Abstract
Leprosy, which is caused by the human pathogen Mycobacterium leprae, causes nerve damage, deformity and disability in over 200,000 people every year. Because of the long doubling time of M. leprae (13 days) and the delayed onset of detectable symptoms, which is estimated to be approximately 3–7 years after infection, there is always a large percentage of subclinically infected individuals in the population who will eventually develop the disease, mainly in endemic countries. piRNAs comprise the largest group of small noncoding RNAs found in humans, and they are distinct from microRNAs (miRNAs) and small interfering RNAs (siRNAs). piRNAs function in transposon silencing, epigenetic regulation, and germline development. The functional role of piRNAs and their associated PIWI proteins have started to emerge in the development of human cancers and viral infections, but their relevance to bacterial diseases has not been investigated. The present study reports the piRNome of human skin, revealing that all but one of the piRNAs examined are downregulated in leprosy skin lesions. Considering that one of the best characterized functions of piRNAs in humans is posttranscriptional mRNA silencing, their functions are similar to what we have described for miRNAs, including acting on apoptosis, M. leprae recognition and engulfment, Schwann cell (SC) demyelination, epithelial–mesenchymal transition (EMT), loss of sensation and neuropathic pain. In addition to new findings on leprosy physiopathology, the discovery of relevant piRNAs involved in disease processes in human skin may provide new clues for therapeutic targets, specifically to control nerve damage, a prominent feature of leprosy that has no currently available pharmaceutical treatment.
Similar content being viewed by others
Introduction
Mycobacterium leprae, the causative agent of leprosy, is the only known bacterium that infects Schwann cells (SCs) of peripheral nerves. In addition to SCs, M. leprae is an obligate intracellular pathogen that infects macrophages and dendritic cells; it mostly affects the skin, mucosa and nerves but may be transported within these cells anywhere in the body1. Over 200,000 people have been diagnosed with leprosy every year for the last 10 years, and approximately half of these people are clinically recognized to have some physical disability that may result in prejudice and stigma2. It is likely that the number of cases is much higher than officially described, and it is thought that there may be up to 4 million hidden cases of leprosy today waiting to be diagnosed, mainly due to loss of medical expertise and lack of access to the health system3,4.
Several piRNomes have been described, and most of these have been related to different forms of cancer. piRNAs are important for gametogenesis, embryogenesis and stem cell maintenance5. Therefore, piRNAs may have high importance for clinical implications for a variety of diseases.
Posttranscriptional gene silencing by piRNA is mediated by two mechanisms as follows: (i) piRNAs target transposon sequences found in the 3-UTRs or 5-UTRs of mRNAs; and (ii) if the piRNA is derived from a pseudogene and antisense transcript transcribed from the opposite strand of the endogenous genes, then the piRNA targets the mRNA of the corresponding endogenous gene5. The piRDisease database 20196 cites 28 diseases with some proven piRNA interference, but none of them are caused by infectious agents. However, there have been reports of piRNA acting on viral diseases7,8. In addition, mapping piRNAs onto specific diseases may increase the availability of possible biomarkers, therapeutic targets9 and chemoresistance evaluation10, which are critical issues for modern leprosy control11,12,13.
Results
After quality control, alignment, and transcript quantitation, several small noncoding RNAs (sncRNAs) and other transcript fragments were identified. From these sequences, an average of 26,000 reads per sample were recognized as piRNAs, identifying a total of 337 differentially expressed (DE) piRNAs in leprosy patients. In total, 15, 86 and 69 piRNAs were exclusively expressed in skin lesions from tuberculoid (TT) tissue, lepromatous (LL) tissue and healthy subject (HS, control) tissue, respectively. In addition, 139 piRNAs were expressed in all tissue samples irrespective of whether from healthy or leprosy patient skin tissue, and 28 piRNAs were significantly expressed in more than one group (Supplementary Fig. 1).
