focused on just three of these because their patterns appear biologically relevant. Gene cluster C1 contained 150 orthogroups exhibiting the same expression pattern in both species, i.e., they were up-regulated with increasing temperature (Fig. 6). Five KEGG pathways were significantly enriched in the C1 group (p < 0.05, FDR = 0.05), among them genes belonging to core metabolic pathways (KEGG pathway: 01100). Comparing these data to the differentially expressed gene dataset, we found in total 3 genes (phosphatidylinositol-glycan-specific phospholipase D, acidic mammalian chitinase-like, transmembrane 144) that belonged to C1 cluster and were differentially expressed between control and mild heat exposure or between control and high heat exposure in both species.
Shared and contrasting co-expression patterns of gene orthogroups (OGs) between heat-tolerant and heat-sensitive Brachionus species. C1, C6, C7 represent the numbers of clusters produced by Clust program69. The total number and co-expression pattern of all clusters are given in Supplementary Fig. 9.
Two clusters (C6 and C7) contained genes with opposing patterns of expression in the two species. In cluster C6, genes from the heat-tolerant B. calyciflorus s.s. had low expression at control conditions, were up-regulated at mild heat, and down-regulated again under high heat. In contrast, for B. fernandoi, genes in C6 cluster had higher expression under control conditions, were down-regulated at mild heat and up-regulated again at high heat. The longevity regulating pathway (KEGG pathway: 04212) was significantly enriched in C6 cluster (p < 0.05, FDR = 0.05). We found two genes that were differentially expressed in both species (BAI1-associated protein, uncharacterized protein LOC105328640) and at the same time co-expressed in this cluster. In cluster C7, for B. calyciflorus s.s. gene expression was high under control and mild heat conditions and down-regulated at high heat. However, for the heat-sensitive B. fernandoi, gene expression was low for control and mild heat conditions and up-regulated under high heat. In this cluster, a total number of 12 pathways were significantly enriched (p < 0.05, FDR = 0.05) among which pathways involved in signal transduction, nervous and endocrine systems, and replication and repair. Comparing to the differentially expressed gene dataset, we found 24 genes being differentially expressed in at least one pairwise comparison and at the same time belonging to C1 cluster for B. calyciflorus s.s. For the species B. fernandoi, 8 genes met the above two criteria. In total one gene (not annotated to any known protein) belonged to C7 cluster and was differentially expressed in mild vs. high heat for both species.
Discussion
In response to stressful environmental conditions, an immediate cellular stress response is activated. Prolonged exposure to these environmental stimuli can additionally initiate a stressor-specific secondary response to re-establish homeostasis70. In contrast to the immediate response, which is triggered mainly by macromolecular damage or the generation of oxidative stress, the homeostasis response is triggered by stressor-specific sensors that identify changes, in particular environmental variables70,71. Studies from copepods, oysters and corals reveal that short-term temperature stress and long-term temperature stress response might involve different genes/pathways since long-term response requires a significant change of expression for many genes to ensure cellular homeostasis28,72,73. However, all of the above studies reported a significant overlap in expression responses to short- and long-term heat stress.
In the present study we combined life history and gene expression data to test variation in the temperature-dependent responses of species belonging to the recently resolved B. calyciflorus species complex20 with documented differences in short-term heat tolerance (heat-tolerant vs. heat-sensitive species)23. Our phenotypic data represent long-term adaptation to temperature, while the RNA-seq data represent short-term temperature responses. Nonetheless, we observed a transcriptomic response that was largely consistent with phenotypic data from life history experiments. This has revealed both shared and species-specific patterns in gene expression in response to heat and identified key functional pathways associated with temperature adaptation in these species. Our study demonstrates the power of testing the transcriptomic response of an organism to an environmental stressor by combining transcriptome with phenotypic data.
Temperature-dependent life history responses in heat-tolerant vs. heat-sensitive Brachionus species
Rotifers living in temporally variable habitats are exposed to frequent changes in their environment that may impact their life history. In both species we found a profound effect of temperature on survival, with increasing temperature reducing their life span. Our results corroborate studies in other rotifer taxa that showed a profound effect of temperature on survival22,74,75,76,77. Due to their ectothermic nature, rotifer body temperature increases with increasing ambient temperature, which accelerates metabolic rates. Given unlimited food resources, juvenile and egg development are accelerated up to a certain critical temperature limit78. Fecundity, on the other hand did not follow the same pattern. Fecundity was significantly reduced only at high heat exposure (B. calyciflorus s.s., 32 °C; B. fernandoi, 26 °C), suggesting that fecundity is maintained across variable temperatures in both species, up to a limit above which a robust response cannot be maintained.
