Abstract
Species delimitation offered by DNA-based approaches can provide important insights into the natural history and diversity of species, but the cogency of such processes is limited without multigene phylogenies. Recent attempts to barcode various Solenopsidini ant taxa (Hymenoptera: Formicidae: Myrmicinae), including the thief ant Solenopsis saudiensis Sharaf & Aldawood, 2011 described from the Kingdom of Saudi Arabia (KSA), were precipitated by the unexpected existence of a closely related species, the Nearctic S. abdita Thompson, 1989 within the S. molesta species complex native to Florida. This finding left the species status of the former uncertain. Here, we investigated the taxonomy and phylogeny of these two species to determine whether or not S. abdita represents a new global tramp species. We inferred a phylogeny of the two species using DNA sequence data from four nuclear genes (Abd-A, EF1α-F1, EF1α-F2, and Wingless) and one mitochondrial gene (COI) sampled from populations in Florida, Guatemala, Hawaii, and Saudi Arabia. Both species clustered into one distinct and robust clade. The taxonomy of S. saudiensis was re‐examined using morphometrics. A reassessment of the morphological characters used to diagnose the worker and queen castes were consistent with molecular evidence. Based on combined morphological and molecular evidences S. saudiensis is declared as a junior synonym of S. abdita (syn. nov.). In addition, our findings indicate that S. abdita is a novel global tramp species which has a far wider distribution than previously thought and has established itself in many new habitats and different geographic realms.
Similar content being viewed by others
Introduction
Ants are a highly adaptive eusocial arthropod group with impressive diversity and abundance and are encountered in most terrestrial ecosystems1,2. The cosmopolitan genus Solenopsis Westwood, 1840 (Formicidae: Myrmicinae) is composed of two subgroups, fire ants and thief ants. While fire ants are infamous for being aggressive and highly invasive (e.g. Solenopsis invicta and S. geminata), the majority of Solenopsis species belong to the thief ants, a group consisting mainly of minute, subterranean species with monomorphic or mildly polymorphic workers3. The genus is speciose, with 196 recognized valid species and 22 subspecies4 widespread in the tropics and warm temperate regions5,6,7, with a majority of species reported from the Neotropical realm8. Several traits, such as nest type, tramp behavior, omnivory, and social polymorphism (monogyny and polygyny in a single species), facilitate their establishment in newly colonized environments9,10. The fire ants of the S. geminata and saevissima species groups11, e.g. S. geminata and S. invicta9, are notorious pests. Among the most damaging invasive ants in the world12, they have spread around the world via human commerce13,14. While the expansion of urban ecosystems together with the extraordinary growth in international trade drive the spread and establishment of many species outside their native ranges15,16, Solenopsis appears particularly well-adapted to urban habitats due to their generalized diet17.
Biological invasions are the indirect outcome of human-mediated drivers of global change. Today, such invasions pose major challenges to agriculture and ecological balance18. The number of invasive species has continued to rise owing to growth in international trade and globalization19,20. Immediate and effective control and management strategies are predicated on accurately identifying invading pest species, placing taxonomy and systematics research at the forefront of invasive species exploration21. However, species identifications are not always easy since many alien insects are morphologically difficult to distinguish from native species. The absence of diagnostic morphological characters, a lack of modern taxonomic revisions and keys, poor taxonomic histories, and unknown species origin can all hamper the identification of invasive species. Morphological data22,23, molecular data 24,25, or a combination thereof26,27 are needed to enable the correct identification of new invasive species.
Recent research highlighted a potentially diverse fauna of Solenopsis on the Arabian Peninsula28,29 with only a single introduced species, S. geminata (Fabricius, 1804), known from the United Arab Emirates (UAE)30,31. Six Solenopsis species have been recorded from the Arabian Peninsula: S. elhawagryi Sharaf and Aldawood, 2012, S. geminata (Fabricius, 1804), S. omana Collingwood and Agosti, 1996, S. saudiensis Sharaf and Aldawood, 2011 (herein treated as a junior synonym of S. abdita), S. sumara Collingwood and Agosti, 1996, and S. zingibara Collingwood and Agosti, 199628,29,30,32. However, only two species were recorded from the KSA, S. elhawagryi and S. saudiensis. It is likely that more species (both native and exotic) will be documented in the future, given the vast areas of the KSA that remain to be explored.
Despite the abundance and ecological significance of some fire ant species, the genus as a whole remains poorly studied. The systematics of this group has been plagued by difficulties in distinguishing species and their relationships. The group presents a paucity of constant and reliable diagnostic morphological characters coupled with evidently common intraspecific variations that go beyond interspecific differences3,33. These difficulties are particularly daunting in the large, polymorphic fire ant group, where the worker caste can provide useful characters for species identification33, but even more dire in the thief ants, which are mainly monomorphic (e.g. S. saudiensis29), and offer even fewer diagnostic characters for species delimitation. This surely represents a major impediment for faunistic inventories and biogeographical studies.
Members of the genus can be recognized by the following character states: masticatory margin of mandibles armed with three or four teeth; palp formula 2,2 or 1,2; clypeus longitudinally bicarinate, with a median area distinctly elevated and deeply inserted posteriorly between the frontal lobes; anterior margin of clypeus with a single long median seta; antennae 10-segmented with a two-segmented club; frontal carinae and antennal scrobes absent; propodeum unarmed34. However, the α-taxonomy of Solenopsis is still confused and identification to a species level is substantially challenging. Regardless of the virtually unknown ecology and cryptic habits of most species, two basic issues may explain the difficulty of identifying specimens of Solenopsis. First, worker caste morphology lacks diagnostic characters, especially for monomorphic species (e.g. S. abdita) that present troublesome intraspecific variation in morphological traits3,33,35. Second, most species were inadequately described due to limited material36. Third, rampant misidentifications, and the tendency to lump difficult-to-identify specimens into “wastebin species” groups causes bias and distorts the possible species lists of Solenopsis fauna for any given area. Finally, the use of numerous trinomials and quadrinomials has caused serious taxonomical ambiguities37.
Such taxonomic complexities have fueled a growing interest in the adoption of DNA-based approaches for ant descriptions and identifications. Character-based DNA barcoding, using short mitochondrial DNA fragments of the cytochrome c oxidase I (COI) gene, was introduced as a tool for rapid species identification or delimitation in ant surveys38,39,40,41,42 and systematic revisions43,44. Additionally, it represents a useful tool to assign different castes to a species. This is particularly useful where morphological differences between workers and sexuals may be insurmountable and only co-occurrence in nests or molecular methods allow robust assignment43. Since its introduction45,46, DNA barcoding has been extensively used47,48,49 and significantly refined50,51,52, but several pitfalls of barcoding approaches remain53. Therefore, species hypotheses based on DNA barcodes should ideally be additionally supported by additional molecular, morphological, geographical, ecological and/ or ethological data54.