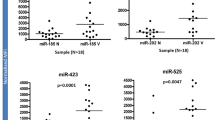
Sample comparisons were followed by the characterization of the involved piRNAs. Comparison of leprosy patient (TT + LL samples) to HS showed that 14 piRNAs were downregulated in leprosy skin (Supplementary Table 1).From these 14 piRNAs, 5 showed the best sensitivity/specificity relationship in ROC analysis (AUC > 0.9—Fig. 1A, Supplementary Fig. 2), and all 5 of these piRNAs were also DE in other analyses (TT vs. HS and LL vs. HS). In the comparison of TT samples to HS samples, 21 piRNAs were DE. Of these, 20 piRNAs were downregulated in TT, but piR-hsa-27283 was upregulated in TT. From the DE piRNAs, 13 showed the best sensitivity/specificity relationship in ROC analysis (AUC > 0.9—Fig. 1B, Supplementary Fig. 3). In the comparison of LL samples to HS samples, 16 piRNAs were downregulated in LL. Of these 16 DE piRNAs, 8 showed the best sensitivity/specificity relationship in ROC analysis (AUC > 0.9—Fig. 1C, Supplementary Fig. 4). To avoid ambiguous or multimapping identification, colocalized piRNAs were merged (Supplementary Fig. 5).
Aiming to better understand the regulatory role of these 25 downregulated piRNAs (DE with AUC > 0.9), we examined potential piRNA targets using the miRanda14 program. Because gene silencing with piRNAs may occur through several mechanisms5, we used both 3′ and 5′ UTR regions of genes in miRanda (Supplementary Fig. 6).
We identified over 5,000 putative target genes, and 955 of these target genes were targeted simultaneously in both their 5′ and 3′ regions. Using gene ontology (GO; Supplementary Fig. 7), we selected 174 genes that were the putative targets of at least three piRNAs simultaneously (Supplementary Table 2, Supplementary Table 3), and we performed GO enrichment aiming to identify biological processes significantly (p-value < 0.05) involved in the leprosy phenotype (Fig. 2, Supplementary Fig. 7).
Red indicates differentially expressed piRNAs, and blue indicates potential simultaneous mRNA targets. Green indicates biological processes significantly associated with leprosy phenotype (p-value < 0.05 in gene ontology enrichment). mRNAs were selected based on the following two criteria: (i) target of at least three DE piRNAs and (ii) involved in at least one biological process significantly associated with leprosy phenotype. (*) Indicates colocalized piRNAs.
DE piRNAs in skin lesions allowed the identification of piRNAs that differentiate LP from HS and also differentiate LP poles, TT and LL from HS. The DE piRNAs were all downregulated in all comparisons (except piR-hsa-27283 in TT vs. HS). In silico analyses of targets and GO enrichment (considering only the posttranscriptional function of piRNAs) revealed genes and pathways important in the leprosy phenotype and the mechanism of immunophysiopathology.
Discussion
A recent finding of an unexpected role for piRNA in somatic cells has revealed that the capacity to promote sensory axon regeneration after injury may be blocked by piRNA. The piRNA pathway acts against axon regrowth independent of its return to the nucleus and transcriptional silencing. However, with posttranscriptional gene silencing, piRNA inhibition of axonal regeneration is blocked, and sensory nerve regeneration occurs15.
Our results demonstrated that, with one exception, all DE piRNAs are downregulated, indicating that targeted genes involved in regeneration of peripheral nerves infected by M. leprae may be silent or expressed at low levels (Fig. 3). Considering that one piRNA, like the upregulated piR-hsa-27283, may have approximately 3 thousand genes as targets, it is difficult to analyse its functions as a single piRNA. On the other hand, as the only upregulated piRNA on leprosy skin lesions, piR-has-27283 may be useful as a biomarker of disease, a feature that can be tested in the future. Another important mechanism of regulating SC maturation and axon regeneration after nerve injury is promoted by pro-regenerative macrophages that function to clear debris within the nerve microenvironment. These pro-regenerative macrophages are essential for axon regeneration, and their presence in the lesional microenvironment promotes remyelination and SC differentiation, reducing immature SC density16. These mechanisms are regulated by two important molecules, namely, growth arrest specific 6 (GAS6) and IL-6, and the pathways of these molecules are regulated by piRNAs as described in (Fig. 3).
