Abstract
Protein malonylation, a reversible post-translational modification of lysine residues, is associated with various biological functions, such as cellular regulation and pathogenesis. In proteomics, to improve our understanding of the mechanisms of malonylation at the molecular level, the identification of malonylation sites via an efficient methodology is essential. However, experimental identification of malonylated substrates via mass spectrometry is time-consuming, labor-intensive, and expensive. Although numerous methods have been developed to predict malonylation sites in mammalian proteins, the computational resource for identifying plant malonylation sites is very limited. In this study, a hybrid model incorporating multiple convolutional neural networks (CNNs) with physicochemical properties, evolutionary information, and sequenced-based features was developed for identifying protein malonylation sites in mammals. For plant malonylation, multiple CNNs and random forests were integrated into a secondary modeling phase using a support vector machine. The independent testing has demonstrated that the mammalian and plant malonylation models can yield the area under the receiver operating characteristic curves (AUC) at 0.943 and 0.772, respectively. The proposed scheme has been implemented as a web-based tool, Kmalo (https://fdblab.csie.ncu.edu.tw/kmalo/home.html), which can help facilitate the functional investigation of protein malonylation on mammals and plants.
Similar content being viewed by others
Introduction
Lysine malonylation (Kmal), a reversible post-translational modifications (PTMs), can be identified by mass spectrometry and database searching1. Several studies have indicated that PTMs play critical roles in regulating cellular functions and are related to a lot of disease progression2,3,4,5. For instance, type 2 diabetes has been reported to be regulated by malonylation, particularly the pathways associated with fatty acid metabolism and the glucose6. Both mitochondrial and enzymes urea cycle have been shown to be regulated by protein malonylation7. Additionally, it is confirmed that lysine malonylation is a new type of histone PTM and the unusual modification of histones induces diseases, such as cancer8. In plants, malonylation has been shown to be a critical reaction in the metabolisms of xenobiotic phenolic glucosides in tobacco and Arabidopsis9. Meanwhile, previous experiments targeting lysine malonylaome in Oryza sativa and Triticum aestivum L. have demonstrated a dominate presence of malonylated proteins in the metabolic processes which include the tricarboxylic acid (TCA) cycle, carbon metabolisms, photosynthesis, and glycolysis/gluconeogenesis10,11.
A number of computational methods have been introduced to predict malonylation sites based on machine learning approaches. Xu et al. developed the first web-server, Mal-Lys, to predict Kmal sites for M. musculus proteins12. Specifically, the support vector machine (SVM) and minimum redundancy maximum relevance (mRMR) technique were adopted to develop the prediction model by considering features from the peptide fragments, including the position specific amino acid propensity, sequence order information, and physicochemical properties. Xiang et al. used pseudo amino acids as features to construct an SVM-based classifier13. Wang et al. took multiple organisms into consideration to build a novel online prediction tool, MaloPred, for the identification of malonylation sites in E.coli, M. musculus, and H. sapiens, separately, by integrating not only protein sequence information and physicochemical properties, but also evolutionarily similar features14. Taherzadeh et al. further investigated whether different species require different prediction models to maximize the accuracy15. By training the models using data from mice and testing them on other species, they found similar underlying physicochemical mechanisms between mice and humans, but not between mice and bacteria. It should be noted that their SVM-based web server, SPRINT-Mal, was the first online malonylation sites prediction tool to take into account the predicted structural properties of the proteins. Zhang et al. provided systematic comparisons of sequence-based features, physicochemical-property-based features, and evolutionary-derived features in the identification of Kmal sites for E. coli, M. musculus, and H. sapiens, respectively16. Random forest (RF), SVM, LightGBM, K-nearest neighbor (KNN), and logistic regression (LR) were adopted to generate optimal feature sets. The integration of the single-method-based models through ensemble learning was found to improve the prediction performance in independent tests. Ahmed et al. proposed a new hybrid resampling method for highly imbalanced data17. Furthermore, deep learning approach has recently been widely applied to biological sequence analysis18,19,20,21. Chen et al. first constructed an integration of the deep learning model based on long short-term memory (LSTM) with an RF classifier for the prediction of mammalian malonylation sites22. Due to the strong capability of the deep learning methodology to learn sparse representation, this methodology showed a superior performance compared to traditional machine learning model. In addition to the malonylation site, many computational methods have been developed for the prediction of various PTM sites based on protein sequences23,24,25,26,27,28.
