Abstract
Leaf senescence and plant aging are traits of great interest for breeders. Senescing cells undergo important physiological and biochemical changes, while cellular structures such as chloroplasts are degraded with dramatic metabolic consequences for the whole plant. The possibility of prolonging the photosynthetic ability of leaves could positively impact the plant’s life span with benefits for biomass production and metabolite accumulation; plants with these characteristics display a stay-green phenotype. A group of plant transcription factors known as NAC play a pivotal role in controlling senescence: here we describe the involvement of the tomato NAC transcription factor Solyc12g036480, which transcript is present in leaves and floral buds. Since its silencing delays leaf senescence and prevents plants from ageing, we renamed Solyc12g0364 HḖBĒ, for the Greek goddess of youth. In this manuscript we describe how HEB downregulation negatively affects the progression of senescence, resulting in changes in transcription of senescence-promoting genes, as well as the activity of enzymes involved in chlorophyll degradation, thereby explaining the stay-green phenotype.
Similar content being viewed by others
Introduction
Senescence is crucial for plant fitness1 and it is a trait of great interest for breeders, since premature senescence can affect crop yield, post-harvest storage and quality2. Plant aging can be induced either by endogenous signals or by environmental stresses triggering controlled disassembly and disintegration at cellular and tissue levels, which ultimately affects the whole organism3. Organ senescence can be achieved through the activity of relevant cell structures and metabolic processes, such as organelle dismantling and chlorophyll breakdown, a phenomenon which causes the macroscopic leaf color changes observed in green plants. Moreover, previously accumulated macromolecules (i.e. proteins, lipids, nucleic acids and pigments) are also degraded and their products are relocated into sink tissues or organs4,5,6. In annual plants, nutrients are transferred to fruits or seeds, in perennial ones to stems and roots 7.
Organ senescence is also accomplished by cell wall modifications, phytohormone fluctuations, dismantling of macromolecules and activation of specific genes; these processes occur during leaf and petal senescence as well as during fruit ripening. The global regulation of these developmental programs involves several players, such as transcription factors, sugars, polyamines and hormones (for reviews see Wojciechowska et al.8 and Forlani et al.9). The genetic program behind senescence is highly complex and regulated at transcriptional, post-transcriptional, translational and post-translational levels3. Several studies have demonstrated the relevance of epigenetic mechanisms in the control of leaf senescence and fruit ripening10,11,12.
NAC transcription factors (NAM No Apical Meristem, ATAF1/2 and CUC Cup-Shaped Cotyledon) play a pivotal role in leaf senescence. This family is one of the largest plant-TFs families and comprises 101 members in Solanum lycopersicum, 138 in Arabidopsis thaliana, 158 in Oryza sativa ssp. indica, and more than 400 in Brassica napus (The PlantTFDB, https://planttfdb.cbi.pku.edu.cn/). NAC proteins are activators and/or repressors of gene expression and modulate plant development, plant defense and stress tolerance processes (for review see Olsen et al.13, Nakano et al.14, Kim et al.15, Ohbayashi and Sugiyama16, Mathew and Agarwal17). NAC proteins have been documented to be involved in leaf, petal and fruit senescence in Arabidopsis thaliana3,15,18,19,20,21,22,23,24,25,26,27,28,29,30, Solanum lycopersicum31,32,33, Oryza sativa34,35,36,37,38, Hordeum vulgare39,40, Glycine max41, Bambusa emeiensis42, Trifolium pratense43, Helianthus annuus44, Gossypium hirsutum45,46,47, Musa x paradisiaca48,49, Vitis vinifera50 and Nicotiana tabacum51. Among those, several NAC genes are linked to a stay-green phenotype. This term is used to indicate cultivars, varieties, transgenic or knock-out lines able to maintain their green color longer than wild-type plants. In these plants, long-lasting leaf coloration is correlated to durable chlorophyll accumulation compared to wild-type plants or standard varieties, and it is often associated with delayed senescence52. In Arabidopsis, senescence mechanisms induce the expression of ORESARA1 (ORE1) and ORE1 SISTER1 (ORS1) genes. ORE1 activates program cell death and ORS1 participates to salt-induced senescence; the corresponding knock-out mutant plants display a stay-green phenotype and delayed senescence19,20,22,26. Conversely, the disruption of VND-INTERACTING2 (VNI2) and JUNGBRUNNEN1 (JUB1)—which also encode for two NAC proteins—causes early senescence while their overexpression induces a stay-green phenotype24,53. Recently, it was demonstrated that transgenic tomato lines, with reduced accumulation of SlNAP2 messenger (Solanum lycopersicum NAC-like, activated by Apetala3/Pistillata), display a stay-green phenotype even upon ABA (abscisic acid) application33.
In this manuscript, we describe the role of Solyc12g036480, which encodes a NAC transcription factor able to modulate leaf senescence in tomato. We demonstrate that Solyc12g036480 downregulation, achieved via Virus-induced gene silencing (VIGS), confers longer life span and delayed overall senescence in tomato plants; for this reason we named this gene HḖBĒ (HEB) after the Greek youth goddess.
Results and discussion
HEB expression analyses
The tomato NAC TFs family counts 101 members and only few of them have been functionally characterized. As yet, tomato NAC proteins have been described as involved in defense responses, stomata opening and closure, drought tolerance, flower-boundary morphogenesis, leaf senescence and fruit ripening33,54,55,56,57. Among these 101 NAC members, we have selected Solyc12g036480/HEB for a deeper characterization.
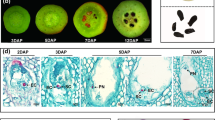
According to the transcriptome data collection of the Tomato Genome Consortium, HEB is equally transcribed in leaves and roots, but from the experimental data of Huang and Schiefelbein, HEB messenger is not detected in roots58,59. In order to define temporally and spatially HEB expression pattern, quantitative Real-Time PCRs (qRT-PCRs) were performed. Expression analyses were carried out using organs dissected by Micro-tom plants; UBIQUITIN 3 (UBI3) and ELONGATION FACTOR 1α (EF1α) were used as reference genes60. HEB transcript was found in young and old leaves and in young floral buds, but its mRNA is barely detected in roots, stem, mature green and red ripe fruits [developmental stages as described in61 (Fig. 1)].
qRT-PCR performed on HEB transcript in different organs at different developmental stages. HEB is transcribed in leaves (young and senescing) and in flower buds. Bars represent the average of three technical replicates and error bars indicate standard deviation. Three independent replicates were performed and a representative experiment is shown.