piRNAs related to the epigenetic control of pathways involved in vesicles transport, phagocytosis, demyelination, apoptosis, inflammation, loss of sensation, and pain are differentially expressed in leprosy patient (LP). DE piRNAs do not act on the same targets of miRNA, but they synergistically regulate the same pathway, e.g., infection route by AKA1, TMEM59 and ITSN1 involved in mycobacterial endocytosis in neurons and anti-apoptotic pathways (CARF). Skin anesthesia or hypoesthesia and pain are characteristic of leprosy and have been related to calcium and potassium channel proteins, including SHISA6, KCNMA1, KCNQ2, KCNH1, CACNA1C, SLC1A2 and KCNK13, which are all targets of piRNAs DE. Inflammation and neuropathic pain are part of leprosy clinical aspects and may be moderated by MAPK, HOMER1, PLEKHG4, SLC4A2, GLI4, and DUSP8 by RHOA pathways. ETV6 and CARF are associated with neural regeneration and differentiation of stem cells into Schwann cells. Other important mechanisms of regulating SC maturation and axon regeneration after nerve injury are promoted by pro-regenerative macrophages and are dependent on GGCX, vitamin K, IL-6R and DUSP8 induced by GAS6 stimulation. Pro-regenerative macrophages clear debris in the nerve microenvironment and are essential for axon regeneration. The permanence of pro-regenerative macrophages in the lesional microenvironment promote remyelination and SC differentiation, reducing immature SC density.
GAS6 is involved in the stimulation of cell proliferation and is associated with a variety of diseases16. GAS6 can act through Tyro3 (TAM family of receptors) and contribute to cell survival, invasion, migration, chemoresistance, and metastasis, and as a result, several classes of TAM inhibitors are being developed in clinical studies17. GAS6 is produced by quiescent fibroblasts, osteoblasts, and macrophages through GGCX with vitamin K as a coenzyme to the completion of GAS6 carboxylation16. Thus, the regenerative function of macrophages in SCs infected with M. leprae may depend on the expression of vitamin K-dependent genes (such as GGCX) that produce GAS6 within the microenvironment to promote regrowth of damaged nerves15.
In the present study, the downregulation of piRNAs target to IL6R can allow the expression of IL6R in the membrane of pro-regenerative macrophages. This receptor is necessary for the effective action of IL-6 that has been described in patients with higher titers18,19 and as a marker of neuropathic pain in patients with leprosy20. Therefore, IL-6 may be related to bette nerve regeneration in addition to the stimulation of a more efficient cellular response under the auspices of a downregulated piRNA profile.
Corticosteroids are used to treat acute nerve damage in leprosy, but they have moderate efficacy in treating nerve function impairment. It has been shown that corticosteroids do not have a superior effect to placebo in improving nerve function21. After a demyelinating injury, oligodendrocyte precursor cells (OPCs) become activated and subsequently proliferate and migrate to the lesion site22, but they have limited action related to corticosteroid treatment due to inhibition of pro-regenerative macrophage uptake23,24. Pro-regenerative macrophages persist at the site of the injured nerve and regulate SC maturation and conduction velocity postinjury by a complex GAS615,16-dependent transcriptional profile via GGCX and vitamin K pathways. Increased GAS6 has been described in type 2 reactional episodes of erythema nodosum leprosum (ENL)25. The downregulated expression of the piRNAs that control GAS6 production in LP may be related to higher levels of GAS6 in ENL.
Three studies published between 1950 to 1953 showed regression of leprosy nodules in patients treated with vitamin K3 derivatives, which was decades prior to the use of multidrug therapy (MDT) to treat leprosy patients26,27,28. These studies evaluated the daily injection of 50 mg of vitamin K into patients experiencing leprosy reaction, and they reported an extraordinarily rapid improvement within five days with a lowering of fever, alleviation of pain symptoms and full recovery of the patient. Within fifteen days of this treatment, all symptoms of the reactional episode had disappeared without any infiltration. After eight months, the skin lesions disappeared completely and did not persist, without any tingling. Later examination of nasal swab and skin biopsies for the presence of acid-fast bacilli were found to be negative. Based on the interaction of GAS6 and vitamin K pathways, these reports of relief from the often debilitating symptoms of ENL and rapid improvement of skin lesions prior to the use of MDT and corticosteroids suggest the possibility that piRNAs may be involved in posttranscriptional mechanisms promoting migration of pro-regenerating macrophages to the site of nerve injury followed by regrowth of damaged nerves.