To improve our understanding of the mechanism of malonylation, it is necessary to identify the malonylation sites accurately in advance. However, the experimental identification was mainly performed using mass spectrometry, which is time-consuming, labor-intensive, and expensive. Computational approaches could be used to effectively and accurately identify malonylation sites. Currently, existing computational approaches mostly rely on feature engineering, while deep learning is capable of excavating the underlying characteristics from a large-scale training dataset. Additionally, although the biological functions of malonylation in plants require attention, the currently existing tools have only taken into account malonylation sites in humans, mice, and bacteria. An efficient methodology for the identification of malonylation sites in more organisms would greatly improve the understanding of the mechanisms of malonylation. Therefore, the primary purpose of this study was to develop hybrid models combining CNN and machine learning algorithms for the prediction of malonylation sites in mammals and plants, respectively. Meanwhile, a user-friendly web tool, which includes an optimal classifier, was established for individual use in the identification of malonylation sites.
Results
Sequence analysis
As shown in Fig. 1, the amino acid compositions (AACs) of the malonylation and non-malonylation peptides varied between Glutamic acid (E), Glycine (G), Serine (S), and Valine (V) in mammalian proteins. On the other hand, the AACs in the plant proteins varied between Glutamic acid (E), Aspartic acid (D), Arginine (R), and Tryptophan (W). Only the composition of Glutamic acid (E) varied in both mammalian and plant proteins.
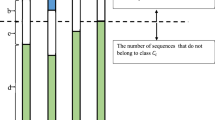
WebLogo was mainly used to analyze the frequencies of occurrence of every position around the malonylation sites29. In Supplementary Fig. S1, mouse and human, both mammals, tended to have similar patterns. More specifically, the malonylation peptides had higher frequencies in the cases of Lysine (K), Leucine (L), and Glutamic acid (E) in mammalian proteins. As shown in Fig. 2, using the TwoSampleLogo30 of malonylation and non-malonylation peptides, the enrichment of amino acids neighboring the malonylation sites across species was observed. Lysine (K) was found to be significantly enriched at multiple positions in both H. sapiens and M. musculus proteins, particularly at positions from 1 to 19 and positions 27, 28, and 29. Meanwhile, leucine (L) was found to be depleted at positions 9, 10, and 13. On the other hand, T. aestivum proteins showed a very different pattern when compared to H. sapiens and M. musculus proteins for arginine (R) enrichment at positions 10, 12, 13, 14, 21, 28, and 29 and no evidence of serine (S) depletion. The two sample logo for H. sapiens to M. musculus, H. sapiens to T. aestivum, and M. musculus to T. aestivum is shown in Fig. 3, which indicates that T. aestivum is different from the mammals. Subsequently, the data of the H. sapiens and M. musculus proteins could be combined to build a prediction model for mammals, thereby building another prediction model for T. aestivum.
Feature analysis
Pearson’s correlation coefficient (PCC) was employed to evaluate the dependency between the label and the feature in the training dataset. It should be noted that the PCC values always range from + 1 to –1. A value greater than zero denotes a positive association, whereas a value less than zero denotes a negative association. A value equal to zero indicates that there is no correlation. Therefore, the larger the absolute value of the PCC value, the stronger the correlation.