Identification of HEB putative orthologues
To identify HEB putative orthologues in other plant species, we generated a phylogenetic tree using the first 50 sequences selected using Phytozome 12 (https://phytozome.jgi.doe.gov/pz/portal.html#). Since in the Phytozome database only Solanum lycopersicum and S. tuberosum genomes are included, we additionally screened the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The phylogenetic tree (Supplemental Fig. S1) revealed the presence of a close NAC protein of tomato (Solyc06g069100). This might be caused by a recent duplication event since two NAC transcription factors are retrieved in other Solanaceae species, but are not found in Asteraceae. In-silico analysis demonstrates that Solyc06g069100 is expressed only in flower buds before anthesis59. To confirm Solyc06g069100 expression pattern, we performed qRT-PCRs on the dissected Micro-tom organs previously used (Supplemental Fig. S2a). Solyc06g069100 is highly transcribed in floral buds, but its messenger is poorly detected in old leaves and green fruits. We identified a single putative orthologue of HEB in species of the genus Arabidopsis (A. halleri, A. lyrata and A. thaliana) and Capsella (C. grandiflora and C. rubella) that belong to the Brassicaceae lineage I62. However, more than one putative orthologue is retrieved in lineage II62 (Brassica rapa and Eutrema salsugineum). The putative orthologue in Arabidopsis (AtNAC058), is, to date, the only one characterized: it is expressed in fruits and, its protein product participates in controlling silique maturation and senescence63. HEB and its Arabidopsis putative orthologue AtNAC058 are expressed in different tissues, nonetheless sequence identity and/or shared synteny is not sufficient to imply functional similarity. Orthologues as such are strictly the result of speciation, and in this case evolutionary convergence should also be taken into account64.
HEB silencing through VIGS assay
To functionally characterize HEB we transiently silenced its expression in developing leaves by using Virus-induced gene silencing (VIGS). VIGS has been extensively employed in Solanaceae species, such as tobacco plants, that can be infected with an efficiency near 100%65.
VIGS technique exploits the post-transcriptional gene silencing to temporary target a selected gene (for a review see Lange et al.66). VIGS benefits of RNAi-mediated antiviral defense mechanisms that naturally occur in plants. dsRNAs corresponding to the target gene are produced and cleaved by the ribonuclease DICER into siRNAs oligonucleotides of 21 to 24 bp, that are used to drive the RISC complex (RNA-Induced Silencing Complex) to specifically degrade the selected transcript67.
The silencing of the PHYTOENE DESATURASE (PDS) gene has been used as positive control of the infiltration protocol. PDS is an enzyme necessary for carotenoid biosynthesis, therefore the successfully infected plants are recognizable for the photo-bleached leaves and fruits68,69. Agrobacterium-infiltrated tomato plants bear fruits that fail to accumulate lycopene and thus display an altered pigmentation70. We obtained the photo‐bleached phenotype in 6 out of 10 tomato plants (60% efficiency), in agreement with Liu and collaborators69. We were able to silence SlPDS in leaves, flowers and fruits (Supplemental Fig. S3).
The fragment used to silence HEB was identified using the SGN VIGS Tool on the Sol Genomics Network website (https://vigs.solgenomics.net/). A 499 bp target region was selected, spanning from 392–891 bp of the coding sequence, therefore excluding the 5′ region which contains the highly conserved NAC domain (Supplemental Fig. S2b). Such a fragment silences specifically HEB and does not affect the expression of the close NAC gene Solyc06g069100, as shown by the alignment in Supplemental Fig. S2b. The target region was amplified using cDNA from 15 days old tomato seedlings and cloned into pTRV2-Gw plasmid71. The silencing of HEB in tomato leaves was performed in two biological replicates. Plants were infected with both pTRV1 (coding for viral functions such as replication and movement) and pTRV2-HEB (which encodes the coat protein and the sequence of interest). Furthermore, control groups were set: plants co-infiltrated with pTRV1 and pTRV2-GFP, and not infected plants. As negative control, we decided to replace the gateway cassette with a reporter gene, in our case GFP (Green Fluorescent Protein) from jellyfish. Indeed the Gw cassette71 aligns with the tomato genome from nucleotide 207 to 896.
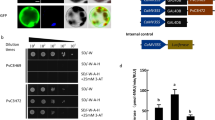
Infiltrations were performed on young leaves at 28 days after sowing (n = 20). In the first infection, leaves were collected at 46 dpi (days post infection), in the second infection at 53 dpi since the two groups of plants germinated and grew differently. To confirm the ability of the construct to downregulate HEB, we extracted total RNA from leaves before the appearance of any visible phenotype, using leaves at 24 dpi. The terminal leaflet from the third node of each plant have been collected. The analysis of HEB expression by qRT-PCR confirmed a reduction of HEB transcripts only in leaves infected with pTRV1 and pTRV2-HEB (Fig. 2a). These data clearly indicate that, as expected, silencing of HEB occurred only when pTRV1 and pTRV2-HEB are Agrobacterium-infiltrated and HEB downregulation anticipates the appearance of any visible phenotype.
(a) qRT-PCR analysis to evaluate HEB downregulation, cDNA of terminal leaflets at 24 dpi has been used; HEB silencing occurs only in plants co-infiltrated with both pTRV1 and pTRV2-HEB. Bars represent the average of three technical replicates and error bars indicate standard deviation. Three independent replicates were performed and a representative experiment is shown. (b) From the left: not infected plants, pTRV1 + pTRV2-GFP and pTRV1 + pTRV2-HEB. HEB downregulation prevents senescence, pTRV1 + pTRV2-HEB co-infected plants display a stay-green phenotype. (c–j) Quantification of the phenotype and VIGS evaluation over the two infections performed: (c,e,g,i) refer to the first infection; (d,f,h,j) refer to the second infection. (c,d) Percentage of yellow leaves (number of yellow leaves/number of total leaves), data were collected by two different infections (n = 10 plants for each infection). (e,f) HEB silencing frequency: percentage of plants that exhibited green leaves. 10 plants were infected per each replica. (g,h) HEB silencing effectiveness, percentage of green leaves (number of green leaves/number of total leaves); data were collected by two independent infections [n = 310 leaves for the first infection (g), n = 600 leaves for the second infection (h)]. (i,j) HEB silencing efficiency: percentage of green leaf area on the total leaf area, referred to two different infections (n = 5 for each infection). Letters above or below the bars (g-j) display statistical difference based on Tukey HSD test at P ≤ 0.05.
Silencing of HEB leads to a forever-green phenotype in tomato leaves
Plants infiltrated with both pTRV1 and pTRV2-HEB plasmids delayed senescence compared to control plants and displayed a global aging arrest (Fig. 2b). Such phenotype is particularly striking in older plants: although control plants are approaching the end of their life cycle, the pTRV1 and pTRV2-HEB infected plants still have leaves with extended greenness (Supplemental Fig. S4,b). To quantify the phenotype, we calculated the percentage of senescing leaves on total leaves (Supplemental Fig. S5). The yellow leaves were rated visually, we counted the number of yellow leaves approximately every 3 days for 20 days after the appearance of the first yellow spots (Fig. 2c,d). In both the infections, thae not infected control individuals showed the highest percentage of senescing leaves, while the pTRV1 + pTRV2-HEB plants the lowest (Fig. 2c,d). Such a trend is maintained during the entire time frame considered.
As suggested by Broderick and collaborators72, we estimated the quality of the VIGS analysis calculating (i) the silencing frequency, the percentage of plants with visible silencing on total plants; (ii) the silencing effectiveness, the percentage of leaves with visible silencing on total leaves; (iii) the silencing efficiency, that is the percentage of green leaf areas on total leaf surface (Fig. 2e–j). pTRV1 + pTRV2-HEB co-infected plants showed the highest percentages in frequency, effectiveness and efficiency compared to control plants, meaning that the silencing occurred in a significant and stable way. On the contrary, not infected plants displayed the lowest frequency, effectiveness and efficiency. The number of green leaves of pTRV1 + pTRV2-GFP co-infected plants was similar to the one estimated for pTRV1 + pTRV2-HEB plants but, in each leaf, the green areas were smaller compared to the pTRV1 + pTRV2-HEB ones (Fig. 2i,j). These data indicate that HEB downregulation mainly delays leaf senescence thus, in the end, plant life span is prolonged.