Population growth rate (r) is considered a proxy to evaluate environmental specializations and stress response, representing the ability of rotifers to grow in a particular environment78,79,80,81. Previous work in the rotifer complex of Brachionus plicatilis used population growth as a proxy to evaluate the salinity constrains between sibling species and found that—while species tolerated a wide range of salinities—their population growth rates responded differentially to this environmental factor81,82,83. It was hence suggested that optimal growth rate indicates adaptation to the respective environmental conditions. Consequently, environmentally triggered growth rate variation among species implies specializations, which facilitate dominance of the respective species in different periods of the year and makes sympatric co-occurrence possible15. In rotifers, broad temperature tolerance has been found7,17,22 which might reflect an adaptation to temperature fluctuations occurring in aquatic habitats. Both B. calyciflorus s.s. and B. fernandoi experience temperature fluctuations in their natural habitats and can survive a broad range of temperatures, however, their densities vary considerably, relative to ambient temperature7,17,22. High densities of B. calyciflorus s.s. have been reported in the summer up to 32 °C, while high densities of B. fernandoi have been reported during spring and winter even down to 4 °C7,17. We have shown that although both species can survive a relatively wide range of temperatures, both their population growth rate and expression of representative heat-stress genes is markedly different. Our results further corroborate that these species are specialized in their temperature tolerance, which might translate into habitat specializations and/or seasonal successions.
Differences in life histories between sibling species of the B. calyciflorus species complex have been found in response to competition and/or predation risk84. A recent study85 performed under stable food and temperature conditions (24 °C) showed differences in life history traits such as egg and juvenile developmental times, and egg production between these species. As a corollary, they demonstrated that observed differences are consistent across tested clones within species, i.e., they really represent differentiation between the species. According to our fitness results, our two species have evolved different strategies to respond to increased temperature, with B. calyciflorus adopting a life strategy of high population growth and low survival, as opposed to B. fernandoi with a strategy of low population growth and high survival. Our findings indicate that life histories of these two sibling species are differentially adapted, supporting the idea that the species are ecologically diverged and specialized for different environmental conditions, in particular with regard to temperature.
Heat shock response in heat-tolerant vs. heat-sensitive Brachionus species
Heat shock response, which involves the induction of heat shock protein (hsp) genes, is a well known and evolutionary conserved mechanism present in both prokaryotes and eukaryotes40. Induction of hsps has been connected to several stress conditions such as exposure to extreme temperatures, heavy metals, pathogens, and osmotic stress39. In the present study, expression of hsp related genes mirrored measured changes in the population growth rate across a temperature gradient. Population growth rate was low when hsp genes were up-regulated. This pattern was consistent in both species, showing that hsp genes are indeed part of a species’ stress response, when environmental conditions (here, temperature) are outside the ‘comfort zone’ for optimal growth.
Closely related species differentially adapted to cold vs. warm habitats have also been found to express hsp genes differently in other aquatic organisms such as the amphipod Eulimnogammarus. In these amphipods, species originated from a cold habitat when exposed to heat, up-regulated hsp genes at lower temperatures compared to species from a warmer habitat86. In the present study, closely related Brachionus species with differences in their ability to tolerate heat have been found to express hsp genes differently. More specifically, hsp genes were induced outside the temperature of optimal growth: in the heat-sensitive B. fernandoi, hsp genes were induced by heat, while in heat-tolerant B. calyciflorus s.s. the majority of hsp genes were induced at the lower end of temperature exposure (20 °C), indicating that 20 °C may already be cold stress for the heat-tolerant species.
Proteins of the hsp90 family serve to increase the available chaperons in the cells in order to recover from cellular stress and maintain structural integrity at high temperatures. In contrast to other hsp genes, their expression patterns supported a specific involvement in heat response, as genes encoding for hsp90 were up-regulated towards the higher temperature regime in both species. The expression of hsp90 gene has also been found to be temperature dependent in other aquatic organisms, including copepods and oysters87,88,89. This suggests that induction of hsp90 gene along with heat might be a common mechanism in aquatic organisms. In B. calyciflorus s.s., genes encoding hsp20 were also induced by high heat. Up-regulation of hsp20 genes has been reported previously from other Brachionus species and copepods as a response to elevated temperatures36,90. Transformed bacteria (Escherichia coli) expressing the Brachionus hsp20 (Br-hsps20) gene had a 100 fold increase in survival compared to the non transformed ones under high heat-stress, indicating that Br-hsp20 specifically contributes to increased thermal tolerance36. Up-regulation of hsp20 genes was found to also increase resistance to oxidative stress36. It is possible that an increase in expression of hsp20 reflects a cellular defense mechanism in response to different stressors that might be common among Brachionus species.
A reversed expression pattern between our two species was found in genes encoding for hsp70 and hsp27. In heat-tolerant B. calyciflorus s.s., genes encoding for hsp70 were significantly up-regulated towards the lower temperature, while in B. fernandoi towards the higher temperature. It seems that temperatures such as 20 °C might constitute cold stress for warm adapted species and a stress response might be initiated under these conditions. In B. calyciflorus s.s., genes encoding for hsp40 and hsp60 followed the same induction pattern as hsp70 genes, pointing towards a common mechanism regulating the expression of these three genes as reported also in B. manjavacas37. A synergistic relationship has been reported among hsp40 and hsp70 proteins, as hsp40 regulates the ATPase activity of hsp7091. These genes were induced in B. manjavacas with increasing heat37. Apparently, these genes are induced under any condition that constitutes temperature stress for a particular Brachionus species.