The top ten PCC values of each feature in the mammalian dataset are shown in Supplementary Table S1 and Fig. 4, respectively. The AACs of “E”, “G”, “V”, and “S” are relatively correlated to malonylation, as shown in Fig. 4. Furthermore, the attributes related to amino acids “E”, “G”, and “S” are also found in the top 5 PCC values of the pseudo-amino acid composition (PAAC) group, indicating that these amino acid may be highly correlated with malonylation in mammalian proteins. As for the position-specific features, the top 3 PCC values in the one hot encoding group were as follows: (i) whether the residue next to the central “K” from the downstream was “G”; (ii) whether the residue next to the central “K” from the downstream was also “K”; (3) whether the residue next to the central “K” from the upstream was “G”. All three attributes focus on the position next to the central “K”. The same position tendency can be observed in the AAindex group. All of the highest PCC values focused on position 18 (the residue adjacent to central “K” in the downstream position). In the position-specific scoring matrix (PSSM) group, position 13 (the 4th residue adjacent to the central “K” in the upstream position) had higher PCC values.
In terms of plant proteins, the top ten PCC values of each feature are shown in Supplementary Table S2, and Fig. 4. The listed features are very different from those of the mammalian proteins. The amino acids “R”, “D”, and “W” are only relatively correlated to the plant protein labels. The amino acid “E” is the only one that appears in both organism groups. The positional importance in the plant group is not significant compared to the mammalian group. However, the top PCC values in the AAindex group indicate that malonylation may be positively correlated with side chain hydropathy (ROSM880102) and the transfer of free energy from oct to wat (RADA880102).
Functional analysis
We used the classification system PANTHER to analyze the functional distribution of malonylated proteins31. Figure 5 shows that H. sapiens and M. musculus shared a highly common functional distribution in terms of biological processes. Both were statistically enriched in the cytosol (GO:005829), intracellular (GO:0005622), intracellular part (GO:00044424), cytoplasm (GO:0005737), and cytoplasmic part (GO:0044444). In terms of the cellular components, both H. sapiens and M. musculus were statistically enriched in the cellular metabolic process (GO:0044237). The common processes in which all three species were statistically enriched were the cytoplasm (GO:005737) and the organic substance metabolic process (GO:0071704).
Determination of the window size and the model of each feature
Supplementary Fig. S2, shows the AUC performance of each feature with a different window size of peptides. For the mammalian model, the feature AAINDEX, one hot vector, and PSSM resulted in the best AUC using CNN models with a window size of 37, 33, and 35, respectively. Feature AAC and PAAC showed their best AUC using RF models with a window size of 33. Details of the performance are shown in Table 1. Importantly, the models were only tested after the models were chosen. In other words, the test results did not play any role in the model selection.
The performance of our proposed hybrid model in mammalian proteins is shown in Table 1. After determining the best window size of each feature and the aggregation methods, we used a tenfold cross-validation method to compare the performance after the addition of more features. AAINDEX, PSSM, and One hot encoding were found to result in the best AUC. The performance of our proposed hybrid model in plant proteins is shown in Table 2. The ensemble model resulted in a great improvement.
Comparison with other existing malonylation site prediction tools in terms of predictive performance
We compared our model with other proposed computational prediction models. Table 3 and Fig. 6 demonstrates the comparative results. It should be noted that the peptides were removed from the independent dataset if the peptides were in the training dataset of the proposed computational prediction models. In total, 19,212 (683 positive sites, 18,529 negative sites) and 20,312 (1,251 positive sites, 19,061 negative sites) peptides were identified for the Kmal-sp16 H. sapiens model and LEMP22, respectively. These results indicate that the model proposed here was comparable to existing other tools.
Web interface of Kmalo
A web-based tool, Kmalo, has been developed to perform the prediction of protein malonylation sites. Users can predict malonylation sites in either mammalian or plant proteins. After submitting a protein in FASTA format via uploading a file or pasting the sequences into the tool, users will receive a job ID with which they can retrieve their results once the prediction process is finished. The results can be conveniently copied, printed directly, or downloaded in several formats, including CSV, XMSL, PDF. Snapshots of the tool’s website are shown in Supplementary Fig. S3.