A recent barcoding study of Saudi Arabian S. saudiensis unfortunately fell prone to such limitations55. Briefly, the authors used biased taxonomic sampling, heavy reliance on the Barcode of Life DataSystems (BOLD) data lacking solid taxonomic identifications, and inappropriate interpretation of analyses to arrive at misleading conclusions. Rasool et al.55 interpreted the finding of a single COI haplotype as proof that all tested S. saudiensis populations constituted a single and strong gene pool adapted to a specific habitat (palm trunk nesting) that was genetically isolated by significant natural barriers. Their analysis further clustered S. saudiensis with other morphologically unrelated species (e.g., the Malagasy S. mameti and the Neotropical S. saevissima) and placed S. elhawagryi with other Solenopsis species from The Americas based on claims of genetic similarities.
The aims of this study are (1) to add to ongoing efforts to develop a barcode reference library of Solenopsis species, (2) to combine both morphological and molecular evidences to investigate the phylogenetic relationship between S. saudiensis and S. abdita, (3) to place the two Saudi Arabian species (S. saudiensis and S. elhawagryi) into a larger biogeographic context using mitochondrial and nuclear gene sequences, (4) to test the conclusions of Rasool et al.55, and (5) to support and verify conclusions of our molecular analyses using morphological observations.
Material and methods
Institutional abbreviations
- BMNH:
-
The Natural History Museum (British Museum, Natural History), London, U.K.
- FMNH:
-
The Field Museum of Natural History, Chicago, IL, U.S.A.
- KSMA:
-
King Saud University Museum of Arthropods, Plant Protection Department, College of Food and Agriculture Sciences, King Saud University, Riyadh, Kingdom of Saudi Arabia.
- NHMB:
-
Naturhistorisches Museum, Basel, Switzerland.
- NMNH:
-
National Museum of Natural History, Smithsonian Institution, Washington, DC, U.S.A.
Throughout the work “w” is used to indicate worker, “m” male or males, and “q” queen.
Sample collection and information
Samples physically accessible for use in our study are listed in Supplementary Table S1. We had access to 12 nominal S. saudiensis samples, one S. abdita sample, and two samples unidentified to species, but which we aligned with S. abdita (S. cf. abdita) based on morphological and molecular data. Additional material examined is listed below. In addition, we included representative samples from regions allowing us to identify the native biogeographic areas of S. abdita/S. saudiensis (i.e., New World, Afrotropics, Eurasia). Our sampling was informed by Shreve et al.56. We further obtained the seven COI sequences from Rasool et al.55, nuclear and COI sequence data from Shreve et al.56, and high-resolution automontage images of S. abdita and S. saudiensis from AntWeb57. In addition, we had access to Rasool’s voucher material for morphological examination. Voucher specimens are deposited at the KSMA and NMNH.
Measurements and indices
Measurements and indices were performed as previously described3,29,58. All measurements are in millimeters.
- TL:
-
Total Length; the outstretched length of the ant from the mandibular apex to the gastral apex.
- HW:
-
Head width; the maximum width of the head behind eyes in full-face view.
- HL:
-
Head length; the maximum length of the head, excluding the mandibles.
- CI :
-
Cephalic Index (HW × 100/HL).
- SL:
-
Scape length, excluding basal neck.
- SI :
-
Scape Index (SL × 100/HW).
- EL:
-
Eye Length; the maximum diameter of the eye.
- ML:
-
Mesosoma length; the length of the mesosoma in lateral view, from the point at which the pronotum meets the cervical shield to the posterior base of the propodeal lobes or teeth.
- PL:
-
Petiole length; the maximum length measured in dorsal view, from the anterior margin to the posterior margin.
- PW:
-
Petiole width; maximum width measured in dorsal view.
- PPL:
-
Postpetiole length; maximum length measured in dorsal view.
- PPW:
-
Postpetiole width; maximum width measured in dorsal view.
Molecular data generation
The phylogenetic relationships among our samples were inferred using molecular data from Shreve et al.56, who sequenced four nuclear genes (see below) and COI to estimate a global phylogeny of Solenopsis. Their data was subsampled to include representative New World species discussed by Rasool et al.55 as well as Old World species. In addition, we generated two new S. saudiensis COI barcodes from Riyadh, which were identical. Finally, all S. saudiensis, S. abdita, and S. cf. abdita samples used by Shreve et al.56 were re-extracted and sequenced in a different laboratory to prevent contamination and ensure that no samples were mixed up. Molecular methods follow Brady et al.59 and Moreau et al.60 Briefly, total genomic DNA was isolated from whole single workers with the Qiagen DNeasy Blood and Tissue kit (Qiagen Inc., Valencia, CA, USA). Only a single individual from each collection event was used to avoid subsampling colonies. DNA sequence data were generated from four nuclear protein‐coding genes (Abdominal-A (Abd-A), elongation factor 1-alpha F1 (EF1α-F1), elongation factor 1-alpha F2 (EF1α-F2), and Wingless (Wg)), and the mitochondrial protein‐coding gene cytochrome c oxidase I (COI). Primer sequences, PCR amplification, and Sanger sequencing protocols are given in Brady et al.59 and Moreau et al.60 We only deviated from the given protocols by adding BSA (0.08 mg/mL final concentration) to the final PCR reaction mix and using a touchdown PCR procedure to increase specificity, which started 5 °C above the published annealing temperatures and decreasing by 0.4 °C/cycle for 12 cycles. PCR amplicons were Sanger sequenced in both directions using PCR primers and the BigDye Terminator 3.1 kit on an ABI 3730xl capillary sequencer (Applied Biosystems, Carlsbad, CA, USA). Sequence traces were assembled in Geneious Prime 2020.05 (https://www.geneious.com) and deposited in GenBank (GenBank accession numbers MT550038–MT550618; see Supplementary Table S2). For comparison, Rasool et al.55 COI sequence data from S. saudiensis collected from the Riyadh region, KSA, were included (GenBank accession numbers KR916796–KR916802; see Supplementary Table S2).
Molecular data analysis
We assembled two molecular datasets: a multilocus dataset derived from Shreve et al.56 consisting of four nuclear loci, and COI, to better place S. saudiensis within a global biogeographic framework. We also assembled a COI barcoding dataset to compare against the S. saudiensis haploytype described by Rasool et al55.
Each locus was globally aligned using the global iterative refinement method (G-INSI-i) implemented in MAFFT 7.402 (Katoh and Standley61,62;—globalpair—maxiterate 1000—retree 100). The concatenated multilocus dataset produced a 2,546 bp alignment, of which 457 nucleotides were variable and 338 were parsimony informative. Use of other alignment algorithms did not impact phylogenetic tree estimation. For each dataset, we estimated maximum likelihood trees using IQTREE 1.6.1263, simultaneously estimating the optimal model of nucleotide substitution using ModelFinder64 (multilocus: SYM + R3, COI: TIM2 + F + I + G4) on an unpartitioned dataset. We estimated branch support using ultrafast bootstraps65,66 (1,000 replicates) and two approximate likelihood-based measures (aLRT) (Shimodaira—Hasegawa—aLRT [SH-aLRT] with 1,000 replicates and the Bayesian-like transformation aLRT [aBayes]67. Bayesian trees were estimated using MrBayes 3.2.6568. For the COI dataset, we applied the best fitting model of nucleotide substitution determined by IQTREE. For the multilocus dataset we used the partitioning scheme and model of nucleotide substitution estimated by Shreve et al.56.