The absence of piRNAs allows peripheral nerve regeneration29,30,31,32. According to the present data, all DE piRNAs in leprosy lesions are downregulated, suggesting that nerve regeneration is not inhibited by piRNAs in leprosy patients. Furthermore, the present work discloses new mechanisms involved on leprosy physiopathology and reveals novel therapeutic targets involved in neurodegeneration and neuropathic pain.
Methods
Biological material and clinical data collection
In total, 17 lesion tissue samples were collected as follows: (i) 6 from healthy subjects (HS); (ii) 6 from lepromatous leprosy patients (LL), and (iii) 5 from tuberculoid leprosy patients (TT). All samples were obtained prior to MDT at URE Dr. Marcello Candia (URE) located in the city of Marituba (Pará, Brazil). Immediately after collection, all samples were frozen and stored in RNAlater (SIGMA R0901). Informed consent was obtained from all individual participants.
RNA extraction and piRNA library preparation
Total RNA was extracted using TRIzol (Thermo Fisher Scientific, Waltham, MA, USA). After isolation, total RNA was stored at − 80 °C until further analysis. Total RNA amount and integrity were determined using the Qubit R 2.0 (Life Technologies, Foster City, CA, USA) and Agilent 2,200 TapeStation (Agilent Technologies, USA) according to the manufacturer’s specifications. Samples with concentrations above 100 μg/uL and RIN (RNA Integrity Number) > 5 were used for sequencing. The concentration of 1 μg/5 μL of sample was used as input to library preparation. We synthesized 17 libraries using the TruSeq Small RNA Library Preparation Kit (Illumina R, San Diego, CA, USA) according to the manufacturer’s instructions. The pool of libraries was quantified using ABI 7,500 equipment (Applied Biosystem, CA, USA) and a KAPA Library Quantification Kit (KAPA Biosystems, Woburn, MA). The libraries were sequenced using a MiSeq Sequencing System (Illumina R, San Diego, CA, USA) and the MiSeq Reagent Kit v3 150 cycle (Illumina R, San Diego, CA, USA).
NGS read quality control, alignment and quantitation
After sequencing, resulting reads were preprocessed and quality filtered (QV > 25). STAR Aligner (v. 2.4.0.1) was used to map the reads to the human genome (v. hg19)33. Htseq-count software34 with piRbase annotation35 was used to quantify transcripts. Before piRNA quantitation, we grouped colocalized piRNAs to avoid ambiguous counting. Differential expression analysis. To identify the differentially expressed (DE) piRNAs, the following four analyses were performed: (i) leprosy vs. HS samples, (ii) TT vs. HS samples, (iii) LL vs. SH and (iv) TT vs. LL samples. For these analyses, raw read count (read counts 10 in, at least, one sample) were used with DESeq2 library34 in the statistical platform R (R Core Team, 2017). piRNAs satisfying the following criteria were tagged as differentially expressed: |Log2 (fold-change)|> 2 and p-value < 0.05. For graphical analysis of piRNAs, expression data were normalized to RPKM. The area under the curve (AUC > 0.9) from the Receiver Operating Characteristic (ROC) analysis was used to identify potential biomarkers among DE piRNAs. All statistical analyses were performed on the platform R.
Identification of DE piRNA target RNAs
piRNA mRNA targets were identified by predicting the complementarity sequence between each piRNA and the 5-UTRs or 3-UTRs regions of all known human mRNAs (RefSeq gene annotations, Human genome assembly, hg19) with miRanda [v3.3a], applying a stringent alignment score (sc; 170), energy threshold (free energy-30.0 kcal/mol) and at least 80% of complementarity. Functional analysis was performed using cluster profiler library36 in R37 to identify all biological processes significantly associated (p 0.05) to piRNA-targeted mRNAs5. PCAs and heatmaps were created in R, and the piRNA-gene interaction network was constructed using Cytoscape software v 3.2.138.