Discussions and conclusion
An increasing amount of studies are currently working towards improving our understanding of the mechanism of protein lysine malonylation. The role of this post-translational modification in a wide range of cell functions has drawn the attention of many research groups. In certain biological processes, malonylation is associated with the development of disease. However, laboratory experiments for the validation of malonylation sites are often time-consuming and expensive. In this study, we proposed a machine learning-based methodology for the prediction of malonylation sites, in hopes of reducing the economic and temporal efforts required by traditional methods. To obtain potential hidden information in the sequences, we applied a deep learning-based methodology to help extract interactive information from the evolutionary information, physicochemical properties, or simply the protein fragments, which were encoded by PSSM, AAindex, and one hot vector. Then, the two commonly used features for the identification of PTM sites, AAC, and PAAC were combined with the extracted information to obtain the final features.
Some previous studies have suggested building a malonylation sites prediction model for mammals using mouse and human proteins. Taherzadeh et al. proposed three machine learning-based models for predicting malonylation sites in humans, mice, and bacteria separately15. They used mouse proteins to test the model trained using human proteins, and used human proteins to test the model trained using mouse proteins. Both resulted in comparatively similar performances. This suggested that malonylation in mouse and human proteins have similar physicochemical mechanisms, which is consistent with the feature analyses shown in Fig. 2, Fig. 3 and Supplementary Fig. S1. Therefore, we constructed a predictor for the identification of malonylation sites using both mouse and human proteins in a single model.
Recent studies have revealed that malonylation sites impact the functional processes not only in mouse and human cells, but also in plant cells10,11. In fact, this study is the first to build a computational method for the identification of malonylation sites in plant proteins. We found that a complicated deep learning-based approach could lead to an overfitting problem. This could be due to the limited amount of data. As such, we modified our framework to make it more suitable for training only a small amount of data. After a series of experiments, we discovered that assembling the prediction results from the models trained by different features resulted in a robust model. Compared to the majority votes strategy, when combined with another SVM model, the former ensemble method resulted in a better performance.
In this study, we constructed hybrid models combining CNN and machine learning algorithms, including RF and SVM, in order to predict malonylation sites in mammals and plants, respectively. The competitive performance compared to the existed tools showed that the proposed hybrid scheme indeed generated useful information from raw protein sequence data. Therefore, the framework we proposed is expected to inspire others to develop novel computational methods on the related issues.
Materials and methods
We created a useful identification system for the identification of malonylation sites, represented by a flow chart of steps, as shown in Supplementary Fig. S4,. Detailed explanations for each process described in the chart are provided in the following sections.
Dataset preparation
In this study, we collected the mammalian proteins from protein lysine modifications database (PLMD)32 and LSTM-based ensemble malonylation predictor (LEMP)22. PLMD is an online database consisting of integrated protein lysine modifications, which includes 5,013 and 4,390 validated malonylation sites from 1841 H. sapiens and 1,466 M. musculus proteins, respectively. LEMP is a newer web tool and so far contains 5,288 malonylation sites and 88,636 non-malonylation sites for the prediction of malonylation sites in mammalian proteins. On the other hand, Liu et al.10 derived plant proteins from 342 malonylation sites in 233 T. aestivum proteins.