Principal component analyses were conducted on the COI dataset in R 3.6.269 using the adegenet 2.1.270 and ade471 packages.
Worth mentioning, searching the BOLD Identification System (IDS) for COI barcodes similar to the S. saudiensis COI haplotype of Rasool et al.55 returned a species-level identification entry of 100% similarity identified as Solenopsis sp. HI01 (sample ID: PKSP5221; deposited in: Harvard Museum of Comparative Zoology (MCZ); sequence ID: ASPNA1425-10.COI-5P; BIN ID: BOLD:AAN0050) collected from Hawaii (20°56′09.6″N 156°30′50.4″W) during 2010 and back to BOLD Jul-2011 historical records (https://v3.boldsystems.org/index.php/IDS_OpenIdEngine?historical=Jul-2011). This observation outlines the utility of the BLAST search tool in the BOLD identification engine and GenBank in fast and appropriate species-level assignment, in case of presence of similar data, and highlights how audit effort of molecular datasets affects interpretations.
Scanning electron microscopy
The mounted specimens were coated with platinum and imaged using a scanning electron microscope, model JSM-6380 LA, located at the College of Science, King Saud University, at a resolution 3.0 nm (30KV, WD8 mm, SEI), accelerating voltage 0.5–30 kV, and a magnification of 85 ×–400 ×.
Results
Molecular phylogenetic analysis
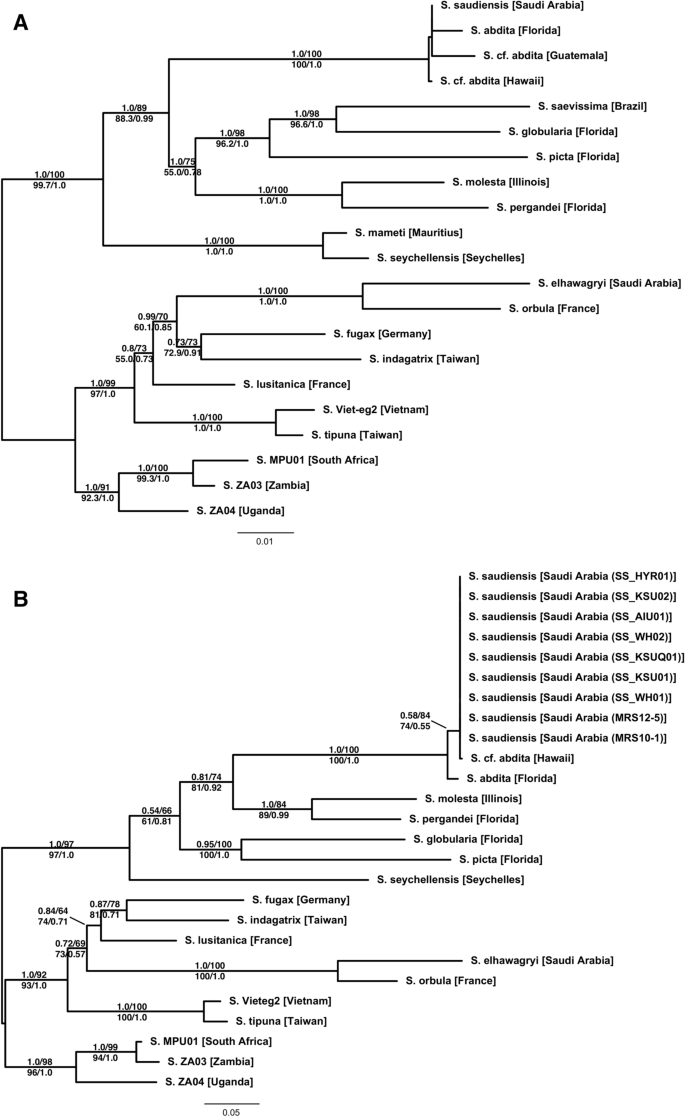
Our molecular phylogenetic analyses of the multilocus dataset based on Bayesian and maximum likelihood (ML) methods (Fig. 1A) show clear and well-supported biogeographic patterns. The African, Eurasian, Nearctic, and Malagasy samples each fall into clades. The only exception is S. saudiensis, which forms a strongly supported (PP = 100, BS = 100, SH-aLRT = 1.0, aBayes = 1.0) clade with S. abdita from Florida and the two S. cf. abdita samples from Guatemala and Hawaii. Each locus individually supports the same pattern (not shown). However, nodes within this clade are all very poorly supported (PP < 10, BS < 60, SH-aLRT = 0.0, aBayes < 0.4). The analyses of only the COI data (Fig. 1B), which includes the Rasool et al.55 data, confirm the overall biogeographic pattern recovered with the multilocus dataset. Importantly, it also shows that all Saudi Arabian S. saudiensis samples and the single Hawaiian S. cf. abdita sample all share an identical COI haplotype.
Phylogenetic trees of the multilocus (A) and the (B) mitochondrial COI datasets estimated using maximum likelihood (ML). The Bayesian phylogenies are fully compatible with the ML trees. Branch support is derived from ML and Bayesian posterior probabilities (above branch: posterior probability/ultra-fast bootstrap; below branch: SH-aLRT/aBayes).
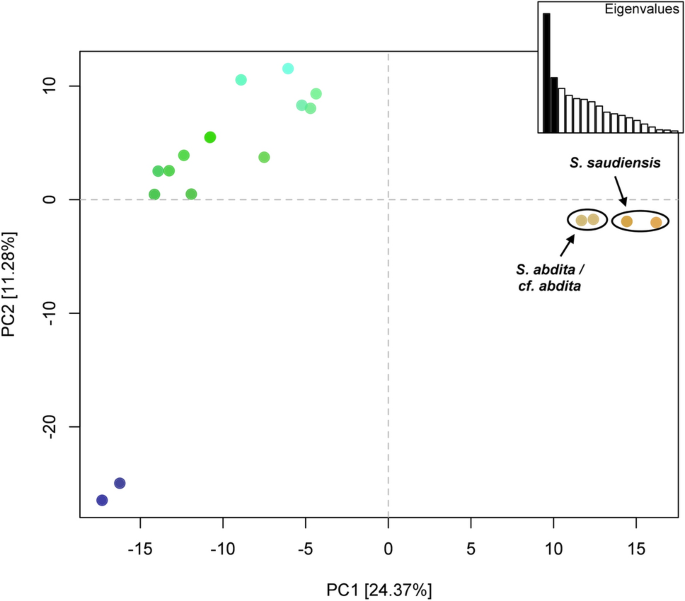
The principal component analysis of the COI dataset shows three clusters (Fig. 2). The first principal component, which explains 24.37% of the variation and more than twice that of the second principal component, clearly separates the S. saudiensis and S. abdita samples from the other Solenopsis species. The S. saudiensis and S. abdita samples are poorly separated, and their differentiation is mainly derived by the first principal component.
Principal component analysis (PCA) of COI dataset. The first two principal components are depicted, which together explain 35.65% of the variance. Points representing S. abdita and S. saudiensis haplotypes are labeled in brown and primarily differentiated by the first principal component. Colors are assigned by projecting the loadings of the first three principal components onto the RGB color channels.