The tomato primary shoot meristem produces 7–12 leaves, then it undergoes to the reproductive transition and turn into an inflorescence73. The sympodial axillary meristem develops in the axil of the last leaf giving rise to only three leaves, but again in the axil of the last leaf a new sympodial meristem develops. Although HEB prolongs plant life span, it does not affect the ability of the plant to produce new leaves; indeed the number of leaves produced by the different groups of plants did not change significantly (Supplemental Fig. S4c). Furthermore, also the number of flowers and fruits is not stricken by HEB downregulation (Supplemental Fig. S4d). These evidences suggest that HEB is able to control leaves life span without affecting the meristem fate.
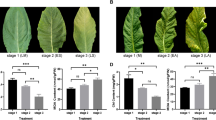
To better characterize the effects of HEB downregulation, we measured the maximum quantum yield (Fv/Fm74) of the PSII (Photosystem II) as indicator of photosystem integrity. For each group of plants, we analyzed three biological replicates per each infection. Visual comparisons of the leaves already indicated that the silencing of HEB delays leaf senescence (Fig. 3a,b), these observations were further confirmed by the IMAGING PAM fluorometer (Fig. 3c,d).
(a–f,j,k) Analyses of the phenotype over the two infections performed: (a,c,e,j) refer to the first infection; (b,d,f, k) refer to the second infection. (a,b) Comparison between pTRV1 + pTRV2-HEB infected leaves and controls (No infection and pTRV1 + pTRV2-GFP) revealed that HEB silencing delays senescence. (c,d) Visual aspect of PSII quantum yields (Fv/Fm, Imaging PAM) of pTRV1 + pTRV2-HEB infected leaves and controls. The tissue color indicates the maximum quantum yield of PSII, ranging from black (no efficiency) to violet (maximum efficiency) as shown by the colored bar on the top. (e,f) Y(II) of pTRV1 + pTRV2-HEB infected leaves and controls. Dots represent the average of 3 technical replicates and error bars indicate standard error. In each time point, statistical differences between pTRV1 + pTRV2-HEB and the controls was assessed with Tukey HSD test at P ≤ 0.05. A representative result from three independent experiments is shown. (g) pTRV1 + pTRV2-HEB older leaves (65 dpi) display yellow and light green spots. (h) Visual aspect and PSII quantum yield (Fv/Fm, Imaging PAM) of pTRV1 + pTRV2-HEB older leaves. (i) Measurement of Y(II) in pTRV1 + pTRV2-HEB older leaves. Dots represent the average of three technical replicates and error bars indicate standard error. In each time point, statistical differences between pTRV1 + pTRV2-HEB green and yellow sectors was assessed with Tukey HSD test at P ≤ 0.05. A representative result from three independent experiments is shown. (j,k) Chlorophyll content quantification in green/yellow sectors of pTRV1 + pTRV2-HEB infected leaves and in controls at 53 (j) and 60 (k) dpi. Bars represent the average of 3 technical replicates and error bars indicate standard error. Letters above the bars (j,k) display statistical difference based on Tukey HSD test at P ≤ 0.01. A representative result from three independent experiments is shown.
As expected, pTRV1 + pTRV2-HEB showed the highest effective quantum yield (Y(II)) after 280 s of actinic light exposition (Fig. 3e,f). pTRV1 + pTRV2-HEB retained an optimal photosynthetic capacity, like younger leaves, in a statistically significant way in all the considered time points.
HEB controls senescence in tomato leaves
Leaf senescence is a progressive process; in adult leaves, yellowing first appears in discrete spots which progressively enlarge. In pTRV1 + pTRV2-HEB leaves previously analyzed (Fig. 3a,b), yellowing was not present differently from the controls. In order to evaluate how the silencing of HEB affects senescence, we sampled older leaves, at 65 dpi, when yellow spots appeared also in pTRV1 + pTRV2-HEB leaves. Total RNA was extracted from pTRV1 + pTRV2-HEB, pTRV1 + pTRV2-GFP and not infected leaves. Quantification of HEB transcripts by qRT-PCR confirmed a downregulation in pTRV1 + pTRV2-HEB leaves compared to pTRV1 + pTRV2-GFP and not infected leaves (Fig. 4).
Quantification by qRT-PCR of the expression of HEB, SlSAG12, SlSAG113, SlNYC1, SlSGR1 and SlPaO from pTRV1 + pTRV2-HEB, pTRV1 + pTRV2-GFP and not infected leaves at the same developmental stage (65 dpi). HEB and all the senescence-related genes resulted to be downregulated in pTRV1 + pTRV2-HEB leaves compared to the controls. Bars represent the average of three technical replicates and error bars indicate standard deviation. Two independent replicates were performed and a representative experiment is shown.
To understand how HEB expression impacts leaf senescence, we quantified the transcript accumulation of a number of genes known to be involved in this process. In particular, we measured the relative expression of two Solanum lycopersicum SENESCENCE ASSOCIATED GENES (SlSAGs)—SlSAG12 (Solyc02g076910) and SlSAG113 (Solyc05g052980)—as well as three other genes involved in chlorophyll degradation, NON-YELLOW COLORING 1 (SlNYC1, Solyc07g024000), PHEOPHORBIDE a OXYGENASE (SlPaO, Solyc11g066440) and STAY-GREEN 1 (SlSGR1, Solyc08g080090).
SAG12 encodes a cysteine protease and it has been widely used as senescence-associated reference gene; in Arabidopsis it is abundant in senescent leaves75, when the yellowing is clearly visible76. SAG12 protein localizes in the senescence-associated vacuoles and participates to RuBisCO degradation77,78. However, the Arabidopsis sag12 mutant does not show any phenotype and leaf senescence progression appears normal79. Conversely, two homologs of SAG12 in rice (OsSAG1-2 and OsSAG2-2) negatively regulate senescence-related cell death80. In tomato, SlSAG12 has been extensively used as senescence-associated reference gen33,81,82. The second candidate gene, SAG113, encodes a phosphatase 2C expressed in ageing tissues and has already been reported as a senescence marker32,33. In Arabidopsis, SAG113 is a negative regulator of stomatal movement, its disruption causes tissue dehydration followed by senescence, and sag113 mutant shows delayed leaf senescence23,83.
qRT-PCR results showed downregulation of SlSAG12 and SlSAG113 in pTRV1 + pTRV2-HEB leaves (Fig. 4) compared to the controls, thus suggesting a delay in the onset of ageing.
During fruit ripening and the establishment of leaf senescence, Chls are massively degraded by plastid proteins. In higher plants, Chl degradation begins with the reduction of Chlb to Chla, mediated by Chlb reductase and 7-hydroxymethyl-chlorophyll a reductase84,85. Following this event, Chl degradation occurs in two steps, firstly, the pigments are converted into a colourless, blue-fluorescing product named primary fluorescent Chl catabolites (pFCC). This step is catalysed by chlorophyllase (Chlase), Mg-dechelatase, pheophorbide a oxygenase and red chlorophyll catabolite reductase. Afterwards, pFCCs are modified and exported into the vacuole, leading to their non-enzymatic isomerization into non-fluorescent chlorophyll catabolites, called NCCs86.