In order to prevent potential bias, a cluster database at a high identity with tolerance (CD-HIT)33 was used to reduce the redundancy at the cutoff threshold of 40% sequence identity. Then, the proteins were segmented into a fixed-length peptide fragment with a lysine (K) located at the center, denoted as:
where R is any of the 20 amino acids, and n represents the distance between the residue and the central K. The number of residues upstream and downstream of the center remained equal. In other words, the window size of each peptide was 2n + 1. If the length of the upstream peptide or downstream peptide was less than n, the dummy residue “X” would be used to fill the lacking residues. In this study, we considered window sizes from 15 to 35 (n = 7, 8, …, 16, 17). Then, we used CD-HIT-2D on positive and negative data to eliminate redundancy. Since the mammalian peptides collected from LEMP were processed with a window size 31, UniProt34 was employed to map the peptides to determine their residues for window sizes 33 and 35. Therefore, the mammalian proteins were composed of LEMP and the processed PLMD data; the redundant peptides were removed. After processing, 80% of the data were randomly selected to form the training dataset for the construction of the prediction model of the malonylation sites in mammalian peptides, and the remaining 20% of the data were selected for an independent testing dataset. Similarly, the same steps were used to process the wheat peptides in PLMD data. We randomly separated the wheat data with a ratio of 7:3 for training and test data for the prediction of malonylation sites in plants. Related works suggest a window size around 25 is suitable for predicting malonylation sites in mammalian proteins. However, little related works have suggested that a varying window sizes for the development of a malonylation site prediction model in plants. Consequently, we considered window sizes ranging from 11 to 39, a wider range than that was considered for mammalian peptides. The experimental datasets are summarized in Table 4 and Supplementary Fig. S5,.
Features extraction
Amino acid composition (AAC) encoding calculates the frequency of the residues (20 standard amino acids and one dummy amino acid “X”) surrounding the modification sites and has been widely used in various prediction works14. Here, the site itself is not counted. As a result, each segment will be encoded as a 21-dimention vector.
Additionally, one hot encoding was used to express the sequence features. Specifically, the peptide was encoded as a two-dimensional matrix. The rows of the matrix were the amino acids in the peptide, and the columns were the 21 amino acids (with 1 dummy amino acid “X”). Every position in each row was filled with “0”, except the one that corresponds to the amino acid in the column, which was filled with “1”35 . This encoding method has been applied in many kinds of PTM site prediction19,20.
Pseudo-amino acid composition (PAAC) encoding was considered in this study. This group of descriptors was first proposed by Shen and Chou (2008)36. iFeature37 in Python was used to generate one of the encoding methods. The descriptor was used to integrate the information of the original hydrophobicity values, the original hydrophilicity values, and the original side chain masses of the 20 natural amino acids with the sequence-order information. As a result, PAAC not only considered the most adjacent residues, but also the adjacent plus λ residues. In this study, λ = 13.
The AAindex database38 collects indices of the representing physicochemical properties of amino acids. In this study, we used iFeature37 to encode each sample peptide. For each sample, the amino acid at each position was represented by 531 physicochemical properties, such that the vector of each encoding peptide was 531 × (L–1) dimensional (where L is the window size of the peptide; only the upstream and downstream were employed). Then, a feature-selection method was applied to remove the redundant properties and obtain the optimal physicochemical property sets. iFeature was used to calculate the information gain of our AAindex-based feature, which was then added to the value of information gain for each physicochemical property at each position. With the highest summed-up value of information gain, 46 physicochemical properties were selected for mammalian proteins and 30 physicochemical properties for plant proteins. After reshaping the features, a two-dimensional matrix with 46 (or 30) columns and (L–1) rows for each sample fragment was obtained.
Position-specific scoring matrix (PSSM) profiles have been used in several related works14,15,16. In this study, PSI-BLAST was performed with a default E-value cutoff in three iterations. The size of the profile was L × 20, where L represents the window size and 20 denotes the number of the standard amino acids. It should be noted that the PSSM profile of the fragment was directly used as the input of the deep learning model, without reshaping.