According to the present work, S. elhawagryi is clearly associated with other Eurasian species and quite distinct from S. saudiensis, which falls out in a clade of Nearctic species. The molecular results are supported by our morphological analyses. The two species are easily separated based on the possession of the presence/absence of postpetiolar teeth and polymorphy/monomorphy of the worker castes. The former species has a postpetiole process in all castes and is polymorphic, whereas the latter lacks a postpetiole process and is clearly monomorphic.
Morphological reassessments/new synonymy
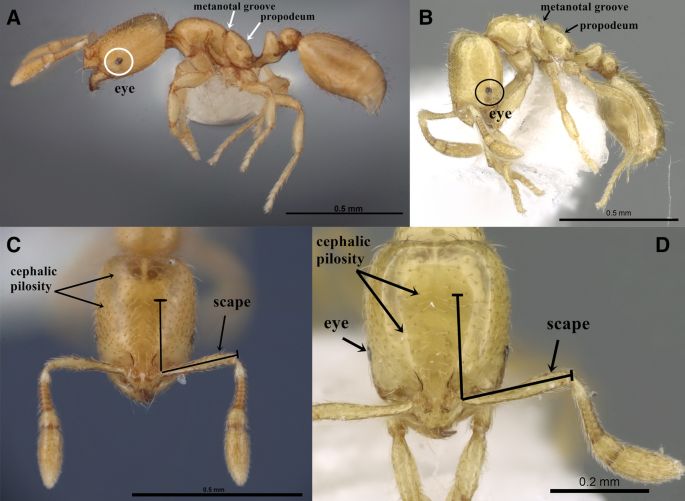
The genus Solenopsis, comprised of some of the smallest ants in the subfamily Myrmicinae, includes numerous minute species with minor workers less than 2 mm length. The material of S. abdita and S. saudiensis are ideally studied with high magnification microscopes, the Leica M205 C Stereomicroscope with a magnification zoom range of 20.5 × to examine and detect diagnostic characters that demonstrate clear morphological similarities between the two species. These similarities can be summarized in the following diagnosis (Fig. S1A–F, Fig. S2A–F) (Fig. 3): monomorphic species.
(A–D) Automontage images of S. abdita and S. saudiensis, (A, C) S. abdita, (A) body in profile, (C) head in full-face view, (FMNH-INS0000078522), Florida, (Photographer: Gracen Brilmyer); (B, D) paratype worker of S. saudiensis, (B) body in profile, (D) head in full-face view, (CASENT0249866), Saudi Arabia, (Photographer: Ryan Perry), from https://www.antweb.org/.
Head. Eyes minute with two to five ommatidia only seen with higher magnification, more frequently two; funicular segments 3–8 about twice as broad as long; anterior clypeal margin with a central pair of stout projecting teeth and a lateral pair of short, broad, basal, blunt teeth. Mesosoma. Dorsum of mesosoma smoothly curved, not flattened before and after the metanotal groove; metanotal groove acutely impressed in profile. Postpetiole. Postpetiole about 1.3 × broader than long in dorsal view; nearly hexagonal in profile with a distinctly convex dorsal surface. Pilosity. Relatively abundant and long hairs sparse on mesosoma, petiole, postpetiole, and gaster; more than 10 erect hairs on the dorsum of promesonotum; posterior tibial hairs mostly appressed. Sculpture. All body surfaces smooth and shining. Color. Uniform yellow or golden yellow, with coarse punctures present on the dorsum of the head.
The similarities include the body size and measurements of different body parts as follows: S. abdita: TL 1.02–1.20; HL 0.34–0.40; HW 0.28–0.34; EL 0.03; SL 0.22–0.24; ML 0.24; PW 0.07–0.09; PPL 0.072–0.084; PPW 0.10; Indices: CI 75–78; SI 58–67 (n = 18)3,36.
S. saudiensis: TL 1.2–1.3; HL 0.31–0.40; HW 0.28–0.31; EL 0.02; SL 0.21–0.27; ML 0.31–0.38; PL 0.10; PW 0.10; PPL 0.10; PPW 0.10–0.13; Indices: CI 75–87; SI 70–90; (n = 12)28,29. The previously mentioned morphological and molecular similarities of the two species support the synonymization of S. saudiensis with S. abdita syn. nov.
Diagnostic and comparison with other Solenopsis species
Our molecular data reflects an apparent degree of similarity between S. saudiensis and S. molesta. The two species are similar in size and color, but the latter can be separated by the relatively larger eyes consisting of three to five ommatidia whereas S. saudiensis has distinctly smaller eyes with only two ommatidia. In addition, S. molesta has a head that is both longer and broader (HL 0.42–0.51, HW 0.36–0.43) than S. saudiensis.
Comparing the queen caste of S. saudiensis described by Sharaf et al.72 with the queen of S. molesta (Say, 1836) confirms that the two species are morphologically unrelated. The queen of S. saudiensis is uniform dark brown or black-brown, consistently smaller (HL 0.53–0.55, HW 0.46–0.50, EL 0.12–0.17), with a pointed petiole node profile and lacks a postpetiolar pair of teeth, whereas the queen of S. molesta is uniform yellow with darker mesosomal dorsum, distinctly larger (HL 0.72–0.84, HW 0.64–0.78, EL 0.24), a broad petiolar node rounded in profile, and postpetiole with a distinct subpetiolar pair of teeth.
Comparing S. abdita/saudiensis to S. pergandei (Forel, 1901), the three species are uniform yellow with eyes consisting of two ommatidia and present a distinct acute metanotal groove. However, S. abdita/saudiensis can be easily separated by the distinctly longer head when seen in full-face view (CI 75–87); abundant, short, subdecumbent or appressed body pilosity; and well-developed anterior central and lateral pairs of clypeal teeth. By contrast, S. pergandei has a nearly quadrate head (CI 89–93); profuse, suberect, and longer body pilosity; and a blunt central pair of anterior clypeal teeth while the lateral pair is absent.
The analysis of Rasool et al.55 shows a sister cluster of two unrelated species, S. mameti Donisthorpe, 1946 from Mauritius and S. saevissima (Smith, 1855) from Brazil. Morphologically, S. abdita/saudiensis and S. mameti are clearly distinct, as the former is uniformly yellow, with a shallow metanotal groove and less abundant, short, scattered body pilosity, whereas the latter is unicolorous dark brown with a deep metanotal groove and abundant, long body pilosity. Solenopsis saevissima is completely different from S. abdita / saudiensis and easily separated by numerous sets of characters including brown color, strong polymorphism in any nest series, profuse, stiff, and long body pilosity, conspicuously large eyes that contains about 12 ommatidia in the longest row, and an emarginated posterior margin of head seen in full-face view.
Solenopsis abdita/S. carolinensis
Among the Nearctic species, S. abdita can be confused with S. carolinensis Forel, 1901 and Thompson36 was not able to present a practical differential diagnosis between the two species. However, Pacheco and Mackay3 successfully recognized the former species by the shorter scape, the broader petiole, and the appressed hairs on the tibiae whereas S. carolinensis has the tibiae with suberect hairs. In addition, the queen caste can be useful in the identification where the queen of S. abdita is dark brown and has smaller eyes, while the queen of S. carolinensis is yellow.
