Abstract
Pancreatic neuroendocrine neoplasms (PanNENs) are rare tumours that compose 1–2% of all pancreatic tumours. Patients with metastatic grade 3 neoplasia are usually treated with chemotherapy but have a poor progression-free and overall survival. According to the WHO 2017 classification, they are divided into neuroendocrine tumours (NETs) G3 and neuroendocrine carcinomas (NECs). Despite the new classification, new diagnostic and prognostic biomarkers are needed to sub-categorise the patients and to help guide therapy decisions. Blood from 42 patients and 42 healthy controls were screened for the presence of 92 proteins with the Immuno-Oncology panel using the Proximity Extension Assay provided by Olink Biosciences. Immunohistochemical staining of FAS ligand (FASLG) was performed on 16 patient tumour specimens using a commercial antibody. Fifty-four out of 87 evaluable proteins differed significantly in concentration between blood from patients and blood from healthy controls. FASLG was the only protein for which the concentration in blood was significantly lower in patients compared to controls and the levels correlated negatively to Ki-67 index. Seven of 14 evaluable PanNEN G3 specimens showed FASLG immunoreactivity in the tumour cells while there was scattered immunoreactivity in immune cells. Positive FASLG immunoreactivity correlated to well-differentiated morphology. FASLG concentration in blood was significantly lower in patients with pancreatic NENs G3 compared to controls, and the expression in tumour tissue was variable. Furthermore, FASLG was negatively correlated to Ki-67 and was more frequently expressed in well-differentiated tumours. Taken together, these results may suggest a role of FASLG in PanNENs.
Similar content being viewed by others
Introduction
Pancreatic neuroendocrine neoplasms (PanNENs) are rare tumours and comprise approximately 1–2% of tumours in the pancreas, with a rising incidence in the last decade1,2. There is no evidence of differences in incidence between races, gender or geographical location3. To determine the neuroendocrine feature of tumours originating from the pancreas, the biomarkers chromogranin A (CgA) and synaptophysin (Syn) are used for immunohistochemical staining4.
The histopathological differentiation of tumours into well- or poorly differentiated has in recent years become an important aspect for the diagnosis, therapy choice and outcome assessment of NENs. No difference among PanNENs has been seen concerning location at head versus body/tail and tumour size. Recent publications has shown the presence of a group of tumours that are well-differentiated, but with Ki-67 index > 20%. In these studies, well-differentiated tumours were illustrated to have a better prognosis than poorly differentiated NENs G35,6,7,8. However, there are not many head to head comparisons of these two disease entities. In 2017, an update of the WHO classification was published with regards to PanNENs, where both proliferation, genetic background and differentiation are factors taken into consideration. Here, PanNENs that belong to the G3 group are defined as either well-differentiated with a Ki-67 > 20%, pancreatic neuroendocrine tumour grade 3 (PanNET G3) or as poorly differentiated pancreatic neuroendocrine carcinoma (PanNEC) with Ki-67 > 20%. The latter is also divided into small cell and large cell morphology9.
Treatments of PanNENs are different, pertaining their proliferation and differentiation. Metastatic PanNECs are traditionally treated with platinum based chemotherapy in combination with etoposide8,10,11,12 while PanNET G3 may be treated more like PanNETs G2 with surgery, peptide receptor radionucleotide therapy, everolimus, temozolomide/capecitabine or sunitinib8,13. Genetically, PanNENs differ with regards to grade and differentiation. A study has shown that PanNETs G3 commonly harbour mutations in the MEN1, ATRX and DAXX genes while PanNECs have alterations in TP53 and RB114. However, there are very few studies that address the question of how these two entities should be handled in the clinic.
Until now, very little is known about the presence of biomarkers in blood in PanNEN G3 patients15 and if there are any biomarkers that might be useful for diagnosis, prediction of therapy or prognosis. In the interest of understanding PanNENs G3 and their complexity, we need to understand their molecular composition. Detection of protein biomarkers in blood is a commonly used way to identify such molecules, and methods in the area have advanced. A recently developed technique is the Proximity Extension Assay (PEA), a sensitive and specific method, which uses a DNA polymerisation step for protein detection16.