Hybrid model construction
The proposed hybrid models are composed of two parts. In the first part, a series of experiments were performed to develop the best prediction model for each single-feature category. Specifically, RF, SVM, and CNN were implemented for the one hot encoding features, including the AAC, PAAC, AAindex, and PSSM profiles, respectively. It should be noted that each model was trained with peptides ranging from window size 15 to 35 in order to determine the proper length. RF models were constructed using the Python package “Scikit-learn”39 , where the number of decision trees was 500. Radial basis function kernel was chosen as the default kernel function for our SVM models. The window size of the optimal model with the highest area under the receiver operating characteristic curve (AUC) score was selected as the most suitable window size. A CNN model including two rounds of the convolutional layers and max pooling layers followed by three fully connected layers was designed. At this stage, 5 single-feature models for mammalian proteins and 5 single-feature models for plant proteins were obtained. Herein, the ‘Tensorflow’ package in Python was used.
The second step involved the aggregation of the separated models trained with different features in order to make a final prediction. Three methodologies for aggregation were used: another neural network, SVM, and majority votes. For the mammalian proteins, considering the CNN model as a feature extractor and combined with the raw AAC, the feeding of the PAAC feature into another neural network obtained the best AUC. On the other hand, for the plant proteins, feeding the positive probability of each feature into a SVM model resulted in the best performance. The schemes of proposed model for mammalian and plant proteins are shown in Supplementary Fig. S6, and Supplementary Fig. S7,, respectively. The cutoff values are 0.046 and 0.195 for mammalian and plant models, respectively.
Metrics of model evaluation
The following metrics were used to evaluate the performance of our models: sensitivity (SEN), specificity (SPE), accuracy (ACC), and Matthews correlation coefficient (MCC). The detailed definitions of these metrics are given below.
where TP denotes the true positives and refers to the number of positive labels that were correctly predicted by the classifier, TN denotes the true negatives and refers to the number of negative labels that were correctly predicted by the classifier, FP denotes the false positives and refers to the number of positive labels that were incorrectly predicted by the classifier, and FN denotes the false negatives and refers to the number of negative labels that were incorrectly predicted by the classifier. Additionally, the area under the receiver operating characteristic curve (AUC) was also considered in this study. It should be noted that a receiver operating characteristic curve (ROC) is a widely used visual tool for comparing predicted performances. It is able to demonstrate the trade-off between the true positive rate (TPR) and the false positive rate (FPR) at various threshold settings. Therefore, the AUC is frequently used when evaluating the overall predictive performance of a wide range of biological systems.
Development of the web-based prediction tool
The web-based prediction tool was mainly written in HyperText Markup Language (HTML), Cascading Style Sheets (CSS), JavaScript (JS), and HyperText Preprocessor (PHP). When a user submits data, this data will be saved as a text file named with a combination of the date, time, and five random characters or numbers in an automatically created folder in the server computer. A series of feature extractions will be performed and every feature will be saved as a text file in the same folder for further use. For the same reason, the prediction result will also be saved as a text file. If the user waits on the same page after having submitted their data, they will be automatically redirected to the results page. Otherwise, the user can use the job ID, with the same name as the automatically-saved input file, to retrieve his or her results.
Data availability
The datasets used and analyzed during the current study are available from the corresponding authors on reasonable request.
References
Peng, C. et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell. Proteom. MCP 10, M111 012658. https://doi.org/10.1074/mcp.M111.012658 (2011).
Nørregaard Jensen, O. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr. Opin. Chem. Biol. 8, 33–41. https://doi.org/10.1016/j.cbpa.2003.12.009 (2004).
Wang, Y.-C., Peterson, S. E. & Loring, J. F. Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res. 24, 143. https://doi.org/10.1038/cr.2013.151 (2013).
Ahearn, I. M., Haigis, K., Bar-Sagi, D. & Philips, M. R. Regulating the regulator: post-translational modification of RAS. Nat. Rev. Mol. Cell Biol. 13, 39. https://doi.org/10.1038/nrm3255 (2011).
Gong, C. X., Liu, F., Grundke-Iqbal, I. & Iqbal, K. Post-translational modifications of tau protein in Alzheimer’s disease. J. Neural Transm. 112, 813–838. https://doi.org/10.1007/s00702-004-0221-0 (2005).