Additional material examined
Solenopsis abdita (Fig. S1A,C, E; Fig. S2A,C, E; Fig. 3A, C): USA, Florida, Monroe Co., Key Largo, Hammock Botanical S.P., 25°10.524ʹN, 080°22.120ʹW, 10 m, 10.x.2010, (Corrie S. Moreau), (CSM1918), FMNH-INS 0000078522, 1 w, [FMNH]; USA, Florida, Osceola Nat. For., Baker Co., 10.07.1993, M. Deyrup, CASENT0104494, 1 w, (image examined).
Solenopsis mameti: MAURITIUS, 26.xii.1946, (R. Mamet), holotype w, (CASENT0102281), [BMNH].
Solenopsis molesta: USA, Virginia, (Pergande), 1015389, 1 w, CASENT0902336, [BMNH].
Solenopsis saevissima: BRAZIL: Syntype w, CASENT0902353, [BMNH]; syntype w, Blumenau, (Mme. V. Steiger), [NHMB].
Solenopsis saudiensis (Fig. S1B,D,F; Fig. S2 4B,D,F; Fig. 3B,D): SAUDI ARABIA: Riyadh, 24°43ʹN, 46°37ʹE, 9.VII.2009, 612 m (Mostafa R. Sharaf & Abdulrahman S. Aldawood), holotype w; two paratype workers with same data as the holotype, CASENT0217364; 117 paratype w, Riyadh, Wadi Hanifa, 24°39ʹN, 46°36ʹE, 15.I.2010, 633 m (Mostafa R. Sharaf & Abdulrahman S. Aldawood), CASENT0249866; 2 dealated q, Riyadh, King Saud University campus, 24°42.832′N, 46°37.534′E, 660 m, 04.iv.2014, (S. Salman), CASENT091433; Riyadh, King Saud University campus, 24.71383°N, 46.62557°E, 660 m, 02.ii.2014, (S. Salman) (2 w); Riyadh, King Saud University campus, 24.71383°N, 46.62557°E, 660 m, 06.ii.2014, (S. Salman) (3 w); Riyadh, Al Emam University, 24.81658 N, 46.71162E, 650 m, 08.ix.2014, (S. Salman) (2 w); Riyadh, King Saud University campus, 24.71383°N, 46.62557°E, 660 m, 10.iii.2014, (S. Salman) (3 w); Riyadh, Rhawdet Khorim, 25.383100°N, 47.278533°E, 559 m, 18.ii.2014, (Al Dhafer et al.) (3 w); Riyadh, Wadi Hanifa, 24.73507°N, 46.57518°E, 674 m, 18.ix.2014, (S. Salman) (2 w); Riyadh, Al Qawayiyah. R-Al Harmaliyah, 24.29773°N, 45.14577°E, 786 m, 20.iv.2015, (Al Dhafer et al.) (2 w); Riyadh, Azulfi, Rowdhat, Alsabalah, 26.36760°N, 44.98560°E, 671 m, 20.v.2015, (Al Dhafer et al.) (1 w); Riyadh, Rhawdet Khorim, 25.383100°N, 47.278533°E, 559 m, 26.v.2012, (Al Dhafer et al.) (2 w); Riyadh, Al Emam University, 24.817056°N, 46.701842°E, 657 m, 07.v.2014, (Mostafa R. Sharaf) (3 w) [KSMA].
Discussion
The field study conducted by Rasool et al.55 in the Riyadh region did not turn up evidence of Solenopsis species other than S. saudiensis. However, the sampling methods and efforts deployed in their study are insufficient to conclude that other Solenopsis are absent, as many localities, habitats, and microhabitats in the province, which has a high diversity of natural and agricultural habitats, were left unexplored.
Our molecular results are clearly consistent with the Arabian revision of the Solenopsis fauna29 and the morphological traits used in species recognition. Based on molecular data as well as a morphological reevaluation of both S. abdita and S. saudiensis, our results indicate that S. saudiensis, described in 2011, represents a junior synonym of S. abdita. Thompson36 states that types of S. abdita were deposited at the Museum of Comparative Zoology (MCZ), the Florida State Arthropod Collection in Gainesville (FSCA), and the Natural History Museum of Los Angeles County (LACM), but extensive searches in these museums were unable to locate the type materials. The absence of S. abdita types has been observed before by Pacheco and Mackay3. In many Solenopsis species, however, the morphological distinction of species on the basis of the worker caste is arduous (e.g. S. iheringi Forel, 1908 and S. bicolor (Emery, 1906); S. johnsoni Pacheco et al., 2013 and S. melina Pacheco et al., 2013; and S. azteca Forel, 1893 and minor workers of wasmannii-group3,11), therefore, the study of the sexual castes including queens and males represent a useful addition for species delimitation11. Here, the comparison of the reproductive female caste of S. saudiensis described by Sharaf et al.72 with the original description of the queen of S. abdita (Thompson, 1989) revealed that most of their taxonomic characters match, including body size, sculpture, pilosity and reflected a straightforward synonymy. The few minor exceptions include body color, which is dark brown in the former species and reddish brown to almost black in the latter species. However, coloration in ants presents wide variation within and between populations3,11.
Ecological similarities are also found in the nesting habits of the two species, since both species were encountered nesting in palm logs (family Arecaceae)3,36. Solenopsis saudiensis has been collected in or near date palm plantations, Phoenix dactylifera L., on the Arabian Peninsula28 and S. abdita has been reported to be commonly found in rotten palm logs in the USA36. The nesting preference of S. abdita includes a broad range of habitats that are either moist or mesic niches in Florida including sandhill, swamp forest, grass tussocks of seasonal ponds, bases of pines in flatwoods, hammocks, rotten wood and palm logs36,73, or bases of date palm trees in the KSA28 where nests are built near the soil surface73.
These results also demonstrate the non-native status of the populations of S. abdita within KSA and represent the first known record of this species in the Old World. Introduced populations are also characterized by a reduced genetic pool as a consequence of a bottleneck effect following their introduction; which we observed in the form of populations from Saudi Arabia and Hawaii presenting identical COI sequences. The presence of this species in two regions outside its native range (the Arabian Peninsula and Hawaii), coupled with particular morphological and ecological traits such as small body size, polygyny36, lestobiotic lifestyle, and association with disturbed environments, supports the tramp status of this species74. Indeed, individuals of S. abdita in KSA were commonly encountered in date palm groves28 and highly disturbed urban habitats (one of the two type series was found nesting under a discarded carpet next to a human settlement28) but also in more natural habitats such as nature reserves (e.g. Rawdhat Khorim75). However, nature reserves are not necessarily disturbance-free and sometimes even the most pristine reserve can have exotic species along roads or buildings. Together, these results contrast with the conclusion of Rasool et al.55 of S. abdita being strongly associated with and specialized to colonize date palm groves following an adaptive process involving a large and strong gene pool.