The aim of this study was to screen for biomarkers of clinical interest in blood from patients with PanNENs G3 and compare the concentrations with healthy controls using the Olink PEA technique.
Results
Biomarker expression in blood
Patient characteristics are summarised in Table 1. In total, 79 out of 84 (94%) blood samples where included in the statistical analysis, five patient blood samples did not meet the quality control guidelines of the Olink Biosciences lab. Proteins with a lower detectability than 15% were removed, resulting in 87 out of 92 (95%) proteins included in the statistical analysis. Fifty-four (62%) proteins had significantly different blood levels when comparing patients and controls (p value < 0.05). All were proteins expressed in higher concentrations in tumour samples than healthy controls, except for FASLG. Blood samples from PanNEN patients had significantly lower concentrations of FASLG than blood samples from healthy controls (Fig. 1).
In a regression analysis to associate blood proteins with Ki-67 cut off levels of < 55% and > 55%, none of the proteins was significantly correlated to Ki-67, when adjusted for multiple testing with the Benjamini–Hochberg approach. Five proteins [Angiopoietin-1 (ANGPT1), CXC chemokine ligand 5 (CXCL5), FASLG, Natural cytotoxicity triggering receptor 1 (NCR1) and Tumour necrosis factor-like weak inducer of apoptosis (TWEAK)] correlated negatively to Ki-67 using unadjusted p values < 0.05 (Fig. 2), with FASLG being the protein of highest interest for G3 PanNENs.
There were no proteins associated to earlier death, neither when adjusted for long time and short time survivors (data not shown).
Correlation of protein levels to treatment response according to the RECIST 1.0 criteria resulted in six proteins [Chemokine (C–C motif) ligand 23 (CCL23), Chemokine (C–C motif) ligand 8 (CCL8), Chemokine (C–C motif) ligand 4 (CCL4), Class I-restricted T cell-associated molecule (CRTAM), Chemokine (C–X–C motif) ligand 1 (CXCL1) and Interleukin-18 (IL18)] that differed significantly between responders and non-responders, although only in the unadjusted assay. The proteins did not seem to differ significantly when corrected for multiple testing.
Immunohistochemical analysis of FASLG
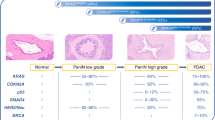
FASLG was the only protein that was present in lower levels in patients than in healthy controls. To further investigate this, immunohistochemical staining of FASLG was performed. Of the 16 tumour samples included in the immunohistochemical analysis, one patient’s tumour sample was excluded due to poor sample quality and another due to missing clinical data, resulting in a total of 14 specimens. Among these samples, 10 had poorly differentiated morphology and four had well-differentiated morphology. Seven out of 14 (50%) specimens were immunoreactive for FASLG in the tumour cells. All FASLG immunoreactive tumour specimens exhibited an organoid structure, with diffuse cytoplasmic and a more evident peripheral membranous staining pattern. Representative images of the immunostainings are shown in Fig. 3. Out of the seven specimens with immunoreactive tumour cells, three were from poorly differentiated and four from well-differentiated tumours, data presented in Table 2.
The immunostaining on the positive control (tonsil tissue) resulted in membranous and cytoplasmic immunoreactivity in a subset of non-germinal centre cells as reported in the Human Protein Atlas (https://www.proteinatlas.org) (data not shown).
Statistical analysis showed that protein expression of FASLG was associated with differentiation. In a chi-square test, well-differentiated tumours were significantly more immunoreactive for FASLG compared to poorly differentiated (p value = 0.018).