Ethics statement. This study was performed in accordance with the recommendations of the Brazilian National Ethics Committee (CONEP) guidelines approved by Pará Federal University Ethics Committee number CAAE 26765414.0.0000.0018 with written informed consent from all subjects, in accordance with the Declaration of Helsinki. The protocol was approved by the Pará Federal University Ethics Committee.
Data availability
The small RNAseq number register is ERP105473 on European Nucleotide Archive database.
References
Scollard, D. M. et al. The continuing challenges of leprosy. Clin. Microbiol. Rev. 19, 338–381 (2006).
World Health Organization. Global leprosy update, 2018: moving towards a leprosy-free world–situation de la lèpre dans le monde, 2018: parvenir à un monde exempt de lèpre. Wkly. Epidemiol. Rec. Relev. épidémiologique hebdomadaire 94, 389–411 (2019).
Salgado, C. G. et al. Are leprosy case numbers reliable?. Lancet Infect. Dis. 18, 135–137 (2018).
Smith, W. C., van Brakel, W., Gillis, T., Saunderson, P. & Richardus, J. H. The missing millions: a threat to the elimination of leprosy. PLoS Neglect. Trop. Dis. 9, 1 (2015).
Watanabe, T. & Lin, H. Posttranscriptional regulation of gene expression by piwi proteins and pirnas. Mol. cell 56, 18–27 (2014).
Muhammad, A., Waheed, R., Khan, N. A., Jiang, H. & Song, X. Pirdisease v1.0: A manually curated database for pirna associated diseases. Database 20, 19 (2019).
Wang, Y. et al. Pirna profiling of dengue virus type 2-infected asian tiger mosquito and midgut tissues. Viruses 10, 213 (2018).
Varjak, M. et al. Characterization of the zika virus induced small rna response in aedes aegypti cells. PLoS Neglect. Trop. Dis. 11, e0006010 (2017).
Chalbatani, G. M. et al. Biological function and molecular mechanism of pirna in cancer. Pract. Lab. Med. 13, e00113 (2019).
Wang, Y. et al. A pirna-like small rna induces chemoresistance to cisplatin-based therapy by inhibiting apoptosis in lung squamous cell carcinoma. Mol. Ther. Acids 6, 269–278 (2017).
Bahmanyar, E. R. et al. Leprosy diagnostic test development as a prerequisite towards elimination: requirements from the user’s perspective. PLoS Neglect Trop Dis 10, 1 (2016).
Prasad, P. & Kaviarasan, P. Leprosy therapy, past and present: can we hope to eliminate it?. Indian J. Dermatol. 55, 316 (2010).
Rosa, P. S. et al. Emergence and transmission of drug-/multidrug-resistant mycobacterium leprae in a former leprosy colony in the Brazilian amazon. Clin. Infect. Dis. 70, 2054–2061 (2020).
Enright, A. J. et al. Microrna targets in drosophila. Genome Biol. 5, R1 (2003).
Roque, C. G. & Hengst, U. Wimpy nerves: Pirna pathway curbs axon regrowth after injury. Neuron 97, 477–478 (2018).
Stratton, J. A. et al. Macrophages regulate schwann cell maturation after nerve injury. Cell Rep. 24, 2561–2572 (2018).
Davra, V., Kimani, S. G., Calianese, D. & Birge, R. B. Ligand activation of tam family receptors-implications for tumor biology and therapeutic response. Cancers 8, 107 (2016).
Pires, C. A. A. et al. Expression of interleukin-1β and interleukin-6 in leprosy reactions in patients with human immunodeficiency virus coinfection. Acta Trop. 172, 213–216 (2017).
Nada, E. et al. Serum interleukin-6 and interferon-γ in patients with leprosy. Egypt. J. Dermatol. Venerol. 38, 80 (2018).
Jardim, M. et al. Il-6 is a marker of neuropathic pain in patients with leprosy. Neurology 88, 314 (2017).
Van Veen, N. H., Nicholls, P. G., Smith, W. C. S. & Richardus, J. H. Corticosteroids for treating nerve damage in leprosy. Cochrane Database Syst. Rev. 5, 2 (2016).