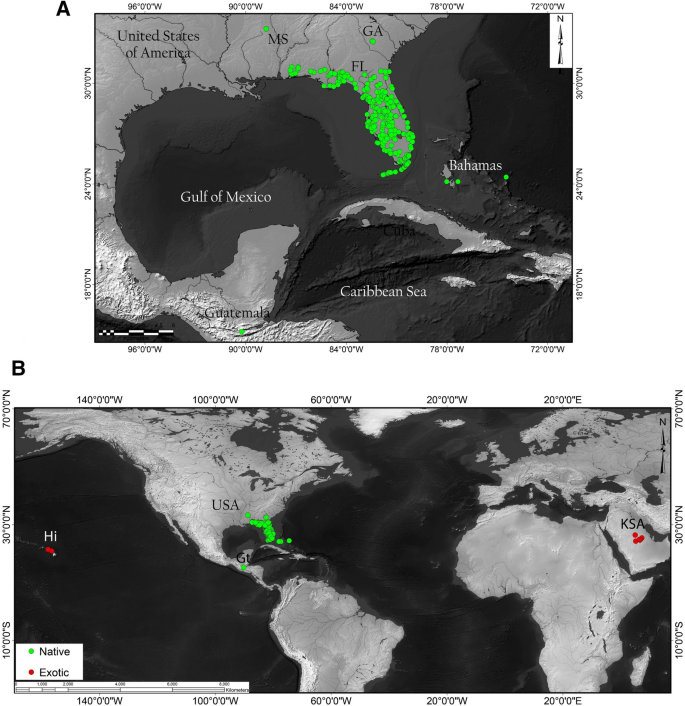
While limited by the extent of the sampling used in our study, the results tend to indicate a New World origin for S. abdita potentially spanning the Nearctic and Neotropical realms. Given that the two samples from the Nearctic and the Neotropical regions (Florida and Guatemala, respectively) are genetically distinct and the species falls out in the New World clade, it seems likely that the native range is also somewhere in the New World (possibly circum-Caribbean). Currently, S. abdita is predominantly recorded from Florida and surrounding states (Fig. 4A; based on data from AntMaps76), which may entirely be an artifact of the geographic focus of the species keys used to identify thief ants.
Worldwide distribution records of S. abdita. (A) Reported distribution in the United States (in green, native) with verified occurrences based mainly on http://antmaps.org7,76 and collection data from Dr. James Wetterer (Florida Atlantic University, USA). (B) Worldwide spread. Red dots indicate collection sites for samples used outside USA (exotic), including Guatemala, Hawaii, and Kingdom of Saudi Arabia (strict sense S. saudiensis). Maps were constructed using ArcGIS 10.3 software (Esri; Redlands, CA, United States).
If species of Solenopsis known as fire ants from the geminata (e.g. S. aurea, S. geminata, and S. xyloni) and saevissima (e.g. S. invicta, S. richteri, and saevissima) groups are notorious invaders in tropical to warm temperate climates regions3,11,14,77, this contrasts with the few successful introductions of the thief ants (previously referred as Diplorhoptrum) that have been recorded and their limited distribution within the introduced range (e.g. S. globularia, S. papuana, and potentially S. terricola to be confirmed as introductions in Florida). While an argument might be made for a potential candidate synonymy of these species with S. abdita, this possibility can be readily excluded by morphological examination. Solenopsis globularia (Smith, F., 1858) is easily distinguished by the greatly dilated/enlarged postpetiole seen in dorsal view, S. papuana Emery, 1900 has larger eyes that consist of three ommatidia plus a high profile of the propodeal dorsum, and S. terricola Menozzi, 1931 is a uniform dark brown species.
The identification of S. abdita as a new introduction within two distinct regions of the world [the Arabian Peninsula and Hawaii (Fig. 4B)] raises several questions. First, what is the extent of the native range of this species? And are populations from Guatemala part of the native or exotic range of this species? Second, due to the challenges of identifying of S. abdita and other thief ants, how many unidentified records of these species exist that potentially demonstrate a wider introduced range? Our study represents a case that could be expanded to more parts of the world to identify both Solenopsis specimens and other challenging ant taxa known to include major tramp species (e.g. Cardiocondyla78, Pheidole79, Tetramorium80).
Several of these taxa are widespread tramp species frequently involved in human‐mediated dispersal. Invasive and tramp species tend to have far-reaching geographical distributions and share life history traits including foraging behavior, nest structure, and queen number9,16.
Our phylogenetic and morphometric results indicate that invasive characteristics evolved within monomorphic S. abdita, such as its small size, lestobiotic lifeway, and phenotypic plasticity, could potentially confound taxonomists. Increased phylogenetic taxon sampling and improved species‐level taxonomy using ultrastructural tools will be necessary to explore the issue of invasive origins in further detail.
Data availability
The specimens used in this study have been databased and the data are freely accessible on AntWeb (https://www.antweb.org). Main data needed to evaluate the conclusions in the paper are present in the paper. Additional data that support the findings of this study are available from the corresponding authors upon reasonable request.
References
Hölldobler, B. & Wilson, E. O. The ants. (Cambridge: Harvard University Press, xii + 732 pp., 1990).
Guénard, B., Weiser, M. D. & Dunn, R. R. Global models of ant diversity suggest regions where new discoveries are most likely are under disproportionate deforestation threat. Proc. Natl. Acad. Sci. U. S. A. 109, 7368–7373 (2012).
Pacheco, J. A. & Mackay, W. P. The Systematics and Biology of the New World Thief Ants of the Genus Solenopsis (Hymenoptera: Formicidae) ( Edwin Mellen Press, Lewiston, 2013).
Bolton, B. An online catalog of the ants of the world. https://www.antcat.org/ (last accessed 19 January 2020) (2020).
Brown, W. L. Jr. Diversity of ants. In Ants. Standard Methods for Measuring and Monitoring. Biodiversity Biological diversity Handbook Series (eds Agosti, D. et al.) 45–79 (Smithsonian Institution Press, Washington, D.C., 2000).
Bolton, B. Synopsis and classification of Formicidae. Mem. Am. Entomol. Inst. 71, 1–370 (2003).
Guénard, B., Weiser, M. D., Gomez, K., Narula, N. & Economo, E. P. The Global Ant Biodiversity Informatics (GABI) database: synthesizing data on ant species geographic distribution. Myrmecol. News 24, 83–89 (2017).
Fernández, F. & Sendoya, S. List of Neotropical ants (Hymenoptera: Formicidae). Biota Colomb. 5, 3–93 (2004).
Holway, D. A., Lach, L., Suarez, A. V., Tsutsui, N. D. & Case, T. J. The causes and consequences of ant invasions. Annu. Rev. Ecol. Evol. Syst. 33, 181–233 (2002).
Morrow, M. E., Chester, R. E., Lehnen, S. E., Drees, B. M. & Toepfer, J. E. Indirect effects of red imported fire ants on Attwater’s prairie-chicken brood survival. J. Wildl. Manag. 79, 898–906 (2015).
Trager, J. C. A revision of the fire ants, Solenopsis geminata group (Hymenoptera: Formicidae: Myrmicinae). J. N. Y. Entomol. Soc. 99, 141–198 (1991).
Luque, G. M. et al. The 100th of the world’s worst invasive alien species. Biol. Invasions 16, 981–985 (2014).