Discussion
In this study, we aimed to identify proteins in blood from PanNEN G3 patients that differed in concentration compared to healthy controls. The Olink Bioscience chips revealed that 54 of 87 proteins differed significantly and amongst these proteins were many interleukins and cytokines. All 54 proteins showed higher blood concentrations in tumour samples compared to samples from healthy controls, except for FASLG, which was reversed with lower concentrations in patients. FASLG is a protein mainly present on the membrane of cytotoxic T lymphocytes. It belongs to the tumour necrosis factor (TNF) family, and binds to its receptor FAS on different cells and tissues, including tumour cells and immune cells, i.e. both FASLG and FAS can be expressed by the cytotoxic T lymphocytes17,18. FASLG is one of the key components expressed in cytotoxic T lymphocytes19,20 and is predominantly found in activated T lymphocytes and natural killer cells (NK-cells)21. Binding of FAS to FASLG triggers an apoptotic reaction via conversion of 8 zymogen into an active form22. Both FAS and FASLG are important in cancer cell immunity as they have been seen to have both tumorigenic and tumour suppressive roles23,24. Circulating FASLG in cancer has been seen before, where a correlation with thyroid cancer recurrence was demonstrated. This suggests that FASLG may be used as a biomarker for disease recurrence25. It is unclear what the exact clinical relevance of FASLG in NENs is, but in this cohort, FASLG seems to be present in lower concentrations in blood from patients compared to controls. Interestingly, FASLG correlated negatively to Ki-67, in an unadjusted test. The higher Ki-67 index among the patients, the less concentrations of FASLG were seen in tumour samples, i.e. FASLG seems to be less present in patients with presumed poorer prognosis26.
The FAS–FASLG interaction has been shown to be a mechanism by which tumours can escape the immune system. An immune cell that encounters a cell with FASLG will undergo apoptosis. Tumour cells have adapted the expression of FASLG on their cell membranes to deactivate immune cells and hence, eliminate them from entering the tumour microenvironment27. Many studies have reported that FAS expression is aberrant in various pancreatic tumours while FASLG is seen in both normal pancreatic cells and pancreatic neoplasia. This may suggest that the aberration of FAS in tumours may be yielding the immune-evasion mechanism and that FASLG expression on tumour cells results in the inactivation of killer T cells and hence becoming resistant to FAS–FASLG mediated apoptosis28,29,30. In a study on autoimmune diabetes, in non-obese mice, soluble FASLG did not result in apoptosis of beta cells and was not a risk factor for developing autoimmune diabetes31. FASLG in sera is reported to be increased in many cancers eluding to the mechanism of FASLG as a “counterattack” protein in different tumours. In some studies it has been claimed that membrane bound FASLG induces apoptosis while soluble FASLG promotes tumour cell survival32,33. Nevertheless, too little has been studied on the differences between soluble (i.e. sera concentrations of FASLG) and tissue expressed FASLG and which of the two that may be the driving factor for tumour progression24.
In our study the immunohistochemical results showed that 7/14 of the PanNEN G3 tumours expressed FASLG in the tumour cells. A correlation between FASLG and differentiation was seen in the immunohistochemical results where well-differentiated tumours were significantly more immunoreactive for FASLG compared to poorly differentiated tumours.
The clinical relevance of FASLG protein expression in tumour cells is not fully understood. The discovery of FASLG and its mechanism in apoptosis in the immune system has given rise to the possibility of a counter-attack mechanism adapted by tumours34. Studies have shown that activated T-cells can be de-activated by FASLG expressed on tumour cells35. Overexpression of FASLG by tumour cells has also been demonstrated to decrease the number of tumour infiltrating immune cells and increase apoptosis of immune cells36,37. A study showed that ovarian carcinomas that expressed FASLG had a significantly poorer prognosis38, however contradictory results have also been reported where one study suggests that apoptosis of colon tumour cells and better prognosis, is independent of FASLG expression39. Another report with colon cancer patients showed that FASLG expression is not correlated to any clinicopathological parameters i.e. FASLG would not be an immunoprotective factor40. In our cohort, FASLG was inversely correlated to Ki-67 where patients with highly proliferative tumours had significantly lower blood concentrations of FASLG. Also, patients with well-differentiated tumours had a significantly higher expression of FASLG compared to patients with poorly differentiated tumours, 100% vs. 30% of tumours being positive. Differentiation and Ki-67 are two well-known factors in disease progression for PanNENs G3, and the association of to both these factors may therefore be of interest for further elucidation.