Metabolism response in heat-tolerant vs. heat-sensitive Brachionus species
Metabolism in ectotherms is inextricably linked to environmental temperature and its rate is accelerated by temperature increase. Genes belonging to core metabolic pathways had, in general, the same expression pattern in both the heat-tolerant B. calyciflorus s.s. and the heat sensitive B. fernandoi, showing an increased expression with temperature induction. However, there were significant differences between the species in genes involved in oxidative phosphorylation, lipid metabolism, and carbohydrate metabolism.
Oxidative stress is related to the production of toxic compounds that are called reactive oxygen species (ROS) which contribute to cellular damage and cause cellular response by modifying proteins and nucleic acids92. Exposure of ectothermic organisms to elevated temperatures accelerates mitochondrial respiration and potentially increases ROS formation93,94. ROS formation blocks heat shock response and refolding activity under heat stress, thereby leading to increasing cellular stress and ultimately heat sensitivity95. Genes related to oxidative stress were significantly up-regulated over temperature increase in heat-sensitive B. fernandoi. In contrast, these genes were significantly down-regulated over temperature increase in the heat-tolerant B. calysiflorus s.s.. Here, oxidative stress response was induced at the lowest temperature tested (20 °C). Among the differentially expressed genes were NADH dehydrogenase and glutathione S-transferase (GST). For the B. calyciflorus s.s., a significant induction (7 × fold change) of NADH dehydrogonase has been reported after sustained cold stress on 14 °C for 30 days51. Transcriptional regulation of GST genes also differed in two congeneric copepod species of the genus Tigriopus. For the species T. japonicus,GST genes were significantly down-regulated in response to temperature elevation up to 35 °C96. In the Pacific oyster, both of the above genes were down-regulated, dependent on the duration of heat exposure89. Furthermore, expression of the GST genes was either up- or down- regulated dependent on temperature intensity and duration of exposure in Brachionus plicatilis34. Transformed bacteria (Escherichia coli) expressing the BrachionusGST zeta gene were significantly protected against the oxidative stress induced by metals such as mercury and cadmium97. This indicates that repression of oxidation stress likely acts as a protective mechanism of the cells and potentially enhances heat tolerance89.
Lipid and carbohydrate metabolism are highly conserved processes that affect nearly all aspects of an organism’s biology. The consumed lipids and carbohydrates are broken down during digestion into fatty acids and simple sugars, providing the essentials to produce a wide range of metabolites that are required for development and survival. Genes related to lipid and carbohydrate metabolism were up-regulated from mild to high heat in heat-tolerant, B. calyciflorus s.s. and down-regulated in heat-sensitive B. fernandoi. Up-regulation with heat of genes related to carbohydrate metabolism has been identified after acute heat exposure in a teleost fish, Gillichthys mirabilis98 and in the Pacific oyster, Crassostrea gigas99. This suggests a need for rapid production of ATP under increasing temperatures. Apparently, B. calyciflorus s.s. has adapted to maintain its metabolism under high heat, while the heat-sensitive species B. fernandoi apparently shuts down costly metabolic processes (indicated by down-regulation of the majority of metabolic related pathways) in order to allocate available resources to survival.
Ribosomal response in heat-tolerant vs. heat-sensitive Brachionus species
Ribosome biogenesis is a complex and energy-demanding process requiring coordination of ribosomal RNA (rRNA) and ribosomal protein production. Genes encoding for ribosomal proteins have been identified several times as a part of stress response and they have been either induced or suppressed upon temperature increase87,89,100,101. Ribosomal protein related genes were up-regulated towards the lower imposed temperature of 20 °C in the heat tolerant B. calyciflorus s.s., indicating again that 20 °C likely comprise stressful conditions for this species. In contrast, in the heat-sensitive B. fernandoi, ribosomal protein related genes were up-regulated under mild heat stress (23 °C), suggesting an increased translation capacity or a protection of ribosomal function through the addition or replacement of ribosomal proteins72. However, further temperature increase up to 26 °C, resulted in down-regulation of ribosomal related genes. This suppression of protein biosynthesis may reflect cellular homeostasis or an energy saving mechanism to cope with thermal stress, as protein metabolism consumes a large amount of ATP.
Other molecular mechanisms of heat response in heat-tolerant and heat-sensitive Brachionus species
Brachionus, as most monogonont rotifers, have two reproductive modes, one asexual allowing for fast population growth and one sexual to promote recombination under unfavorable environmental conditions102,103. The sexual phase of reproduction is generally induced by environmental factors such as photoperiod, population density and food composition102,103,104. Rotifer species are capable of abandoning either the sexual or the asexual phase. Abandoning sexual reproduction is very rare in nature, however, it is a common phenomenon in clones that have been under laboratory cultivation over a long period of time and it relies on a recessive allele105,106. In B. fernandoi, increase of temperature resulted in significant up-regulation of genes related to meiosis, indicating that temperature exposure above 23 °C triggered sexual reproduction. In B. calyciflorus s.s., there was no significant up-regulation of meiosis-related genes, neither at high nor at low temperatures. Possible explanations are that this clone has lost the ability of sexual reproduction or that sexual reproduction is triggered by temperatures beyond the range tested here or stimuli other than temperature.