To determine whether the Chl breakdown is affected by HEB downregulation, we selected three different genes involved in different stages of Chl breakdown. SlNYC1 encodes the Chlb reductase which converts Chlb to Chla84, while SlPaO codes for a pheophorbide a oxygenase that cleaves the porphyrin ring of Pheide a, producing oxidized red Chl catabolite. PaO enzymatic activity participates as well in the de-greening process87,88,89,90. Finally, SlSGR1 was selected as it is implicated in the regulation of all the above-mentioned genes, via translational or post translational regulation84,91,92.
The data generated by our expression quantitation study (Fig. 4) suggest that HEB downregulation prevents the activation of chlorophyll degradation pathways. Indeed, in pTRV1 + pTRV2-HEB leaves, SlNYC1, SlSGR1 and SlPaO transcripts are reduced compared to control leaves, implying a more efficient photosynthetic performance due to chlorophyll integrity.
Taken together, these data suggest that HEB is a positive regulator of senescence mechanisms in tomato leaves, since HEB transient silencing in leaves fails to activate the correct aging pathways, leading to a stay-green phenotype.
HEB downregulation defers the ageing program
To better evaluate HEB involvement in senescence progression, we measured the maximum and effective quantum yield and the chlorophyll (Chl) content of yellow and green sectors of HEB silenced leaves and control leaves at 65 dpi. As expected from the visual output of the IMAGING PAM fluorometer (Fig. 3g,h), the yellow regions of the leaves displayed a reduced photosynthetic efficiency (Fig. 3i). In particular, the maximum quantum yield is statistically significant comparing yellow and green portions. These observations were also confirmed by the Chl quantification (Fig. 3j,k) since the green regions of pTRV1 + pTRV2-HEB leaves contain markedly higher Chl amount compared to the yellow regions and to the controls.
Many NAC transcription factors are known to be involved in the control of leaves senescence in tomato, such as SlNAP2, NOR (NON-RIPENING) and SlORE1S0231,32,33. Interestingly, slnap2 knockdown mutants and nor mutants display a delayed senescence only when excised leaves undergo dark induced senescence, however the physiological senescence is comparable to the wild-type plants. Moreover, in nor mutants or NOR overexpressing lines, Chl content is similar to the control plants, when grown in light, and differences appear only after 14 days of continuous darkness32. A similar phenotype has been described for slnap2 mutant and SlNAP2 overexpressing lines: a difference in Chl content compared to the control is measurable only after 14 days of darkness33. In our experiments, leaves were not subjected to dark adaptation before measurement of Chl content, but rather exposed to canonical long day conditions (16 h light/8 h dark). This suggests that HEB positively modulates leaf senescence and is a strong promoter of the physiologic aging programs.
We also evaluated the expression of SlSAG12, SlSAG113, SlNYC1, SlSGR1 and SlPaO in the different areas of the same pTRV1 + pTRV2-HEB leaf. Total RNA was extracted from three independent replicates, carefully separating dark and pale regions of the leaves, and first used to quantify HEB transcript accumulation. A strong downregulation of HEB was detected in the dark portions of the leaves (Fig. 5a). The senescence-associated genes and the chlorophyll-related genes transcripts were then quantified by qRT-PCR, results showed downregulation of all the targets in the green sectors of the leaves (Fig. 5b).
(a) qRT-PCR analysis on mRNAs from green and yellow portions of pTRV1 + pTRV2-HEB infected leaves. The expression of HEB is strongly reduced in the green sections. Bars represent the average of three technical replicates and error bars indicate standard deviation. Three independent replicates were performed and a representative experiment is shown. (b) Quantification by qRT-PCR of the expression of senescence-related genes SlSAG12, SlSAG113, SlNYC1, SlSGR1 and SlPaO, in green/yellow regions of pTRV1 + pTRV2-HEB infected leaves. A general downregulation of senescence-associated genes is recorded in the green sectors, especially SlSAG12 is strongly reduced. Bars represent the average of three technical replicates and error bars indicate standard deviation. Three independent replicates were performed and a representative experiment is shown. (c) Immunoblot analyses were performed to evaluate the presence and the amount of chlorophyll associated proteins in green/yellow regions. While the chlorophyll-binding proteins Lhca1, Lhca4, Lhcb1 and Lhcb4 accumulation is slightly reduced in the yellow sectors, compared to the green ones, the catalytic enzyme PaO, involved in chlorophyll degradation, is more abundant in the yellow areas. The histone protein H3 was used as loading control, together with a Coomassie Brilliant Blue (C.B.B.) staining of the SDS-PAGE. A representative result from three independent experiments is shown.
To clarify whether HEB silencing can also impact SlPaO protein accumulation, we performed an immunoblot analyses. The protein HISTONE 3 (H3) was used as control (Fig. 5c). In our analysis, PaO poorly accumulated in the green sectors, while in the senescing yellow sectors was more abundant. To corroborate the role of HEB, and to establish the level of accumulation of photosystems I and II, we investigated the accumulation of the chlorophyll binding proteins of the light-harvesting complexes of Photosystem I and II. We detected a decreased level of Lhca1, Lhca4, Lhcb1 and Lhcb4 proteins in the yellow sectors of the leaves. This analysis indirectly supports the hypothesis that HEB is necessary to trigger the senescence process in leaf: indeed with the Lhca1, Lhca4, Lhcb1 and Lhcb4 antibodies we detected a reduced level of antenna proteins, directly associated to Chl. These data corroborate the findings that the silencing of HEB in green leaf sectors led to a marked delay in the ageing program, preventing the accumulation of PaO protein, thus maintaining high level of Lhc proteins.
Can HEB bind PaO promoter?
It was previously described that SlNAP2 controls organ ageing directly binding the regulative regions of SlSGR1 and SlPaO33. Similarly, NOR is able to directly bind the promoter of SlSAG113 and SlSGR1 but not SlPaO32. According to our data, HEB mechanism is comparable to NOR and SlNAP2 ones, since its expression positively correlates with the expression of such genes. As of yet, due to a lack of structural characterization of HEB binding site, we cannot predict whether these differently regulated genes are direct target of HEB. In-silico research of the promoter region of SlSAG12, SlSAG113, SlNYC1, SlSGR1 and SlPaO returned putative binding sites for several NAC genes. We detected a putative binding site for AtNAC058 on the promoter region of SlSAG113, SlSGR1, SlPaO and SlNYC1 (Supplemental Fig. S6). To verify the ability of HEB to bind the regulative regions of SlPaO, we used a yeast 1-Hybrid assay (Supplemental Fig. S7). HEB was fused with the GAL4-AD (Activation Domain) whilst a fragment of 1.45 kb that include the 5′ region of SlPaO was cloned into pHis2 vector (see Materials and methods). Both constructs were introduced into a diploid yeast via mating and colonies were selected on a medium lacking histidine to assay protein–DNA interaction. No clear yeast growth was observable, implying that no interaction occurs among HEB and SlPaO promoter.