The occurrences of mutations in TP53 and RB1 are common in PanNECs distinguishing them from their well-differentiated counterparts14. PanNECs have been shown to harbour mutations in TP53 and RB1 while 20–26% of PanNETs harboured mutations in ATRX and DAXX, a mutation that does not occur in PanNECs41. Interestingly, p53 plays a role in the FAS–FASLG apoptosis axis. p53 can induce apoptosis through an intrinsic pathway and an extrinsic pathway where the FAS–FASLG axis belongs to the later. p53 induces FAS expression when it binds to elements in the FAS gene promoter region or by recruiting FAS receptor from the Golgi apparatus of the cell for FASLG recognition42. Furthermore, p53 can sensitise cancer cells by upregulating their FAS receptor, to the apoptotic attack by cytotoxic T cells or NK-cells. These would recognise the cancer cell by binding its FAS receptor to the FASLG expressed on their membrane and hence induce apoptosis in a cancer cell43. Hence, mutation in the TP53 gene will affect the apoptotic pathway of FAS–FASLG.
One limitation of this study is the rather small number of samples, which may have resulted in lost significance in some of the analyses, after performing multiple testing e.g. in the Ki-67 correlation test. In addition to FASLG, four other proteins were negatively correlated to Ki-67, including ANGPT1, NCR1, CXCL5, and TWEAK. These are proteins which are generally associated with angiogenesis, as well as cytotoxicity and induction of apoptosis44,45,46,47,48. Furthermore, six proteins (CCL4, CCL8, CCL23, CRTAM, CXCL1 and IL18) differed between the treatment responder and non-responder group, in an unadjusted way, but none of them were significant when corrected for multiple testing. A less prominent concentration of some proteins in patients with faster growing tumours, and difference between protein levels in treatment groups may prove to be of interest. These results may therefore warrant further investigation, preferably in larger materials.
In conclusion, we have identified proteins that may be of interest for further investigation but there was not one specific biomarker in which diagnostic or prognostic value could be placed on. The correlation of FASLG to Ki-67 and differentiation may be indicative of a possible new mechanism for FASLG in PanNENs G3 through an anti-tumour effect, although further studies are needed to confirm these findings. More studies in larger patient cohorts are needed to fully understand the FAS and FASLG pathways and their possible role in cancer progression or suppression.
Materials and methods
Patient characteristics
Blood (serum or plasma, based on availability) was collected from 42 PanNEN G3 patients from two Nordic centres and stored in − 80 °C until use. All patients had a PanNEN G3 diagnosed. All tumour specimens were positive for either one or both of the neuroendocrine biomarkers (CgA and Syn), and the median Ki-67 was 40% with missing data from two patients. The majority of patients were male (64%) and the median age was 59 years. Median overall survival was 16.5 months (range 0.6–65.3) with missing data from six patients. Blood (serum) from 42 healthy controls were matched to the best extent to patient’s characteristics regarding age and gender. We searched our pathology biobank for PanNEN G3 specimens and identified 16 samples available for immunohistochemistry. These were re-examined by an experienced pathologist (AP) with regards to differentiation.
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments, or comparable ethical standards. Informed consent was obtained from patients for participating in the registry and for biobanking of tumour tissue and blood samples. Ethical approval for the study was granted, and the need for new informed consent for this sub-study was waived for all participants by the ethical committee in Uppsala (2008/397/1, amendment approved 2017-08-24) and from the ethics committee in Norway.
Proximity extension assay
The Olink Immuno-Oncology panel (Olink ONCOLOGY III, Olink Biosciences, Uppsala, Sweden) used in this study includes 92 proteins and for each sample 1 µL of blood was used. The PEA method is based on binding of PEA probes on paired antibodies, with affinity for each other. Binding of antibody to the target protein will bring the PEA probes closer together and DNA polymerisation starts with DNA polymerase. This results in a unique new DNA sequence which is a marker for the targeted protein. Detection of targeted proteins is obtained through quantitative real-time PCR16. The PCR results are analysed as Normalized Protein eXpression (NPX) values, which are arbitrary units on a log2-scale. NPX is calculated from Ct values and data pre-processing is performed to minimize both intra- and inter-assay variation. A high NPX value corresponds to a high protein concentration and expresses relative quantification between samples but represents no absolute quantification. Assay characteristics including quality control, detection limits and measurements of assay performance and validation can be obtained via Olink’s website (https://www.olink.com). The analysis was performed by Olink Biosciences, Uppsala, Sweden.