Ascunce, M. S. et al. Global invasion history of the fire ant Solenopsis invicta. Science 331, 1066–1068 (2011).
Gotzek, D., Axen, H. J., Suarez, A. V., Helms Cahan, S. & Shoemaker, D. Global invasion history of the tropical fire ant: a stowaway on the first global trade routes. Mol. Ecol. 24, 374–388 (2015).
Bellard, C., Cassey, P. & Blackburn, T. M. Alien species as a driver of recent extinctions. Biol. Lett. 12, 20150623 (2016).
Bertelsmeier, C., Ollier, S., Liebhold, A. & Keller, L. Recent human history governs global ant invasion dynamics. Nat. Ecol. Evol. 1, 0184 (2017).
Guénard, B., Cardinal-De Casas, A. & Dunn, R. R. High diversity in an urban habitat: are some animal assemblages resilient to long-term anthropogenic change?. Urban Ecosyst. 18, 449–463 (2015).
Simberloff, D. et al. Impacts of biological invasions: what’s what and the way forward. Trends Ecol. Evol. 28, 58–66 (2013).
Seebens, H. et al. No saturation in the accumulation of alien species worldwide. Nat. Commun. 8, 14435 (2017).
Seebens, H. et al. Global rise in emerging alien species results from increased accessibility of new source pools. Proc. Natl. Acad. Sci. U. S. A. 115, E2264–E2273 (2018).
Wittenberg, R. & Cock, M. J. W. Invasive Alien Species. How to Address One of the Greatest Threats to Biodiversity: A Toolkit of Best Prevention and Management Practices (CAB International, Wallingford, 2001).
Sarnat, E. M. PIAkey: Identification guide to invasive ants of the Pacific Islands, Edition 2.0, Lucid v. 3.4. USDA/APHIS/PPQ Center for Plant Health Science and Technology, University of California, Davis. https://www.piakey.com (2008).
Hoffmann, B. D., Andersen, A. N. & Zhang, X. Taxonomic confusion of two tramp ant species: Iridomyrmex anceps and Ochetellus glaber are really species complexes. Curr. Zool. 57, 662–667 (2011).
Armstrong, K. F. & Ball, S. L. DNA barcodes for biosecurity: invasive species identification. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 1813–1823 (2005).
Kim, K. H. et al. Identification of key genes for the precise classification between Solenopsis invicta and S. geminata facilitating the quarantine process. Genes 10, 812 (2019).
Jacobson, A. L., Thompson, D. C., Murray, L. & Hanson, S. F. Establishing guidelines to improve identification of fire ants Solenopsis xyloni and Solenopsis invicta. J. Econ. Entomol. 99, 313–322 (2006).
Gotzek, D., Brady, S. G., Kallal, R. J. & LaPolla, J. S. The importance of using multiple approaches for identifying emerging invasive species: the case of the Rasberry Crazy Ant in the United States. PLoS ONE 7, e45314 (2012).
Sharaf, M. R. & Aldawood, A. S. First occurrence of Solenopsis Westwood 1840 (Hymenoptera: Formicidae), in the Kingdom of Saudi Arabia, with description of a new species S. saudiensis n. sp. Ann. Soc. Entomol. Fr. 47, 474–479 (2011).
Sharaf, M. R. & Aldawood, A. S. Ants of the genus Solenopsis Westwood 1840 (Hymenoptera: Formicidae) in the Arabian Peninsula with description of a new species, Solenopsis elhawagryi. PLoS ONE 7, e49485 (2012).
Collingwood, C. A., Tigar, B. J. & Agosti, D. Introduced ants in the United Arab Emirates. J. Arid Environ. 37, 505–512 (1997).
Collingwood, C. A., Agosti, D., Sharaf, M. R. & van Harten, A. Order Hymenoptera, family Formicidae. Arthr. Fauna UAE 4, 405–474 (2011).
Collingwood, C. A. & Agosti, D. Formicidae (Insecta: Hymenoptera) of Saudi Arabia (part 2). Fauna Saudi Arabia 15, 300–385 (1996).
Pitts, J. P., Camacho, G. P., Gotzek, D., McHugh, J. V. & Ross, K. G. Revision of the fire ants of the Solenopsis saevissima species-group (Hymenoptera: Formicidae). Proc. Entomol. Soc. Wash. 120, 308–411 (2018).
Bolton, B. Identification Guide to the Ant Genera of the World (Harvard University Press, Cambridge, 1994).
Pitts, J. P., Mchugh, J. V. & Ross, K. G. Cladistic analysis of the fire ants of the Solenopsis saevissima species-group (Hymenoptera: Formicidae). Zool. Scr. 34, 493–505 (2005).
Thompson, C. R. The thief ants, Solenopsis molesta group, of Florida (Hymenoptera: Formicidae). Fla. Entomol. 72, 268–283 (1989).
Pacheco, J. The New World Thief Ants of the Genus Solenopsis (Hymenoptera, Formicidae) (Dissertation, The University of Texas at El Paso, 2007).
Steiner, F. M. et al. Towards DNA-aided biogeography: ant example from Tetramorium ants (Hymenoptera: Formicidae). Ann. Zool. Fenn. 42, 23–35 (2005).
Alex Smith, M. & Fisher, B. L. Invasions, DNA barcodes, and rapid biodiversity assessment using ants of Mauritius. Front. Zool. 6, 31 (2009).
Ross, K. G., Gotzek, D., Ascunce, M. S. & DeWayne Shoemaker, D. Species delimitation: a case study in a problematic ant taxon. Syst. Biol. 59, 162–184 (2010).
Szalanski, A. L., McKern, J. A., Solorzano, C. & Austin, J. W. Genetic Diversity of Ants (Hymenoptera: Formicidae) from the Ozark-St. Francis National Forest, Arkansas, USA. Sociobiology 56, 1–10 (2010).
Martins, C., Fernando de Souza, R. & Bueno, O. C. Molecular characterization of fire ants, Solenopsis spp., from Brazil based on analysis of mtDNA gene cytochrome oxidase I. J. Insect Sci. 14, 50 (2014).
Fisher, B. L. & Alex Smith, M. A Revision of Malagasy Species of Anochetus Mayr and Odontomachus Latreille (Hymenoptera: Formicidae). PLoS ONE 3, e1787 (2008).
Jansen, G., Savolainen, R. & Vepsalainen, K. Phylogeny, divergence-time estimation, biogeography and social parasite–host relationships of the Holarctic ant genus Myrmica (Hymenoptera: Formicidae). Mol. Phylogenet. Evol. 56, 294–304 (2010).
Hebert, P. D. N., Ratnasingham, S. & deWaard, J. R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. Biol. Sci. 270(Suppl 1), S96–S99 (2003).
Hebert, P. D. N., Cywinska, A., Ball, S. L. & deWaard, J. R. Biological identification through DNA barcodes. Proc. R. Soc. Lond. B 270, 313–321 (2003).
Smith, M. A. & Fisher, B. L. Invasions, DNA barcodes, and rapid biodiversity assessment using ants of Mauritius. Front. Zool. 6, 31. https://doi.org/10.1186/1742-9994-6-31 (2009).