HEB overexpression doesn’t lead to transcript accumulation
In order to confirm that HEB is a senescence positive regulator, we generated transient gain of function plants. We used Agrobacterium to infect tomato leaves with a construct bearing 35S::HEB (n = 2). Each leaf was ideally divided in two parts, separated by the midrib; one half was infected with 35S::HEB construct while the other half with a control construct bearing the 35S::GUS (β-glucuronidase) reporter (Supplemental Fig. S8a). 10 days after the infection leaves were collected (Supplemental Fig. S8b) and the IMAGING PAM fluorometer was used to evaluate the photosynthetic efficiency of the two halves of the leaves (Supplemental Fig. S8c). The portions of the leaves infected with the 35S::HEB construct revealed a slight increase in the photosynthetic yield compared to the control region (Supplemental Fig. S8d). In order to evaluate the expression level of β-glucuronidase and HEB, the total RNA was extracted from the two halves of the leaves. qRT-PCRs revealed that the β-glucuronidase gene was overexpressed in the 35S::GUS half of the leaves (Supplemental Fig. S8e). A small amount of β-glucuronidase was also detected in the 35S::HEB half of the leaves, and this might be a collateral effect of the procedure. Conversely, HEB was not overexpressed in both the leaves portions (Supplemental Fig. S8e), this suggests that HEB transcript is difficult to accumulate and might be quite unstable thus preventing the possibility to run analysis that request transient over- or mis-expression.
The fountain of youth: possible application of a forever young plant
One of the main goals of crop science is the improvement of traits that can increase plant yield and biomass. This can be achieved by several ways, one of which is the extension of the photosynthetic ability by prolonging a plant’s lifecycle. Keeping a plant in a stay-green status can be reached in different manners, known as “the five ways to stay green”: (i) delaying the initiation of leaf senescence, (ii) slowing down the rate of leaf senescence, (iii) delaying chlorophyll degradation, (iv) causing tissue death (for example by freezing, boiling or drying) or (v) enhancing greenness93. HEB falls in the first category of stay-green phenotype, since it causes a delay in the initiation of leaf senescence, and here we have demonstrated that HEB silencing defers ageing progression leading to a stay-green phenotype and prevents the transcription of genes involved in chlorophyll degradation (Figs. 2, 3, 4, 5).
Interestingly, the longer life span of leaves does not affect flower and fruit yield in the time frame considered (20 days, Supplemental Fig. S4). In cereals, it was shown that the stay-green phenotype has a negative effect on yield since prolonged lifecycle of crops causes nutrient dilution. This phenomenon is known as the “dilution effect”, when the relationship between crop yield and mineral concentrations become unfavorable94. For instance, silencing of the wheat NAC gene NAM-B1 delays senescence, though concurrently, protein content and presence of elements such as zinc and iron in seeds are dramatically compromised95. In tomato nor mutants, fruit ripening and dark-induced senescence are delayed32, albeit at the cost of fruit quality: ethylene and pigment biosynthesis are both downregulated, while cellulose synthase proteins are upregulated, causing an increase of fruit firmness. Nutrient biosynthesis was also found to be impaired, however resistance to pathogens appears enhanced96. Considering these findings, the study of HEB fruits to determine potential impacts of prolonged lifecycle on nutrient biosynthesis and/or pathogen resistance is an extremely compelling future endeavor.
Conclusion
In this work we silenced the putative tomato NAC transcription factor Solyc12g03648/HEB through VIGS assay. The results we uncovered suggest that HEB is involved in the regulation of leaf senescence in tomato, acting as a positive regulator. Reduction of HEB transcript leads to the fail-activation of SENESCENCE ASSOCIATED GENES (SAGs) and lack of chlorophyll degradation mechanisms, ultimately delaying leaf senescence and prolonging the life span of the entire plant. Further research will be focused on identifying the molecular mechanisms through which HEB controls the ageing processes, by identifying its target genes and interactors. With this research we identified a tomato gene involved in delaying leaf senescence, laying the base for future applications which will allow the cultivation of longer-lived crops.
Material and methods
Plant material and growth conditions
Micro-Tom tomato plants were grown on soil under greenhouse condition with a 16 h light/8 h dark cycle at 22/18 °C. For VIGS assay we Agrobacterium-infiltrated young leaves 28 days after sowing.
Phylogenetic analysis
To identify HEB putative orthologues, we screened the Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html) using HEB protein sequence to conduct blastp analyses; the first 50 sequences obtained were then selected for the phylogenetic tree. For the Solanaceae sequences not present in the Phytozome database we screened the NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) using HEB protein sequence to conduct blastp.
We also queried blastp on the Phytozome database using HEB protein sequence against Solanum lycopersicum proteome and AtNAC058 protein against Arabidopsis thaliana proteome. We selected the proteins with the highest score, and we used them as control of the phylogenetic tree. All the selected proteins were aligned with MUSCLE. The phylogenetic tree was constructed with MEGAX (https://www.megasoftware.net/) using a Maximum Likelihood method (JTT protein model, bootstrapping of 100).
RNA extraction, cDNA synthesis and expression analysis
Total tomato RNA was extracted using the cetyl trimethyl ammonium bromide (CTAB) protocol (adapted from Chang et al.97). Genomic DNA was removed using TURBO™ DNase (Invitrogen™) according to the manufacturer’s instructions. The RNA was reverse transcribed using the iScript™ gDNA Clear cDNA Synthesis Kit (Biorad) and the cDNA was used as template in qRT-PCR reactions. qRT-PCR was carried out on a CFX96 Real-Time system (Bio-Rad), using the primer pairs reported in Supplemental Table S1. The UBI3 and EF1α transcripts were used as internal references60. The Bio-Rad CFX Manager software (V3.1) was used to analyze data from three biological and three technical replicates (except where otherwise specified, see Fig. 4).
VIGS assays
In this work we used the pTRV plasmids previously described by Orzaez et al.71. HEB and GFP fragments were cloned from seedling cDNA and pGREENII plasmid respectively, using the primers reported in the Supplemental Table S1. The pTRV2-PDS plasmid was kindly provided by Concha Gómez Mena (Instituto de Biología Molecular y Celular de Plantas, Valencia, Spain).
GV3101 Agrobacterium culture was transformed with pTRV1, containing the viral genes for replication and movement, and a second culture with pTRV2, containing the fragment for PDS or HEB or the GFP fragment. These cultures were used to infiltrate tomato plants (n = 10 for each construct in each infection), while plants without plasmids were used as mock (n = 10 in each infection). For the PDS silencing we Agrobacterium-injected 10 plants.
The infiltration was performed as described in98. Young leaves were infiltrated using syringes without needles, while for fruit infection we infiltrated the pedicels of the flower with a needles syringe. Tomato not infiltrated or infiltrated with pTRV1 and pTRV2-GFP was used as control. Each inoculation was carried out two times.