Du, Y. et al. Lysine malonylation is elevated in type 2 diabetic mouse models and enriched in metabolic associated proteins. Mol. Cell. Proteom. MCP 14, 227–236. https://doi.org/10.1074/mcp.M114.041947 (2015).
Nishida, Y. et al. SIRT5 regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol. Cell 59, 321–332. https://doi.org/10.1016/j.molcel.2015.05.022 (2015).
Xie, Z. et al. Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteom. 11, 100–107. https://doi.org/10.1074/mcp.M111.015875 (2012).
Taguchi, G. et al. Malonylation is a key reaction in the metabolism of xenobiotic phenolic glucosides in Arabidopsis and tobacco. Plant J. 63, 1031–1041. https://doi.org/10.1111/j.1365-313X.2010.04298.x (2010).
Liu, J. et al. Systematic analysis of the lysine malonylome in common wheat. BMC Genom. 19, 209. https://doi.org/10.1186/s12864-018-4535-y (2018).
Mujahid, H. et al. Malonylome analysis in developing rice (Oryza sativa) seeds suggesting that protein lysine malonylation is well-conserved and overlaps with acetylation and succinylation substantially. J. Proteom. 170, 88–98. https://doi.org/10.1016/j.jprot.2017.08.021 (2018).
Xu, Y., Ding, Y.-X., Ding, J., Wu, L.-Y. & Xue, Y. J. S. R. Prediction of lysine malonylation sites in proteins integrated sequence-based features with mRMR feature selection. Sci. Rep. 6, 38318 (2016).
Xiang, Q., Feng, K., Liao, B., Liu, Y. & Huang, G. Prediction of lysine malonylation sites based on pseudo amino acid. Comb. Chem. High Throughput Screen. 20, 622–628. https://doi.org/10.2174/1386207320666170314102647 (2017).
Wang, L.-N., Shi, S.-P., Xu, H.-D., Wen, P.-P. & Qiu, J.-D.J.B. Computational prediction of species-specific malonylation sites via enhanced characteristic strategy. Bioinformatics 33, 1457–1463 (2016).
Taherzadeh, G. et al. Predicting lysine-malonylation sites of proteins using sequence and predicted structural features. J Comput Chem 39, 1757–1763 (2018).
Zhang, Y. et al. Computational analysis and prediction of lysine malonylation sites by exploiting informative features in an integrative machine-learning framework. Brief. Bioinform. https://doi.org/10.1093/bib/bby079 (2018).
Ahmed, A., Sarkar, K., Aziz, Y. & Khan, T. Prediction of Lysine-Malonylation Sites via Sequential and Physicochemical Features. PhD Thesis (2018).
Huang, Y., He, N., Chen, Y., Chen, Z. & Li, L. BERMP: a cross-species classifier for predicting m(6)A sites by integrating a deep learning algorithm and a random forest approach. Int. J. Biol. Sci. 14, 1669–1677. https://doi.org/10.7150/ijbs.27819 (2018).
He, F. et al. In 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). 108–113.
Zhao, X. et al. General and Species-specific Lysine Acetylation Site Prediction Using a Bi-modal Deep Architecture. Vol. PP (2018).
Xie, Y. et al. DeepNitro: prediction of protein nitration and nitrosylation sites by deep learning. Genom. Proteom. Bioinform. 16, 294–306. https://doi.org/10.1016/j.gpb.2018.04.007 (2018).
Chen, Z. et al. Integration of a deep learning classifier with a random forest approach for predicting malonylation sites. Genom. Proteom. Bioinform. 16, 451–459. https://doi.org/10.1016/j.gpb.2018.08.004 (2018).
Khan, Y. D., Batool, A., Rasool, N., Khan, S. A. & Chou, K.-C. Prediction of nitrosocysteine sites using position and composition variant features. Lett. Org. Chem. 16, 283–293 (2019).