Immunohistochemistry
Whole-section tumour samples from 16 PanNEN G3 patients with available paraffin-embedded tissue material were collected and cut into 4 µm sections. Sections were deparaffinised in xylene, hydrated in graded alcohols and blocked for endogenous peroxidase in 0.3% hydrogen peroxide diluted in 95% ethanol. For antigen retrieval, a Decloaking Chamber (Biocare Medical, Pacheco, CA) was used. Slides were immersed and boiled in citrate buffer, pH 6 (Lab Vision, Fremont, CA) for 4 min at 125 °C and then allowed to cool to 90 °C (the total program is approximately 40 min). Automated immunohistochemistry was performed essentially as previously described49,50 using an Autostainer 480 instrument (Thermo Fisher Scientific, Waltham, MA). The primary rabbit polyclonal antibody against FAS ligand (FASLG) (HPA054959, Atlas Antibodies, Stockholm, Sweden) was diluted in 1:800 UltraAb Diluent (Thermo Fisher Scientific) followed by incubation for 30 min at room temperature (RT). The slides were further incubated with the secondary reagent anti-rabbit/mouse horseradish peroxidase-conjugated UltraVision (Thermo Fisher Scientific) for 30 min at RT, and developed for 10 min using Diaminobenzidine (DAB) Quanto (Thermo Fisher Scientific) as chromogen. All incubations were followed by rinse in wash buffer (Thermo Fisher Scientific) 2 × 5 min. Slides were counterstained in Mayer’s Hematoxylin (Histolab, Gothenburg, Sweden) and cover slipped using Pertex (Histolab) as mounting medium. Human tonsil was used as a positive control.
The analysis of the FASLG immunostainings was blindly performed by one pathologist (AP), followed by three independent observers (AA, MG and ETJ). H-score was calculated for each sample. The intensity of FASLG staining was graded 0 for negative, 1 for intermediate and 2 for strong, and this was multiplied by the percentage of stained tumour cells. Consistently weak positive FASLG staining was present in the normal pancreatic islets.
Statistical analysis
Statistical analysis was performed by the Olink Biostatistics unit. Four different statistical methods were used. T-test was used to identify differences in protein concentrations between healthy controls and tumour patients. Regression analysis was conducted to search for correlation between protein concentrations and Ki-67 index in tumour patients. Survival analysis was used to investigate associations between protein concentration and survival in tumour patients. Finally, a one-way ANOVA test was performed to compare protein concentrations in different treatment response groups. All p values were adjusted for multiple testing within each test using the Benjamini–Hochberg approach51.
A chi-square test was performed to study correlations between FASLG expression and tumour differentiation (IBM SPSS statistics software, v25, USA).
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Fraenkel, M., Kim, M. K., Faggiano, A. & Valk, G. D. Epidemiology of gastroenteropancreatic neuroendocrine tumours. Best Pract. Res. Clin. Gastroenterol. 26, 691–703. https://doi.org/10.1016/j.bpg.2013.01.006 (2012).
Halfdanarson, T. R., Rubin, J., Farnell, M. B., Grant, C. S. & Petersen, G. M. Pancreatic endocrine neoplasms: epidemiology and prognosis of pancreatic endocrine tumors. Endocr. Relat. Cancer 15, 409–427. https://doi.org/10.1677/ERC-07-0221 (2008).
Yao, J. C. et al. One hundred years after “carcinoid”: epidemiology of and prognostic factors for neuroendocrine tumors in 35,825 cases in the United States. J. Clin. Oncol. 26, 3063–3072. https://doi.org/10.1200/JCO.2007.15.4377 (2008).
Oberg, K. E. Gastrointestinal neuroendocrine tumors. Ann. Oncol. 21(Suppl 7), 72–80. https://doi.org/10.1093/annonc/mdq290 (2010).