Epigenetic control on transcription can be achieved by many mechanisms, including DNA methylation or post-translational modifications to histone tails, including histone methylation and acetylation. It is known from genomic/transcriptomic studies of B. manjavacas and other rotifers that rotifers lack DNA methyltransferases (Dnmt1, Dnmt3) for epigenetic transcriptional regulation31,56. However, they do not lack the molecular machinery for post-translational regulation to histone tails, which play an important role in regulating gene expression. Histone tails are modified by enzymes called histone methyltransferases which catalyze the transfer of one, two or three methyl groups to lysine or arginine residues of histone protein tails107. Thus, there are two main types of histone methyltransferases, the lysine-targeting (e.g., histone-lysine N-methyltransferases) and the arginine-targeting (e.g., histone-arginine N-methyltransferases). The number of methyl groups attached in specific sites of histone tails and the amino acid which is methylated in histones determine the activation or suppression of gene transcription. In B. fernandoi, exposure to high temperatures resulted in up-regulation of histone (H3 and H4) methyltransferase genes. Histone H4K20 methyltransferase, catalyses a single methylation of lysine (20 residue) in histone H4, forming the dimethylated form. This modification has been related to silencing chromatin108. Silencing of chromatin might be related to translation suppression that we found for the heat-sensitive species under high heat exposure. In contrast, histone H3K4 trimethyltransferase catalyses a trimethylation of lysine (4 residue) in histone H3, ultimately generating a trimethylated form. This modification influences the binding of chromatin-associated proteins and in most cases the trimethylation of this position is associated with gene activation109. Transcriptional activation via trimethylation of H3K79 and H3K4 sites might be associated with a numerous environmental-information-processing pathways that were up-regulated in this species under high heat exposure.
Conclusions
We found significantly different responses to heat between heat-tolerant and heat-sensitive Brachionus species. Transcriptomic responses were found to correlate with differences in fitness and especially differences in population growth, indicating underlying mechanisms of phenotypes’ responses to environmental change. Generally, the respective species upregulated metabolism/translation related genes under the temperature where their growth rate was high, while stress related (and—in one species—meiosis related) genes were expressed beyond the temperature regime optimal for growth. What had been historically considered the single species B. calyciflorus actually comprises several closely related rotifer species, which are differentially adapted to different environmental conditions (here, temperature). We have shown that this is driven by differing gene expression profiles and peak performance temperature. These differences allow them to occur in sympatry, but in different seasons. The genes found to be upregulated under heat stress might be targets of selection potentially contributing to the ecological divergence of the two species. Additionally, their expression profiles might be used as biomarkers to assess species vulnerability to environmental conditions and climate changes.
Our experimental setup aimed at minimizing the influence of factors other than temperature. As a general rule in ectotherms, temperature has a strong influence on developmental time by shortening time to maturation and generation time. By applying a short-term heat exposure of 4 h, we minimized a differential age-structure effect among the temperature treatments. Different developmental stages (i.e. juveniles, mature females with and without eggs, no mictic females nor males were observed) were pooled together randomly in all temperatures treatments. Admittedly, while the growth conditions prior to the heat exposure guaranteed substantial population growth rate and a mixture of all life stages, we could not control for exactly equal relative share of all life stages in the experimental treatments. This might have contributed minor variations in our observed expression levels regarding genes connected to oogenesis, mitosis, and cell cycle.
Data availability
All SRA files are available from the Gen Bank database (accession numbers: SRR10426055-76). All other data that support the findings of this study are included in online supplementary material.
References
Paaijmans, K. P. et al. Temperature variation makes ectotherms more sensitive to climate change. Glob. Change Biol.19, 2373–2380 (2013).
Parmesan, C. Ecological and evolutionary responses to recent climate change. Annu. Rev. Ecol. Evol. Syst.37, 637–669 (2006).
Seifert, L., Weithoff, G., Gaedke, U. & Vos, M. Warming-induced changes in predation, extinction and invasion in an ectotherm food web. Oecologia178, 485–496 (2015).
Cullum, A. J. In Encyclopedia of Ecology (eds Jorgensen, S. E. & Fath, B.) 3557–3564 (Elsevier, Amsterdam, 2008).
Hershey, A. E., Lamberti, G. A., Chaloner, D. T. & Northington, R. M. In Ecology and Classification of North American Freshwater Invertebrates (eds Thorp, H. J. & Covich, A. P.) 659–694 (Elsevier, Amsterdam, 2010).
Papakostas, S. N., Michaloudi, E., Triantafyllidis, A., Kappas, I. & Abatzopoulos, T. J. Allochronic divergence and clonal succession: two microevolutionary processes sculpturing population structure of Brachionus rotifers. Hydrobiologia700, 33–45 (2013).
Zhang, Y. et al. Temporal patterns and processes of genetic differentiation of the Brachionus calyciflorus (Rotifera) complex in a subtropical shallow lake. Hydrobiologia807, 313–331 (2017).
Wen, X., Xi, Y., Zhang, G., Xue, Y. & Xiang, X. Coexistence of cryptic Brachionus calyciflorus (Rotifera) species: roles of environmental variables. J. Plankton Res.38, 478–489 (2016).