Butt, A. H. & Khan, Y. D. Prediction of S-sulfenylation sites using statistical moments based features via CHOU’S 5-step rule. Int. J. Peptide Res. Ther. https://doi.org/10.1007/s10989-019-09931-2 (2019).
Huang, K.-Y. et al. dbPTM in 2019: exploring disease association and cross-talk of post-translational modifications. Nucleic Acids Res. 47, D298–D308 (2019).
Huang, C. H. et al. UbiSite: incorporating two-layered machine learning method with substrate motifs to predict ubiquitin-conjugation site on lysines. BMC systems biology 10(Suppl 1), 6. https://doi.org/10.1186/s12918-015-0246-z (2016).
Bui, V. M. et al. SOHSite: incorporating evolutionary information and physicochemical properties to identify protein S-sulfenylation sites. BMC Genom. 17(Suppl 1), 9. https://doi.org/10.1186/s12864-015-2299-1 (2016).
Su, M. G. & Lee, T. Y. Incorporating substrate sequence motifs and spatial amino acid composition to identify kinase-specific phosphorylation sites on protein three-dimensional structures. BMC Bioinform. 14(Suppl 16), S2. https://doi.org/10.1186/1471-2105--14-S16-S2 (2013).
Crooks, G. E., Hon, G., Chandonia, J.-M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Vacic, V., Iakoucheva, L. M. & Radivojac, P. Two sample logo: a graphical representation of the differences between two sets of sequence alignments. Bioinformatics 22, 1536–1537 (2006).
Mi, H., Muruganujan, A., Casagrande, J. T. & Thomas, P. D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551 (2013).
Xu, H. et al. PLMD: An updated data resource of protein lysine modifications. J. Genet. Genom. 44, 243–250. https://doi.org/10.1016/j.jgg.2017.03.007 (2017).
Huang, Y., Niu, B., Gao, Y., Fu, L. & Li, W. CD-HIT suite: a web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682 (2010).
Consortium, U. The universal protein resource (UniProt). Nucleic Acids Res. 36, D190–D195 (2007).
Lin, C.-T. et al. Protein metal binding residue prediction based on neural networks. Int. J. Neural Syst. 15, 71–84 (2005).
Shen, H.-B. & Chou, K.-C. PseAAC: a flexible web server for generating various kinds of protein pseudo amino acid composition. Anal. Biochem. 373, 386–388. https://doi.org/10.1016/j.ab.2007.10.012 (2008).
Chen, Z. et al. iFeature: a Python package and web server for features extraction and selection from protein and peptide sequences. Bioinformatics 34, 2499–2502. https://doi.org/10.1093/bioinformatics/bty140 (2018).
Kawashima, S. et al. AAindex: amino acid index database, progress report 2008. Nucleic Acids Res. 36, D202–D205 (2007).
Pedregosa, F. et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Acknowledgements
This work is supported by Ministry of Science and Technology (108-2221-E-008-043-MY3) and the Warshel Institute for Computational Biology funding from Shenzhen City and Longgang District and by the School of Life and Health Sciences, The Chinese University of Hong Kong, Shenzhen.
Author information
Authors and Affiliations
Contributions
C.R.C., Y.P.C., and Y.L.H. carried out the data collection and curation, participated in the bioinformatics analyses, and drafted the manuscript. Y.P.C. carried out the web tool implementation. C.R.C., L.C.W., and T.Y.L. participated in the design of the study and performed the draft revision. J.T.H. and T.Y.L. conceived of the study and participated in its design and coordination. S.Y.C. helped to revise the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the articleΓÇÖs Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the articleΓÇÖs Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chung, CR., Chang, YP., Hsu, YL. et al. Incorporating hybrid models into lysine malonylation sites prediction on mammalian and plant proteins. Sci Rep 10, 10541 (2020). https://doi.org/10.1038/s41598-020-67384-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-67384-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.