Tang, L. H., Basturk, O., Sue, J. J. & Klimstra, D. S. A practical approach to the classification of WHO Grade 3 (G3) well-differentiated neuroendocrine tumor (WD-NET) and poorly differentiated neuroendocrine carcinoma (PD-NEC) of the pancreas. Am. J. Surg. Pathol. 40, 1192–1202. https://doi.org/10.1097/PAS.0000000000000662 (2016).
Milione, M. et al. The clinicopathologic heterogeneity of grade 3 gastroenteropancreatic neuroendocrine neoplasms: morphological differentiation and proliferation identify different prognostic categories. Neuroendocrinology 104, 85–93. https://doi.org/10.1159/000445165 (2017).
Coriat, R., Walter, T., Terris, B., Couvelard, A. & Ruszniewski, P. Gastroenteropancreatic well-differentiated grade 3 neuroendocrine tumors: review and position statement. Oncologist 21, 1191–1199. https://doi.org/10.1634/theoncologist.2015-0476 (2016).
Sorbye, H., Baudin, E. & Perren, A. The problem of high-grade gastroenteropancreatic neuroendocrine neoplasms: well-differentiated neuroendocrine tumors, neuroendocrine carcinomas, and beyond. Endocrinol. Metab. Clin. N. Am. 47, 683–698. https://doi.org/10.1016/j.ecl.2018.05.001 (2018).
Kloppel, G. et al. Pancreatic neuroendocrine tumors: update on the new World Health Organization Classification. AJSP Rev. Rep. 22, 233–239. https://doi.org/10.1097/Pcr.0000000000000211 (2017).
Strosberg, J. R. et al. The NANETS consensus guidelines for the diagnosis and management of poorly differentiated (high-grade) extrapulmonary neuroendocrine carcinomas. Pancreas 39, 799–800. https://doi.org/10.1097/MPA.0b013e3181ebb56f (2010).
Janson, E. T. et al. Nordic guidelines 2014 for diagnosis and treatment of gastroenteropancreatic neuroendocrine neoplasms. Acta Oncol. 53, 1284–1297. https://doi.org/10.3109/0284186X.2014.941999 (2014).
Garcia-Carbonero, R. et al. ENETS consensus guidelines for high-grade gastroenteropancreatic neuroendocrine tumors and neuroendocrine carcinomas. Neuroendocrinology 103, 186–194. https://doi.org/10.1159/000443172 (2016).
Vinik, A. I. & Raymond, E. Pancreatic neuroendocrine tumors: approach to treatment with focus on sunitinib. Ther. Adv. Gastroenterol. 6, 396–411. https://doi.org/10.1177/1756283X13493878 (2013).
Tang, L. H. et al. Well-differentiated neuroendocrine tumors with a morphologically apparent high-grade component: a pathway distinct from poorly differentiated neuroendocrine carcinomas. Clin. Cancer. Res. 22, 1011–1017. https://doi.org/10.1158/1078-0432.CCR-15-0548 (2016).
Boons, G., Vandamme, T., Peeters, M., Van Camp, G. & Op de Beeck, K. Clinical applications of (epi)genetics in gastroenteropancreatic neuroendocrine neoplasms: moving towards liquid biopsies. Rev. Endocr. Metab. Disord. 20, 333–351. https://doi.org/10.1007/s11154-019-09508-w (2019).
Assarsson, E. et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE 9, e95192. https://doi.org/10.1371/journal.pone.0095192 (2014).
Peter, M. E. et al. The role of CD95 and CD95 ligand in cancer. Cell Death Differ. 22, 549–559. https://doi.org/10.1038/cdd.2015.3 (2015).
Itoh, N. et al. The polypeptide encoded by the cDNA for human cell surface antigen FAS can mediate apoptosis. Cell 66, 233–243 (1991).
Suda, T. et al. Expression of the FAS ligand in cells of T cell lineage. J. Immunol. 154, 3806–3813 (1995).