Kotter, M. R., Stadelmann, C. & Hartung, H.-P. Enhancing remyelination in disease—can we wrap it up?. Brain 134, 1882–1900 (2011).
Wang, Y.-Z. et al. Action mechanism of corticosteroids to aggravate guillain-barré syndrome. Sci. Rep. 5, 13931 (2015).
Fingerle-Rowson, G. et al. Regulation of macrophage migration inhibitory factor expression by glucocorticoids in vivo. Am. J. Pathol. 162, 47–56 (2003).
Pocaterra, L. et al. Clinical course of erythema nodosum leprosum: an 11-year cohort study in Hyderabad, India. Am. J. Trop. Med. Hygiene 74, 868–879 (2006).
Merklen, F. & Riou, M. Regression of hansen’s nodules in a patient treated only with vitamin k3 derivatives. Bull de la Soc francaise de dermatologie et de syphiligraphie 60, 55–56 (1953).
Merklen, F. & Riou, M. Trial of an ester of di-hydrovitamin k3 in leprosy. La Press. Medicale 60, 1570–1572 (1952).
Floch & Sureau. Vitamin k therapy in the leprosy. Bull. de la Soc. de pathologie exotique et de ses filiales 46, 631–8 (1953).
Kim, K. W. et al. A neuronal pirna pathway inhibits axon regeneration in C. elegans. Neuron 97, 511–519 (2018).
Sohn, E. J., Jo, Y. R. & Park, H. T. Downregulation miwi-pirna regulates the migration of schwann cells in peripheral nerve injury. Biochem. Biophys. Res. Commun. 519, 605–612 (2019).
Zhang, C. et al. Pirna-dq541777 contributes to neuropathic pain via targeting cdk5rap1. J. Neurosci. 39, 9028–9039 (2019).
Jung, J. et al. Gas6 prevents epithelial-mesenchymal transition in alveolar epithelial cells via production of pge2, pgd2 and their receptors. Cells 8, 643 (2019).
Dobin, A. et al. Star: ultrafast universal rna-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S., Pyl, P. T. & Huber, W. Htseq—a python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Zhang, P. et al. Pirbase: A web resource assisting pirna functional study. Database 20, 14 (2014).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. Clusterprofiler: An r package for comparing biological themes among gene clusters. Omics 16, 284–287 (2012).
Team, R. C. et al. R: A language and environment for statistical computing. (2013).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Acknowledgements
This work was supported by CNPq (486183/2013-0 CNPq Grant for MBS; 448741/2014-8 Grant for JGB; 428964/2016-8 Grant and 313633/2018-5 scholarship for CGS; and 306815/2018-4 Grant for ARS), CAPES PROAMAZONIA 3288/2013, CAPES Biocomputacional—RPGPH (3381/2013), Ministério da Saúde do Brasil 035527/2017, PROPESP/UFPA, Companhia Vale do Rio Doce 27756/2019, Fulbright Scholar to Brazil 2019–2020 (JSS), and the Heiser Program of the New York Community Trust for Research in Leprosy (JGB, MBS, CGS and JSS) Grants P15-000827, P16-000796 and P18-000250.
Author information
Authors and Affiliations
Contributions
P.P., C.G.S., S.S., and A.R.S. designed research. C.G.S., R.C.B., J.G.B. and A.R.G. enrolled patients, performed, and registered clinical diagnosis. P.P., T.V.S., A.M.R.S. and A.F.V. sequenced piRnome of skin lesions samples. A.M.R.S. and F.C.M. performed bioinformatic analyses. P.P., M.B.S., A.R.S., T.V.S., J.G.B., J.S.S., S.S., C.G.S. and A.R.S. analyzed the data. P.P., M.B.S., F.C.M., J.S.S., S.S., and A.R.S. wrote the article. P.P., M.B.S., R.C.B., A.R.G., T.V.S., A.M.R.S., A.F.V., J.B., J.S.S., S.S., C.G.S. and A.S. agree with manuscript results and conclusions.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pinto, P., da Silva, M.B., Moreira, F.C. et al. Leprosy piRnome: exploring new possibilities for an old disease. Sci Rep 10, 12648 (2020). https://doi.org/10.1038/s41598-020-69355-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-69355-7
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.