Chlorophyll content and chlorophyll fluorescence analysis
Pigments were extracted using 90% (v/v) acetone from different portion of leaves. The chlorophyll a and b contents were measured using a spectrophotometer (Amersham Biosciences) at 663- and 645-nm wavelength. Total chlorophyll (a + b) values were determined as described previously by Arnon99 and normalized relative to tissue fresh weight. The pulse-modulated fluorometer IMAGING-PAM M-Series (Walz) was used to measure in vivo chlorophyll a fluorescence of tomato leaves100. Infected or not infected leaves were placed under the fluorometer and three measurements for each phenotype/infection were performed, and three biological replicates were used. Samples were first dark adapted and the fluorescence was measured as reported in63.
Immunoblot analyses
For immunoblot analyses, tomato leaves were collected from plants infected with both pTRV1 and pTRV2- HEB, at 46 dpi—first infection- and 53 dpi—second infection. Total protein content was extracted according to101. Protein extracts, corresponding to 5 mg of leaves fresh weight, were fractionated by SDS-PAGE gel (12% [w/v] acrylamide102) and then transferred to polyvinylidene difluoride membranes103. Replicate filters were cropped and immunodecorated with antibodies specific for proteins with different molecular weights, Lhca1, Lhca4, Lhcb1, Lhcb4, PaO and Histone H3. Lhca1, Lhca4, Lhcb1, Lhcb4, and PaO antibodies were obtained from Agrisera, Histone H3 antibody from Sigma-Aldrich.
Yeast 1-hybrid assay
The SlPaO promoter region (Solyc11g066440) of 1,434 bp was amplified using a primer pair containing EcoRI restriction sites (Supplemental Table S1), cloned into pHis2 vector (Clontech) previously linearised using EcoRI. The bait plasmid (pPaO-pHis2) was used to transform Saccharomyces cerevisiae Y187 strain (Clontech). The Solyc12g03648/HEB gene (981 bp) cloned into pBlueScript II SK(+) vector was purchased from Biomatik Corporation (Cambridge, Canada), excised with EcoRI and XhoI and ligated into pGADT7 (Clontech) EcoRI/XhoI digested. The prey plasmid (HEB-pGADT7) was introduced into Saccharomyces cerevisiae AH109 strain (Clontech) and transformants mated with Y187 strain containing the bait plasmid as described by Resentini et al.104. Diploids were selected on medium lacking Trp and Leu. Growth diploid colonies were scraped on selective media lacking Trp, Leu and His and supplemented with 0, 1, 2 or 5 mM 3-AT (Sigma-Aldrich). Yeast 1-Hybrid was also performed using as bait and prey plasmids pHis2 and pGADT7 respectively as controls.
Transient expression in tomato leaves
We transformed Agrobacterium strain GV3101 with 35S::HEB and 35S::GUS constructs. These cultures were used to Agrobacterium-infiltrate tomato leaves (n = 2). Briefly, Agrobacterium cultures of 35S::HEB and 35S::GUS were pre-inoculated in 5 ml liquid medium and let grow for 20 h in stirring conditions at 28 °C. OD at 600 nm was then measured, cultures were concentrated and resuspended in infiltration buffer (MgCl2 10 mM, MES pH 5, 6, 10 mM, acetosyringone 150 µM) to the final OD. Cultures were grown for two hours at room temperature in stirring conditions. Each leaf was ideally divided in two halves and infiltrated with 2 ml of 35S::GUS suspension in one half and with 2 ml of 35S::HEB suspension in the other half.
References
Woo, H. R., Masclaux-Daubresse, C. & Lim, P. O. Plant senescence: how plants know when and how to die. J. Exp. Bot. 69, 715–718 (2018).
Buchanan-Wollaston, V. Senescence processes in plants. Annual plant review, volume 26. Ann. Bot. 101, 197 (2008).
Kim, H. J. et al. Time-evolving genetic networks reveal a NAC troika that negatively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. 115, E4930–E4939 (2018).
Gregersen, P. L., Holm, P. B. & Krupinska, K. Leaf senescence and nutrient remobilisation in barley and wheat. Plant Biol. 10(Suppl 1), 37–49 (2008).
Watanabe, M. et al. Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis. Plant Physiol. 162, 1290–1310 (2013).
Avice, J.-C. & Etienne, P. Leaf senescence and nitrogen remobilization efficiency in oilseed rape (Brassica napus L.). J. Exp. Bot. 65, 3813–3824 (2014).
Woo, H. R., Kim, H. J., Nam, H. G. & Lim, P. O. Plant leaf senescence and death: regulation by multiple layers of control and implications for aging in general. J. Cell Sci. 126, 4823–4833 (2013).
Wojciechowska, N., Sobieszczuk-Nowicka, E. & Bagniewska-Zadworna, A. Plant organ senescence: regulation by manifold pathways. Plant Biol. 20, 167–181 (2018).
Forlani, S., Masiero, S. & Mizzotti, C. Fruit ripening: the role of hormones, cell wall modifications, and their relationship with pathogens. J. Exp. Bot. 70, 2993–3006 (2019).
Ay, N., Janack, B. & Humbeck, K. Epigenetic control of plant senescence and linked processes. J. Exp. Bot. 65, 3875–3887 (2014).
Zhong, S. et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31, 154–159 (2013).
Liu, R. et al. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc. Natl. Acad. Sci. USA 112, 10804–10809 (2015).
Olsen, A. N., Ernst, H. A., Leggio, L. L. & Skriver, K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87 (2005).
Nakano, Y., Yamaguchi, M., Endo, H., Rejab, N. A. & Ohtani, M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6, 288 (2015).
Kim, H. J., Nam, H. G. & Lim, P. O. Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 33, 48–56 (2016).
Ohbayashi, I. et al. Evidence for a role of ANAC082 as a ribosomal stress response mediator leading to growth defects and developmental alterations in arabidopsis. Plant Cell 29, 2644–2660 (2017).
Mathew, I. E. & Agarwal, P. May the fittest protein evolve: favoring the plant-specific origin and expansion of NAC transcription factors. BioEssays 40, e1800018 (2018).
Chou, M.-L. et al. The direct involvement of dark-induced Tic55 Protein in chlorophyll catabolism and its indirect role in the MYB108-NAC signaling pathway during leaf senescence in Arabidopsis thaliana. Int. J. Mol. Sci. 19, 1854 (2018).
Balazadeh, S. et al. A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 62, 250–264 (2010).
Balazadeh, S. et al. ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol. Plant 4, 346–360 (2011).
Balazadeh, S., Riano-Pachon, D. M. & Mueller-Roeber, B. Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol. 10(Suppl 1), 63–75 (2008).
Kim, J. H. et al. Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323, 1053–1057 (2009).
Zhang, K. & Gan, S.-S. An abscisic acid-AtNAP transcription factor-SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing arabidopsis leaves. Plant Physiol. 158, 961–969 (2012).
Wu, A. et al. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24, 482–506 (2012).
Sakuraba, Y., Han, S.-H., Lee, S.-H., Hortensteiner, S. & Paek, N.-C. Arabidopsis NAC016 promotes chlorophyll breakdown by directly upregulating STAYGREEN1 transcription. Plant Cell Rep. 35, 155–166 (2016).
Matallana-Ramirez, L. P. et al. NAC transcription factor ORE1 and senescence-induced BIFUNCTIONAL NUCLEASE1 (BFN1) constitute a regulatory cascade in Arabidopsis. Mol. Plant 6, 1438–1452 (2013).