Xiang, X. et al. Patterns and processes in the genetic differentiation of the Brachionus calyciflorus complex, a passively dispersing freshwater zooplankton. Mol. Phylogenet. Evol.59, 386–398 (2011).
Segers, H. Global diversity of rotifers (Rotifera) in freshwater. Hydrobiologia595, 49–59 (2008).
Fontaneto, D., Kaya, M., Herniou, E. A. & Barraclough, T. G. Molecular phylogenetics and evolution extreme levels of hidden diversity in microscopic animals (Rotifera) revealed by DNA taxonomy. Mol. Phylogenet. Evol.53, 182–189 (2009).
Mills, S. et al. Fifteen species in one: deciphering the Brachionus plicatilis species complex (Rotifera, Monogononta) through DNA taxonomy. Hydrobiologia796, 39–58 (2017).
Gabaldón, C., Fontaneto, D., Carmona, M. J., Montero-Pau, J. & Serra, M. Ecological differentiation in cryptic rotifer species: what we can learn from the Brachionus plicatilis complex. Hydrobiologia796, 7–18 (2017).
Gómez, A., Serra, M., Carvalho, G. R. & Lunt, D. H. Speciation in ancient cryptic species complexes : evidence from the molecular phylogeny of Brachionus plicatilis (Rotifera). Evolution56, 1431–1444 (2002).
Serra, M. & Fontaneto, D. In Rotifers (eds Hagiwara, A. & Yoshinaga, T.) 15–32 (Springer, New York, 2017).
Suatoni, E., Vicario, S., Rice, S., Snell, T. W. & Caccone, A. An analysis of species boundaries and biogeographic patterns in a cryptic species complex: the rotifer—Brachionus plicatilis. Mol. Phylogenet. Evol.41, 86–98 (2006).
Xiang, X. L., Xi, Y. L., Wen, X. L. & Ge, Y. L. Molecular phylogeny and population genetic differentiation patterns in Brachionus calyciflorus Pallas (Rotifera) complex from two lakes in China. Ann. Limnol. Int. J. Limnol.53, 401–410 (2017).
Fontaneto, D., Giordani, I., Melone, G. & Serra, M. Disentangling the morphological stasis in two rotifer species of the Brachionus plicatilis species complex. Hydrobiologia583, 297–307 (2007).
Montero-Pau, J., Ramos-Rodríguez, E., Serra, M. & Gómez, A. Long-term coexistence of rotifer cryptic species. PLoS ONE6, e21530. https://doi.org/10.1371/journal.pone.0021530.t001 (2011).
Papakostas, S. et al. Integrative taxonomy recognizes evolutionary units despite widespread mitonuclear discordance: evidence from a rotifer cryptic species complex. Syst. Biol.65, 508–524 (2016).
Michaloudi, E. et al. Reverse taxonomy applied to the Brachionus calyciflorus cryptic species complex: morphometric analysis confirms species delimitations revealed by molecular phylogenetic analysis and allows the (re) description of four species. PLoS ONE13, e0203168. https://doi.org/10.1371/journal.pone.0203168 (2018).
Li, L., Niu, C. & Ma, R. Rapid temporal succession identified by COI of the rotifer Brachionuscalyciflorus Pallas in Xihai Pond, Beijing, China, in relation to ecological traits. J. Plankton Res.32, 951–959 (2010).
Paraskevopoulou, S., Tiedemann, R. & Weithoff, G. Differential response to heat stress among evolutionary lineages of an aquatic invertebrate species complex. Biol. Lett.14, 20180498. https://doi.org/10.1098/rsbl.2018.0498 (2018).
Khang, T. F. & Lau, C. Y. Getting the most out of RNA-seq data analysis. PeerJ3, e1360. https://doi.org/10.7717/peerj.1360 (2015).
Wray, G. A. et al. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol.20, 1377–1419 (2003).
Margres, M. J. et al. Quantity, not quality: rapid adaptation in a polygenic trait proceeded exclusively through expression differentiation. Mol. Biol. Evol.34, 3099–3110 (2017).
Herrmann, M., Ravindran, S. P., Schwenk, K. & Cordellier, M. Population transcriptomics in Daphnia: the role of thermal selection. Mol. Ecol.27, 387–402 (2018).
Smolina, I. et al. Contrasting transcriptome response to thermal stress in two key zooplankton species, Calanusfinmarchicus and C. glacialis. Mar. Ecol. Prog. Ser.534, 79–93 (2015).
Snell, T. W. & Janssen, C. R. Rotifers in ecotoxicology: a review. Hydrobiologia313, 231–247 (1995).
Hanson, S. J., Stelzer, C. P., Mark Welch, D. B. & Logsdon, J. M. Comparative transcriptome analysis of obligately asexual and cyclically sexual rotifers reveals genes with putative functions in sexual reproduction, dormancy, and asexual egg production. BMC Genomics14, 412. https://doi.org/10.1186/1471-2164-14-412 (2013).
Gribble, K. E. & Mark Welch, D. B. Genome-wide transcriptomics of aging in the rotifer Brachionus manjavacas, an emerging model system. BMC Genomics18, 217. https://doi.org/10.1186/s12864-017-3540-x (2017).
Snell, T. W. Rotifers as models for the biology of aging. Int. Rev. Hydrobiol.99, 84–95 (2014).