Ng’endo, R. N., Osiemo, Z. B. & Brandl, R. DNA barcodes for species identification in the hyperdiverse ant genus Pheidole (Formicidae: Myrmicinae). J. Insect Sci. 13, 27 (2013).
DeSalle, R. & Goldstein, P. Review and interpretation of trends in DNA barcoding. Front. Ecol. Evol. 7, 302 (2019).
Liu, J. et al. Multilocus DNA barcoding—species identification with multilocus data. Sci. Rep. 7, 16601 (2017).
Zinger, L. et al. DNA metabarcoding—need for robust experimental designs to draw sound ecological conclusions. Mol. Ecol. 28, 1857–1862 (2019).
Eberle, J., Ahrens, D., Mayer, C., Niehuis, O. & Misolf, B. A plea for standardized nuclear markers in metazoan DNA taxonomy. Trends Ecol. Evol. 35, 336–345 (2020).
Collins, R. A. & Cruickshank, R. H. The seven deadly sins of DNA barcoding. Mol. Ecol. Resour. 13, 969–975 (2013).
Hoy, M. A. Molecular systematics and the evolution of arthropods. In In Insect Molecular Genetics. An Introduction to Principles and Applications 3rd edn, 521–589 (Academic Press, 2013). https://doi.org/10.1016/B978-0-12-415874-0.00012-3.
Rasool, K. G. et al. DNA barcoding of the fire ant genus Solenopsis Westwood (Hymenoptera: Formicidae) from the Riyadh region, the Kingdom of Saudi Arabia. Saudi J. Biol. Sci. 27, 184–188 (2020).
Shreve, S., Achury, R., Johnson, K., Suarez, A. & Gotzek, D. Multi-locus molecular phylogeny of Solenopsis (Hymenoptera: Formicidae). Preprint at: https://www.biorxiv.org/content/10.1101/2020.06.05.136945v1 (2020).
AntWeb v8.24. California Academy of Sciences. https://www.antweb.org (last accessed 6 April 2020) (2020).
Sharaf, M. R., Aldawood, A. S., Mohamed, A. A. & Garcia, F. H. The genus Lepisiota Santschi, 1926 of the Arabian Peninsula with the description of a new species, Lepisiota elbazi sp. Nov. from Oman, an updated species identification key, and assessment of zoogeographic affinities. J. Hymenopt. Res. 76, 127–152 (2020).
Brady, S. G., Schultz, T. R., Fisher, B. L. & Ward, P. S. Evaluating alternative hypotheses for the early evolution and diversification of ants. Proc. Natl. Acad. Sci. U. S. A. 103, 18172–18177 (2006).
Moreau, C. S., Bell, C. D., Vila, R., Archibald, S. B. & Pierce, N. E. Phylogeny of the ants: diversification in the age of angiosperms. Science 312, 101–104 (2006).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Katoh, K. & Standley, D. M. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 32, 1933–1942 (2016).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
Minh, B. Q., Nguyen, M. A. & von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195 (2013).
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018).
Anisimova, M., Gil, M., Dufayard, J. F., Dessimoz, C. & Gascuel, O. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 60, 685–699 (2011).
Ronquist, F. et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012).
R Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org (2019).
Jombart, T. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405 (2008).
Dray, S. & Dufour, A. The ade4 Package: implementing the duality diagram for ecologists. J. Stat. Softw. 22, 1–20 (2007).
Sharaf, M. R., Salman, S. & Aldawood, A. S. First description of the queen of the ant Solenopsis saudiensis Sharaf & Aldawood (Hymenoptera: Formicidae) from Saudi Arabia. Zool. Middle East 61, 50–54 (2014).
Deyrup, M. A. Ants of Florida: Identification and Natural History. (CRC Press, Boca Raton: Taylor and Francis, FL, 2017).
Passera, L. Characteristics of tramp species. In Exotic Ants: Biology, Impact, and Control of Introduced Species (ed. Williams, D. F) 23–43 (Westview, Boulder, 1994).
Sharaf, M. R., Abdeldayem, M. S., Aldhafer, H. & Aldawood, A. S. The ants (Hymenoptera: Formicidae) of Rawdhat Khorim nature preserve, Saudi Arabia, with description of a new species of the genus Tetramorium Mayr. Zootaxa 3709, 565–580 (2013).
Janicki, J. H., Narula, N., Ziegler, M., Guénard, B. & Economo, E. P. Visualizing and interacting with large-volume biodiversity data using client-server web mapping applications: the design and implementation of antmaps.org. Ecol. Inform. 32, 185–193 (2016).
Wetterer, J. K. Worldwide spread of the tropical fire ant, Solenopsis geminata (Hymenoptera: Formicidae). Myrmecol. News 14, 21–35 (2011).
Seifert, B. The ant genus Cardiocondyla (Insecta: Hymenoptera: Formicidae)—a taxonomic revision of the C. elegans, C. bulgarica, C. batesii, C. nuda, C. shuckardi, C. stambuloffii, C. wroughtonii, C. emeryi, and C. minutior species groups. Ann. Nat. Hist. Mus. Wien Ser. B. Bot. Zool. 104, 203–338 (2003).
Sarnat, E. M., Fischer, G., Guénard, B. & Economo, E. P. Introduced Pheidole of the world: taxonomy, biology and distribution. ZooKeys 543, 1–109 (2015).
Bolton, B. The ant tribe Tetramoriini (Hymenoptera: Formicidae) The genus Tetramorium Mayr in the Oriental and Indo-Australian regions, and in Australia. Bull. Br. Mus. Nat. Hist. Entomol. Ser. 36, 67–151 (1977).
Acknowledgements
We are grateful to the following colleagues: Jim Wetterer for useful discussion on S. abdita distribution in Florida; Corrie Moreau (Cornell University) for useful discussion on S. abdita; Ryan Perry for photographing S. saudiensis; Gracen Brilmyer for photographing S. abdita, and Omer Hamid for SEM work. The authors are indebted to Said M. Said (Geology Department, Faculty of Science, Cairo University) for his efforts creating distribution maps, and Ahmed Shams Al ‘Ola for editing SEM images. This work was basically supported by the Deanship of Scientific Research at King Saud University [RG-1438-010]. Mostafa Sharaf thanks Boris Kondratieff (Colorado State University) for continuous support.
Author information
Authors and Affiliations
Contributions
M.R.S., D.G., A.A.M., B.L.F., and A.S.A. coordinated research; D.G., M.R.S., and A.S.A. provided funding; M.R.S., D.G., and A.A.M. conceived the research; D.G., A.A.M., M.R.S., and B.G. conducted experiments; B.L.F., B.G., and H.A. provided technical and intellectual support; D.G., A.A.M., and M.R.S. analyzed data. M.R.S., D.G., A.A.M., and B.G. wrote and reviewed the different drafts of the manuscript. B.L.F. and A.A.M. reviewed and edited the final draft of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sharaf, M.R., Gotzek, D., Guénard, B. et al. Molecular phylogenetic analysis and morphological reassessments of thief ants identify a new potential case of biological invasions. Sci Rep 10, 12040 (2020). https://doi.org/10.1038/s41598-020-69029-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-69029-4
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.