Kagi, D. et al. FAS and perforin pathways as major mechanisms of T cell-mediated cytotoxicity. Science 265, 528–530 (1994).
Suda, T., Takahashi, T., Golstein, P. & Nagata, S. Molecular cloning and expression of the FAS ligand, a novel member of the tumor necrosis factor family. Cell 75, 1169–1178 (1993).
Ashkenazi, A. & Dixit, V. M. Apoptosis control by death and decoy receptors. Curr. Opin. Cell Biol. 11, 255–260 (1999).
Waring, P. & Mullbacher, A. Cell death induced by the FAS/FAS ligand pathway and its role in pathology. Immunol. Cell Biol. 77, 312–317. https://doi.org/10.1046/j.1440-1711.1999.00837.x (1999).
Peter, M. E. et al. The role of CD95 and CD95 ligand in cancer. Cell Death Differ. 22, 885–886. https://doi.org/10.1038/cdd.2015.25 (2015).
Owonikoko, T. K. et al. Soluble FAS ligand as a biomarker of disease recurrence in differentiated thyroid cancer. Cancer 119, 1503–1511. https://doi.org/10.1002/cncr.27937 (2013).
Hamilton, N. A. et al. Ki-67 predicts disease recurrence and poor prognosis in pancreatic neuroendocrine neoplasms. Surgery 152, 107–113. https://doi.org/10.1016/j.surg.2012.02.011 (2012).
Liu, W. H. & Wang, D. G. Apoptosis regulating genes in neuroendocrine tumors. Histol. Histopathol. 15, 851–859. https://doi.org/10.14670/HH-15.851 (2000).
Bernstorff, W. V. et al. FAS (CD95/APO-1) and FAS ligand expression in normal pancreas and pancreatic tumors. Implications for immune privilege and immune escape. Cancer 94, 2552–2560. https://doi.org/10.1002/cncr.10549 (2002).
Kornmann, M., Ishiwata, T., Kleeff, J., Beger, H. G. & Korc, M. FAS and Fas-ligand expression in human pancreatic cancer. Ann. Surg. 231, 368–379. https://doi.org/10.1097/00000658-200003000-00010 (2000).
Kornmann, M., Ishiwata, T., Maruyama, H., Beger, H. G. & Korc, M. Coexpression of FAS and FAS-ligand in chronic pancreatitis: correlation with apoptosis. Pancreas 20, 123–128. https://doi.org/10.1097/00006676-200003000-00003 (2000).
Trivedi, P. M. et al. Soluble FAS ligand is not required for pancreatic islet inflammation or beta-cell destruction in non-obese diabetic mice. Cell Death Discov. 5, 136 (2019).
Lu, C. & Liu, K. Epigenetic regulation of PD-L1 expression and pancreatic cancer response to checkpoint immunotherapy. Transl. Cancer Res. 6, S652–S654 (2017).
Afshar-Sterle, S. et al. FAS ligand-mediated immune surveillance by T cells is essential for the control of spontaneous B cell lymphomas. Nat. Med. 20, 283–290. https://doi.org/10.1038/nm.3442 (2014).
Ryan, A. E., Shanahan, F., O’Connell, J. & Houston, A. M. Addressing the “FAS counterattack” controversy: blocking fas ligand expression suppresses tumor immune evasion of colon cancer in vivo. Cancer Res. 65, 9817–9823. https://doi.org/10.1158/0008-5472.CAN-05-1462 (2005).
Krammer, P. H. CD95’s deadly mission in the immune system. Nature 407, 789–795. https://doi.org/10.1038/35037728 (2000).
Bennett, M. W. et al. The FAS counterattack in vivo: apoptotic depletion of tumor-infiltrating lymphocytes associated with FAS ligand expression by human esophageal carcinoma. J. Immunol. 160, 5669–5675 (1998).
Okada, K. et al. Frequency of apoptosis of tumor-infiltrating lymphocytes induced by fas counterattack in human colorectal carcinoma and its correlation with prognosis. Clin. Cancer. Res. 6, 3560–3564 (2000).