Mahmood, K., El-Kereamy, A., Kim, S.-H., Nambara, E. & Rothstein, S. J. ANAC032 positively regulates age-dependent and stress-induced senescence in Arabidopsis thaliana. Plant Cell Physiol. 57, 2029–2046 (2016).
Kim, H. J. et al. Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 65, 4023–4036 (2014).
Guo, Y. & Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46, 601–612 (2006).
Garapati, P., Xue, G.-P., Munne-Bosch, S. & Balazadeh, S. Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 168, 1122–1139 (2015).
Lira, B. S. et al. Manipulation of a senescence-associated gene improves fleshy fruit yield. Plant Physiol. 175, 77–91 (2017).
Ma, X., Balazadeh, S. & Mueller-Roeber, B. Tomato fruit ripening factor NOR controls leaf senescence. J. Exp. Bot. 70, 2727–2740 (2019).
Ma, X. et al. The NAC transcription factor SlNAP2 regulates leaf senescence and fruit yield in tomato. Plant Physiol. 177, 1286–1302 (2018).
Zhou, Y. et al. Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biol. 13, 132 (2013).
Liang, C. et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. 111, 10013–10018 (2014).
Sakuraba, Y. et al. Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol. 56, 2325–2339 (2015).
El Mannai, Y., Akabane, K., Hiratsu, K., Satoh-Nagasawa, N. & Wabiko, H. The NAC transcription factor gene OsY37 (ONAC011) promotes leaf senescence and accelerates heading time in rice. Int. J. Mol. Sci. 18, 2165 (2017).
Mao, C. et al. A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 174, 1747–1763 (2017).
Kjaersgaard, T. et al. Senescence-associated barley NAC (NAM, ATAF1,2, CUC) transcription factor interacts with radical-induced cell death 1 through a disordered regulatory domain. J. Biol. Chem. 286, 35418–35429 (2011).
Christiansen, M. W. & Gregersen, P. L. Members of the barley NAC transcription factor gene family show differential co-regulation with senescence-associated genes during senescence of flag leaves. J. Exp. Bot. 65, 4009–4022 (2014).
Pimenta, M. R. et al. The stress-induced soybean NAC transcription factor GmNAC81 plays a positive role in developmentally programmed leaf senescence. Plant Cell Physiol. 57, 1098–1114 (2016).
Chen, Y., Qiu, K., Kuai, B. & Ding, Y. Identification of an NAP-like transcription factor BeNAC1 regulating leaf senescence in bamboo (Bambusa emeiensis ‘Viridiflavus’). Physiol. Plant. 142, 361–371 (2011).
Chao, Y. et al. Transcriptome analysis of leaf senescence in red clover (Trifolium pratense L.). Physiol. Mol. Biol. Plants 24, 753–765 (2018).
Moschen, S. et al. Identification of candidate genes associated with leaf senescence in cultivated sunflower (Helianthus annuus L.). PLoS ONE 9, e104379 (2014).
Shah, S. T. et al. Isolation and expression profiling of GhNAC transcription factor genes in cotton (Gossypium hirsutum L.) during leaf senescence and in response to stresses. Gene 531, 220–234 (2013).
Fan, K. et al. A novel NAP member GhNAP is involved in leaf senescence in Gossypium hirsutum. J. Exp. Bot. 66, 4669–4682 (2015).
Zhao, F. et al. GhNAC12, a neutral candidate gene, leads to early aging in cotton (Gossypium hirsutum L.). Gene 576, 268–274 (2016).
Shan, W. et al. Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J. Exp. Bot. 63, 5171–5187 (2012).
Tak, H., Negi, S., Gupta, A. & Ganapathi, T. R. A stress associated NAC transcription factor MpSNAC67 from banana (Musa x paradisiaca) is involved in regulation of chlorophyll catabolic pathway. Plant Physiol. Biochem. PPB 132, 61–71 (2018).
Zhu, Z. et al. DRL1, encoding a NAC transcription factor, is involved in leaf senescence in grapevine. Int. J. Mol. Sci. 20, 2678 (2019).
Li, W. et al. NAC family transcription factors in tobacco and their potential role in regulating leaf senescence. Front. Plant Sci. 9, 1900 (2018).
Kusaba, M., Tanaka, A. & Tanaka, R. Stay-green plants: what do they tell us about the molecular mechanism of leaf senescence. Photosynth. Res. 117, 221–234 (2013).
Yang, S.-D., Seo, P. J., Yoon, H.-K. & Park, C.-M. The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23, 2155–2168 (2011).
Hendelman, A., Stav, R., Zemach, H. & Arazi, T. The tomato NAC transcription factor SlNAM2 is involved in flower-boundary morphogenesis. J. Exp. Bot. 64, 5497–5507 (2013).
Du, M. et al. Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell 26, 3167–3184 (2014).
Gao, Y. et al. A NAC transcription factor, NOR-like1, is a new positive regulator of tomato fruit ripening. Hortic. Res. 5, 75 (2018).
Thirumalaikumar, V. P. et al. NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 16, 354–366 (2018).
Huang, L. & Schiefelbein, J. Conserved gene expression programs in developing roots from diverse plants. Plant Cell 27, 2119–2132 (2015).
Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 (2012).
Lacerda, A. L. M. et al. Reference gene selection for qPCR analysis in tomato-bipartite begomovirus interaction and validation in additional tomato-virus pathosystems. PLoS ONE 10, e0136820–e0136820 (2015).
Pesaresi, P., Mizzotti, C., Colombo, M. & Masiero, S. Genetic regulation and structural changes during tomato fruit development and ripening. Front. Plant Sci. 5, 124 (2014).
Kagale, S. et al. Polyploid evolution of the brassicaceae during the cenozoic era. Plant Cell https://doi.org/10.1105/tpc.114.126391 (2014).
Mizzotti, C. et al. Time-course transcriptome analysis of Arabidopsis siliques discloses genes essential for fruit development and maturation. Plant Physiol. 178, 1249–1268 (2018).
Theissen, G. Secret life of genes. Nature 415, 741 (2002).
Liu, Y., Schiff, M., Marathe, R. & Dinesh-Kumar, S. P. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429 (2002).
Lange, M., Yellina, A. L., Orashakova, S. & Becker, A. Virus-induced gene silencing (VIGS). In Plants: An Overview of Target Species and the Virus-Derived Vector Systems BT - Virus-Induced Gene Silencing: Methods and Protocols (ed. Becker, A.) 1–14 (Humana Press, Totowa, 2013).
Carrington, J. C. & Ambros, V. Role of microRNAs in plant and animal development. Science 301, 336–338 (2003).
Kumagai, M. H. et al. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA. Proc. Natl. Acad. Sci. USA 92, 1679–1683 (1995).
Liu, Y., Schiff, M. & Dinesh-Kumar, S. P. Virus-induced gene silencing in tomato. Plant J. 31, 777–786 (2002).
Orzaez, D., Mirabel, S., Wieland, W. H. & Granell, A. Agroinjection of tomato fruits. A tool for rapid functional analysis of transgenes directly in fruit. Plant Physiol. 140, 3–11 (2006).
Orzaez, D. et al. A visual reporter system for virus-induced gene silencing in tomato fruit based on anthocyanin accumulation. Plant Physiol. 150, 1122–1134 (2009).