Park, J. C., Hagiwara, A., Park, H. G. & Lee, J. S. The glutathione S-transferase genes in marine rotifers and copepods: Identification of GSTs and applications for ecotoxicological studies. Mar. Pollut. Bull.156, 111080. https://doi.org/10.1016/j.marpolbul.2020.111080 (2020).
Han, J. et al. Effects of temperature changes on life parameters, oxidative stress, and antioxidant defense system in the monogonont marine rotifer Brachionus plicatilis. Mar. Pollut. Bull.155, 111062. https://doi.org/10.1016/j.marpolbul.2020.111062 (2020).
Kaneko, G., Kinoshita, S., Yoshinaga, T., Tsukamoto, K. & Watabe, S. Changes in expression patterns of stress protein genes during population growth of the rotifer Brachionus plicatilis. Fish Sci.68, 1317–1323 (2002).
Rhee, J. et al. Molecular and biochemical modulation of heat shock protein 20 (Hsp20) gene by temperature stress and hydrogen peroxide (H2O2) in the monogonont rotifer Brachionus sp. Comp. Biochem. Phys. C154, 19–27 (2011).
Smith, H. A., Burns, A. R., Shearer, T. & Snell, T. W. Three heat shock proteins are essential for rotifer thermotolerance. J. Exp. Mar. Biol. Ecol.413, 1–6 (2012).
Mayer, M. P. & Bukau, B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci.62, 670–684 (2005).
De Jong, L., Moreau, X. & Thiery, A. In Heat shock proteins: International Research (ed. Colombus, F.) 375–392 (Nova Science Publishers, New York, 2008).
Feder, M. E. & Hofmann, G. E. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol.61, 243–282 (1999).
Parsell, D. A. & Lindquist, S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu. Rev. Genet.27, 437–449 (1993).
Rezende, E. L., Castañeda, L. E. & Santos, M. Tolerance landscapes in thermal ecology. Funct. Ecol.19, 799–809 (2014).
Guillard, R. R. L. & Lorenzen, C. J. Yellow-green algae with Chlorophyllide. J. Phycol.8, 10–14 (1972).
Weithoff, G. Vertical niche separation of two consumers (Rotatoria) in an extreme habitat. Oecologia139, 594–603 (2004).
Weithoff, G. Dietary restriction in two rotifer species—the effect of the length of food deprivation on life span and reproduction. Oecologia153, 303–308 (2007).
Poole, R. W. Introduction to Quantitative Ecology (McGraw-Hill, New York, 1974).
Lotka, A. J. Studies on the mode of growth of material aggregates. Am. J. Sci.141, 199–216 (1907).
Zuur, A. F., Leno, E. N., Walker, N. J., Saveliev, A. A. & Smith, G. M. Mixed Effects Models and Extensions in Ecology with R (Springer, New York, 2009).
Weithoff, G. & Wacker, A. The mode of nutrition of mixotrophic flagellates determines the food quality for their consumers. Funct. Ecol.21, 1092–1098 (2007).
R Core Team. R: A Language and Environment For Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2013). https://www.R-projest.org.
Paraskevopoulou, S., Dennis, A. B., Weithoff, G., Hartmann, S. & Tiedemann, R. Within species expressed genetic variability and gene expression response to different temperatures in the rotifer Brachionus calyciflorus sensu stricto. PLoS ONE14, e0223134. https://doi.org/10.1371/journal.pone.0223134 (2019).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120 (2011).
Andrews, S. FastQC: a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc (2010).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol.29, 644–652 (2011).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinform.10, 421. https://doi.org/10.1186/1471-2105-10-421 (2009).
Kim, H. S. et al. The genome of the freshwater monogonont rotifer Brachionus calyciflorus. Mol. Ecol. Res.18, 646–655 (2018).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform.12, 323. https://doi.org/10.1186/1471-2105-12-323 (2011).
Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research4, 1521. https://doi.org/10.12688/f1000research.7563.1 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Babicki, S. et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res.44, 147–153 (2016).
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C. & Kanehisa, M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res.35, 182–185 (2007).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res.28, 27–30 (2000).
Kanehisa, M., Sato, Y., Furumichi, M., Morishima, K. & Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res.47, 590–595 (2019).
Luo, W., Friedman, M., Shedden, K., Hankenson, K. & Woolf, P. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinform.10, 161. https://doi.org/10.1186/1471-2105-10-161 (2009).
Yu, G., Wang, L., Han, Y. & He, Q. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS16, 284–287 (2012).
Luo, W. & Brouwer, C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics29, 1830–1831 (2013).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, New York, 2016).
Emms, D. & Kelly, S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol.16, 157. https://doi.org/10.1186/s13059-015-0721-2 (2015).
Abu-Jamous, B. & Kelly, S. Clust: automatic extraction of optimal co-expressed gene clusters from gene expression data. Genome Biol.19, 172. https://doi.org/10.1186/s13059-018-1536-8 (2018).
Kültz, D. Molecular and evolutionary basis of the cellular stress response. Annu. Rev. Physiol.67, 225–257 (2005).