Munakata, S. et al. Expressions of FAS ligand and other apoptosis-related genes and their prognostic significance in epithelial ovarian neoplasms. Br. J. Cancer 82, 1446–1452 (2000).
Houston, A. et al. FAS ligand expressed in colon cancer is not associated with increased apoptosis of tumor cells in vivo. Int. J. Cancer 107, 209–214. https://doi.org/10.1002/ijc.11392 (2003).
Pryczynicz, A., Guzinska-Ustymowicz, K. & Kemona, A. FAS/FasL expression in colorectal cancer. An immunohistochemical study. Folia Histochem. Cytobiol. 48, 425–429. https://doi.org/10.2478/v10042-010-0058-3 (2010).
Singhi, A. D. & Klimstra, D. S. Well-differentiated pancreatic neuroendocrine tumours (PanNETs) and poorly differentiated pancreatic neuroendocrine carcinomas (PanNECs): concepts, issues and a practical diagnostic approach to high-grade (G3) cases. Histopathology 72, 168–177. https://doi.org/10.1111/his.13408 (2018).
Haupt, S., Berger, M., Goldberg, Z. & Haupt, Y. Apoptosis—the p53 network. J. Cell. Sci. 116, 4077–4085. https://doi.org/10.1242/jcs.00739 (2003).
Aubrey, B. J., Kelly, G. L., Janic, A., Herold, M. J. & Strasser, A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression?. Cell Death Differ. 25, 104–113. https://doi.org/10.1038/cdd.2017.169 (2018).
Metheny-Barlow, L. J. & Li, L. Y. The enigmatic role of angiopoietin-1 in tumor angiogenesis. Cell Res. 13, 309–317. https://doi.org/10.1038/sj.cr.7290176 (2003).
Mandelboim, O. et al. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature 409, 1055–1060. https://doi.org/10.1038/35059110 (2001).
Kawamura, M. et al. CXCL5, a promoter of cell proliferation, migration and invasion, is a novel serum prognostic marker in patients with colorectal cancer. Eur. J. Cancer 48, 2244–2251. https://doi.org/10.1016/j.ejca.2011.11.032 (2012).
Chicheportiche, Y. et al. TWEAK, a new secreted ligand in the tumor necrosis factor family that weakly induces apoptosis. J. Biol. Chem. 272, 32401–32410. https://doi.org/10.1074/jbc.272.51.32401 (1997).
Wiley, S. R. et al. A novel TNF receptor family member binds TWEAK and is implicated in angiogenesis. Immunity 15, 837–846. https://doi.org/10.1016/s1074-7613(01)00232-1 (2001).
Kampf, C., Olsson, I., Ryberg, U., Sjostedt, E. & Ponten, F. Production of tissue microarrays, immunohistochemistry staining and digitalization within the human protein atlas. J. Vis. Exp. https://doi.org/10.3791/3620 (2012).
Uhlen, M. et al. Proteomics tissue-based map of the human proteome. Science 347, 1260419. https://doi.org/10.1126/science.1260419 (2015).
Chen, S. Y., Feng, Z. & Yi, X. A general introduction to adjustment for multiple comparisons. J. Thorac. Dis. 9, 1725–1729. https://doi.org/10.21037/jtd.2017.05.34 (2017).
Acknowledgements
This study was funded by the Swedish Cancer Society (CAN2017/348), the Lions Foundation for Cancer Research at the Uppsala University Hospital, the Selander and Söderberg foundations at Uppsala University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
E.T.J. conceived and designed the research study. E.T.J., S.W., and H.S. conceptualised the project and designed the study. A.A. and C.L. performed the experiments. A.A., E.T.J. and M.G. analysed the data. A.P. re-evaluated the tissue samples and scored the FASLG stainings. A.A. performed statistical analysis. A.A. drafted the manuscript. A.A., M.G., E.T.J. contributed to reviewing the manuscript. All the authors revised the manuscript and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ali, A.S., Perren, A., Lindskog, C. et al. Candidate protein biomarkers in pancreatic neuroendocrine neoplasms grade 3. Sci Rep 10, 10639 (2020). https://doi.org/10.1038/s41598-020-67670-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-67670-7
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.