Broderick, S. R. & Jones, M. L. An optimized protocol to increase virus-induced gene silencing efficiency and minimize viral symptoms in Petunia. Plant Mol. Biol. Report. 32, 219–233 (2014).
Park, S. J., Jiang, K., Schatz, M. C. & Lippman, Z. B. Rate of meristem maturation determines inflorescence architecture in tomato. Proc. Natl. Acad. Sci. 109, 639–644 (2012).
Wingler, A., Marès, M. & Pourtau, N. Spatial patterns and metabolic regulation of photosynthetic parameters during leaf senescence. New Phytol. 161, 781–789 (2004).
Gui, Y., Cai, Z. & Gan, S. Transcriptome of Arabidopsis leaf senescence. Plant. Cell Environ. 27, 521–549 (2004).
Lohman, K. N., Gan, S., John, M. C. & Amasino, R. M. Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol. Plant. 92, 322–328 (1994).
Martinez, D. E., Costa, M. L., Gomez, F. M., Otegui, M. S. & Guiamet, J. J. ‘Senescence-associated vacuoles’ are involved in the degradation of chloroplast proteins in tobacco leaves. Plant J. 56, 196–206 (2008).
Carrion, C. A. et al. In vivo inhibition of cysteine proteases provides evidence for the involvement of ‘senescence-associated vacuoles’ in chloroplast protein degradation during dark-induced senescence of tobacco leaves. J. Exp. Bot. 64, 4967–4980 (2013).
Otegui, M. S. et al. Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J. 41, 831–844 (2005).
Singh, S., Giri, M. K., Singh, P. K., Siddiqui, A. & Nandi, A. K. Down-regulation of OsSAG12-1 results in enhanced senescence and pathogen-induced cell death in transgenic rice plants. J. Biosci. 38, 583–592 (2013).
Ding, F., Wang, M. & Zhang, S. Sedoheptulose-1,7-bisphosphatase is involved in methyl jasmonate- and dark-induced leaf senescence in tomato plants. Int. J. Mol. Sci. 19, 3673 (2018).
Lira, B. S. et al. Plant degreening: evolution and expression of tomato (Solanum lycopersicum) dephytylation enzymes. Gene 546, 359–366 (2014).
Zhang, K., Xia, X., Zhang, Y. & Gan, S.-S. An ABA-regulated and Golgi-localized protein phosphatase controls water loss during leaf senescence in Arabidopsis. Plant J. 69, 667–678 (2012).
Kusaba, M. et al. Rice NON-YELLOW COLORING1 Is Involved in Light-Harvesting Complex II and Grana Degradation during Leaf Senescence. Plant Cell 19, 1362–1375 (2007).
Horie, Y., Ito, H., Kusaba, M., Tanaka, R. & Tanaka, A. Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J. Biol. Chem. 284, 17449–17456 (2009).
Hӧrtensteiner, S. Chlorophyll degradation during senescence. Annu. Rev. Plant Biol. 57, 55–77 (2006).
Tanaka, R., Hirashima, M., Satoh, S. & Tanaka, A. The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a: inhibition of the pheophorbide a oxygenase activity does not lead to the ‘stay-green’ phenotype in Arabidopsis. Plant Cell Physiol. 44, 1266–1274 (2003).
Wuthrich, K. L., Bovet, L., Hunziker, P. E., Donnison, I. S. & Hortensteiner, S. Molecular cloning, functional expression and characterisation of RCC reductase involved in chlorophyll catabolism. Plant J. 21, 189–198 (2000).
Pruzinska, A., Tanner, G., Anders, I., Roca, M. & Hortensteiner, S. Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. USA 100, 15259–15264 (2003).
Pruzinska, A. et al. In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19, 369–387 (2007).
Harpaz-Saad, S. et al. Chlorophyllase is a rate-limiting enzyme in chlorophyll catabolism and is posttranslationally regulated. Plant Cell 19, 1007–1022 (2007).
Ren, G. et al. Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol. 144, 1429–1441 (2007).
Thomas, H. & Howarth, C. J. Five ways to stay green. J. Exp. Bot. 51, 329–337 (2000).
Simmonds, N. W. The relation between yield and protein in cereal grain. J. Sci. Food Agric. 67, 309–315 (1995).
Uauy, C., Distelfeld, A., Fahima, T., Blechl, A. & Dubcovsky, J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314, 1298–1301 (2006).
Yuan, X.-Y., Wang, R.-H., Zhao, X.-D., Luo, Y.-B. & Fu, D.-Q. Role of the tomato non-ripening mutation in regulating fruit quality elucidated using iTRAQ protein profile analysis. PLoS ONE 11, e0164335 (2016).
Chang, S., Puryear, J. & Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11, 113–116 (1993).
Fernandez-Moreno, J.-P., Orzaez, D. & Granell, A. VIGS: A Tool to Study Fruit Development in Solanum lycopersicum BT - Virus-Induced Gene Silencing: Methods and Protocols. in (ed. Becker, A.) 183–196 (Humana Press, 2013). doi:10.1007/978-1-62703-278-0_14.
Arnon, D. I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in beta vulgaris. Plant Physiol. 24, 1–15 (1949).
Tadini, L. et al. Thylakoid redox signals are integrated into organellar-gene-expression-dependent retrograde signaling in the prors1-1 mutant. Front. Plant Sci. 3, 282 (2012).
Martinez-Garcia, J. F., Monte, E. & Quail, P. H. A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 20, 251–257 (1999).
Schagger, H. & von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 (1987).
Ihnatowicz, A. et al. Mutants for photosystem I subunit D of Arabidopsis thaliana: effects on photosynthesis, photosystem I stability and expression of nuclear genes for chloroplast functions. Plant J. 37, 839–852 (2004).
Resentini, F. et al. SUPPRESSOR OF FRIGIDA (SUF4) supports gamete fusion via regulating arabidopsis EC1 gene expression. Plant Physiol. 173, 155–160 (2017).
Acknowledgements
We are thankful to Barbara Ambrose (New York Botanical Garden) and Natalia Pabon-Mora (Universidad de Antioquia, Medellín, Colombia) for their help with the VIGS assay. We thank Concha Gómez Mena (Instituto de Biología Molecular y Celular de Plantas, Valencia, Spain) for providing pTRV plasmids and Luciana Arria and Maria Neve Iodice for their help in plasmids preparation and VIGS analysis. We are thankful to Aureliano Bombarely for his help with the phylogenetic analysis. We also thank Valerio Parravicini and Mario Beretta for their excellent technical assistance and Francesca Lopez for critical reading of the manuscript. This work was supported by a fellowship from the Università degli Studi di Milano [to SF and SR] and by MIUR PRIN [Grant Number 2015BPM9H3_005 to SM].
Author information
Authors and Affiliations
Contributions
S.M. and C.M. conceived the research plan; S.M. supervised the experiments; C.C., S.F., S.R., L.T. and C.M. performed research and analyzed data; S.F., S.M. and C.M. wrote the paper with contributions from all authors. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Forlani, S., Cozzi, C., Rosa, S. et al. HEBE, a novel positive regulator of senescence in Solanum lycopersicum. Sci Rep 10, 11021 (2020). https://doi.org/10.1038/s41598-020-67937-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-67937-z
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.