Kültz, D. Evolution of the cellular stress proteome: from monophyletic origin to ubiquitous function. J. Exp. Biol.206, 3119–3124 (2003).
Meistertzheim, A. L., Tanguy, A., Moraga, D. & Thébault, M. T. Identification of differentially expressed genes of the Pacific oyster Crassostrea gigas exposed to prolonged thermal stress. FEBS J.274, 6392–6402 (2007).
Meyer, E., Aglyamova, G. V. & Matz, M. V. Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-seq procedure. Mol. Ecol.20, 3599–3616 (2011).
Kauler, P. & Enesco, H. E. The effect of temperature on life history parameters and cost of reproduction in the rotifer Brachionus calyciflorus. J. Freshw. Ecol.26, 399–408 (2011).
Ma, Q., Xi, Y. L., Zhang, J. Y., Wen, X. L. & Xiang, X. L. Differences in life table demography among eight geographic populations of Brachionus calyciflorus (Rotifera) from China. Limnologica40, 16–22 (2010).
Xiang, X., Jiang, R., Tao, Y., Chen, Y. & Xi, Y. Differences in life history characteristics among three sympatric evolutionary species of the Rotaria rotatoria complex. J. Freshw. Ecol.31, 351–360 (2016).
Xiang, X., Xi, Y. L., Zhang, J. Y., Ma, Q. & Wen, X. Effects of temperature on survival, reproduction, and morphotype in offspring of two Brachionus calyciflorus (Rotifera) morphotypes. J. Freshw. Ecol.25, 9–18 (2010).
Stelzer, C. P. In Rotifers (eds Hagiwara, A. & Yoshinaga, T.) 88–109 (Springer, New York, 2017).
Weisse, T., Laufenstein, N. & Weithoff, G. Multiple environmental stressors confine the ecological niche of the rotifer Cephalodella acidophila. Freshw. Biol.58, 1008–1015 (2013).
Stelzer, C. P. Evolution of rotifer life histories. Hydrobiologia546, 335–346 (2005).
Lowe, C. D., Kemp, S. J., Díaz-Avalos, C. & Montagnes, D. J. S. How does salinity tolerance influence the distributions of Brachionus plicatilis sibling species?. Mar. Biol.150, 377–386 (2007).
Gabaldón, C., Montero-Pau, J., Serra, M. & Carmona, M. J. Morphological similarity and ecological overlap in two rotifer species. PLoS ONE8, e57087. https://doi.org/10.1371/journal.pone.0057087 (2013).
Walczyńska, A. & Serra, M. Inter- and intraspecific relationships between performance and temperature in a cryptic species complex of the rotifer Brachionus plicatilis. Hydrobiologia734, 17–26 (2014).
Wang, X. L. et al. Differences in life history characteristics between two sibling species in Brachionus calyciflorus complex from tropical shallow lakes. Ann. Limnol. Int. J. Limnol.50, 289–298. https://doi.org/10.1051/limn/2014024 (2014).
Zhang, W., Lemmen, K. D., Zhou, L., Papakostas, S. & Declerck, S. A. J. Patterns of differentiation in the life history and demography of four recently described species of the Brachionus calyciflorus cryptic species complex. Freshw. Biol.64, 1994–2005 (2019).
Bedulina, D. S. et al. Expression patterns and organization of the hsp70 genes correlate with thermotolerance in two congener endemic amphipod species (Eulimnogammaruscyaneus and E. verrucosus) from Lake Baikal. Mol. Ecol.22, 1416–1430 (2013).
Schoville, S. D., Barreto, F. S., Moy, G. W., Wolff, A. & Burton, R. S. Investigating the molecular basis of local adaptation to thermal stress: population differences in gene expression across the transcriptome of the copepod Tigriopus californicus. BMC Evol. Biol.12, 170. https://doi.org/10.1186/1471-2148-12-170 (2012).
Kim, B. M., Kim, K., Choi, I. Y. & Rhee, J. S. Transcriptome response of the Pacific oyster, Crassostrea gigas susceptible to thermal stress: a comparison with the response of tolerant oyster. Mol. Cell. Toxicol.13, 105–113 (2017).
Lim, H. J. et al. Thermal stress induces a distinct transcriptome profile in the Pacific oyster Crassostrea gigas. Comp. Biochem. Phys. D19, 62–70 (2016).
Seo, J. S., Lee, Y. M., Park, H. G. & Lee, J. S. The intertidal copepod Tigriopus japonicus small heat shock protein 20 gene (Hsp20) enhances thermotolerance of transformed Escherichia coli. Biochem. Biophys. Res. Commun.340, 901–908 (2006).
Cintron, N. S. & Toft, D. Defining the requirements for Hsp40 and Hsp70 in the Hsp90 chaperone pathway. J. Biol. Chem.281, 26235–26244 (2006).
Halliwell, B. & Whiteman, M. Measuring reactive species and oxidative damage in vivo and in cell culture: How should you do it and what do the results mean?. Br. J. Pharmacol.142, 231–255 (2004).
Heise, K., Puntarulo, S., Pörtner, H. O. & Abele, D. Production of reactive oxygen species by isolated mitochondria of the Antarctic bivalve Laternula elliptica (King and Broderip) under heat stress. Comp. Biochem. Phys. C.134, 79–90 (2003).
Keller, M., Sommer, A. M., Pörtner, H. O. & Abele, D. Seasonality of energetic functioning and production of reactive oxygen species by lugworm (Arenicola marina) mitochondria exposed to acute temperature changes. J. Exp. Biol.207, 2529–2538 (2004).
Adachi, M. et al. Oxidative stress impairs the heat stress response and delays unfolded protein recovery. PLoS ONE4, e7719. https://doi.org/10.1371/journal.pone.0007719 (2009).
Han, J., Jeong, C. B., Byeon, E. & Lee, J. S. Effects of temperature changes on the generation of reactive oxygen species and the expression and activity of glutathione-S transferases in two congeneric copepods Tigriopus japonicus and Tigriopus kingsejongensis. Fish. Sci.84, 815–823 (2018).
Lee, J. S., Kang, H. M., Park, J. C. & Lee, J. S. Protective role of the freshwater rotifer Brachionus calyciflorus glutathione S-transferase zeta 3 recombinant protein in response to Hg and Cd. Comp. Biochem. Phys. B.243, 110435 (2020).
Buckley, B. A., Gracey, A. Y. & Somero, G. N. The cellular response to heat stress in the goby Gillichthys mirabilis: a cDNA microarray and protein-level analysis. J. Exp. Biol.209, 2660–2677 (2006).
Yang, C., Gao, Q., Liu, C., Wang, L. & Zhou, Z. The transcriptional response of the Pacific oyster Crassostrea gigas against acute heat stress. Fish Shellfish Immun.68, 132–143 (2017).
Podrabsky, J. E. & Somero, G. N. Changes in gene expression associated with acclimation to constant temperatures and fluctuating daily temperatures in an annual killifish Austrofundulus limnaeus. J. Exp. Biol.207, 2237–2254 (2004).
Truebano, M. et al. Transcriptional response to heat stress in the Antarctic bivalve Laternula elliptica. J. Exp. Mar. Biol. Ecol.391, 65–72 (2010).
Gilbert, J. J. Dormancy in Rotifers. Trans. Am. Microsc. Soc.93, 490–513 (1974).
Schröder, T. Diapause in monogonont rotifers. Hydrobiologia546, 291–306 (2005).
Pourriot, R. & Snell, T. W. Resting eggs in rotifers. Hydrobiologia104, 213–224 (1983).
Stelzer, C. P. Obligate asex in a rotifer and the role of sexual signals. J. Evol. Biol.21, 287–293 (2008).
Stelzer, C. P., Schmidt, J., Wiedlroither, A. & Riss, S. Loss of sexual reproduction and dwarfing in a small metazoan. PLoS ONE5, e12854. https://doi.org/10.1371/journal.pone.0012854 (2010).
Wood, A. In Proteins in Eukaryotic Transcription (eds Conaway, J. W. & Conaway, R. C.) 201–222 (Elsevier Academic Press, Amsterdam, 2004).
Hyun, K., Jeon, J., Park, K. & Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med.49, e324. https://doi.org/10.1038/emm.2017.11 (2017).
Ruthenburg, A. J., Allis, C. D. & Wysocka, J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell.25, 15–30 (2007).
Acknowledgements
This work was supported by University of Potsdam, focus area “Adaptation and climate change”. We would like to thank the technician Christina Schirmer for helping in performing the life-table experiments and Caterina Karnhal for helpimg with the preliminary experiments. We additionally want to thank Dr. Alexander Wacker for contributing in R bootstrapping analysis of the population growth data. Finally, we thank Prof. Dr. Michael Lenhard and Dr. Marco Ende for access to their computing servers. We also acknowledge the support of the Deutsche Forschungsgemeinschaft and Open Access Publishing Fund of University of Potsdam. Open access funding provided by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
The project was conceived by R.T., S.P., and G.W. The lab work and the life-table experiments were performed by S.P. Data analysis and interpretation of results was carried out by S.P. and A.B.D., with input from R.T. and G.W. The manuscript was drafted by S.P. Further editing and manuscript finalization was coordinated by S.P., with contributions from all authors. All contributing authors read and agreed to the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Paraskevopoulou, S., Dennis, A.B., Weithoff, G. et al. Temperature-dependent life history and transcriptomic responses in heat-tolerant versus heat-sensitive Brachionus rotifers. Sci Rep 10, 13281 (2020). https://doi.org/10.1038/s41598-020-70173-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-70173-0
This article is cited by
-
A genome-wide screening of the 70 kDa heat shock protein (HSP70) genes in the rotifer Brachionus plicatilis sensu stricto with a characterization of two heat-inducible HSP70 genes
Cell Stress and Chaperones (2023)
-
Synchaeta’s community in the urban coastal area of the Thessaloniki Bay
Hydrobiologia (2023)
-
Niche differentiation in rotifer cryptic species complexes: a review of environmental effects
Hydrobiologia (2023)
-
Differential transcriptomic responses to heat stress in surface and subterranean diving beetles
Scientific Reports (2022)
-
Variation in heat shock protein 40 kDa relates to divergence in thermotolerance among cryptic rotifer species
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.