Abstract
Cr-LAAO, an l-amino acid oxidase isolated from Calloselasma rhodosthoma snake venom, has been demonstrated as a potent stimulus for neutrophil activation and inflammatory mediator production. However, the mechanisms involved in Cr-LAAO induced neutrophil activation has not been well characterized. Here we investigated the mechanisms involved in Cr-LAAO-induced lipid body (also known as lipid droplet) biogenesis and eicosanoid formation in human neutrophils. Using microarray analysis, we show for the first time that Cr-LAAO plays a role in the up-regulation of the expression of genes involved in lipid signalling and metabolism. Those include different members of phospholipase A2, mostly cytosolic phospholipase A2-α (cPLA2-α); and enzymes involved in prostaglandin synthesis including cyclooxygenases 2 (COX-2), and prostaglandin E synthase (PTGES). In addition, genes involved in lipid droplet formation, including perilipin 2 and 3 (PLIN 2 and 3) and diacylglycerol acyltransferase 1 (DGAT1), were also upregulated. Furthermore, increased phosphorylation of cPLA2-α, lipid droplet biogenesis and PGE2 synthesis were observed in human neutrophils stimulated with Cr-LAAO. Treatment with cPLA2-α inhibitor (CAY10650) or DGAT-1 inhibitor (A922500) suppressed lipid droplets formation and PGE2 secretion. In conclusion, we demonstrate for the first time the effects of Cr-LAAO to regulate neutrophil lipid metabolism and signalling.
Similar content being viewed by others
Introduction
Neutrophils are the first leukocytes to migrate to the inflammatory sites in response to chemotactic factors, where they phagocytose pathogens and release lipid mediators that regulate inflammation1,2,3. Among the lipid mediators, prostaglandin E2 (PGE2) acts on blood flow, oedema, and pain4,5,6. Prostaglandins are arachidonic acid metabolites that undergo sequential action by three important enzymes, phospholipase A2 (PLA2), and cyclooxygenases (COX-1 and COX-2) and terminal prostanoid synthase7.
There are several classes of PLA2 in mammals: secreted PLA2, cytosolic PLA2 (cPLA2), calcium-independent PLA2 (iPLA2), lysosomal PLA2, and platelet activation acetylhydrolase (PAF-AH), each of which plays a role in generating eicosanoids8. cPLA2 phosphorylation, especially the α-type cPLA2 (cPLA2-α), cleaves phospholipids, generating free fatty acids, such as arachidonic acid (AA), which is oxidized by COXs9. COXs are responsible for catalyzing the oxygenation and cyclization reactions of fatty acids to form prostaglandin G2 (PGG2). COXs also present a hydroperoxidase activity that converts PGG2 to PGH2, which is then converted to PGE2 by prostaglandin E synthase (PTGES)9,10.
Lipid bodies (LBs; also known as lipid droplets) are dynamic and well regulated organelles that have their formation up-regulated in activated leukocytes and play roles during inflammation11,12. LBs are composed of lipidic organic compounds and diverse functional proteins, such as perilipines (PLINs), which are activated in metabolism, cell signalling, and inflammation12,13,14,15,16. In addition to PLINs, diglyceride acyltransferases (DGATs) are central enzymes for the synthesis of thiacylglycerol from fatty acid substrates derived from lipogenesis17. Accumulating evidence have indicated that LBs act as platforms for the synthesis of inflammatory lipid mediators by compartmentalizing the substrate AA as well as a large number of proteins involved in eicosanoid synthesis, such as PLA2s, COX, PTGES, and 5- and 15-lipoxygenases (5-LO and 15-LO)18,19,20,21,22.
Cr-LAAO, an L-amino acid oxidase (LAAO, EC 1.4.3.2) isolated from Calloselasma rhodosthoma snake (Malaysia viper) venom are flavoenzymes present at relatively high concentrations in most snake venoms. This enzyme has pharmacological effects, including haemolysis and haemorrhage, in addition to the stimulation of apoptosis, inhibition or induction of platelet aggregation, oedema formation, and activation of leukocytes. It also plays an important role in bactericidal, cytotoxic, antiparasitic, antitumor, and antiviral activities23,24,25,26,27. Pontes et al.28,29 showed that Cr-LAAO activates isolated human neutrophils leading to ROS production (superoxide anion and hydrogen peroxide), stimulation of phagocytosis and chemotaxis by p38 mitogen-activated protein kinase (p38MAPK), and activation of phosphoinositide-3 kinase (PI3K). Moreover, Paloschi et al.30 reported that Cr-LAAO can activate the NADPH oxidase complex with PKCα participation. The toxin also induces the release of myeloperoxidase (MPO), cytokines (IL-8, IL-6, and TNF-α), neutrophil extracellular traps (NETs), and lipid mediators (LTB4 and PGE2)28,29.
The biogenesis of LBs during inflammation depends on the cell type and the stimulus that the cell undergoes, since it depends on specific signalling pathways31. Since Cr-LAAO is a molecule capable of stimulating the activation of neutrophils and production of eicosanoids, we hypothesized that Cr-LAAO regulates LBs formation in neutrophils. Here we provide evidence that Cr-LAAO has major effects on human neutrophil lipid metabolism and signaling.
Results
Microarray and gene analysis expression
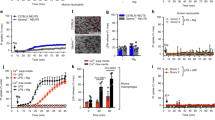
Microarray-based gene expression analysis allows the detection of approximately 22,000 genes, among which genes related to the COX pathway and lipid body were selected. The data were expressed in a heatmap as up-regulation (red) when the expression was higher in Cr-LAAO-stimulated neutrophils versus the negative control, and as down-regulation (green) when the expression was higher in the negative control than in the stimulated neutrophils. PLA2s-expressed genes were divided into cytosolic and secreted forms. The cytosolic forms were selected for this study, while the secreted forms are presented in Table S1. Group IV PLA2 (PLA2G4) subtypes (A, B, C, D), and PLA2 activating protein (PLAA) were predominantly up-regulated in all samples stimulated with Cr-LAAO. The PLA2G4 E and F subtypes, as well as group VI PLA2 (PLA2G6) did not show a consistent expression in all samples. COX-1 (PTGS1), COX-2 (PTGS2), prostaglandin reductase 2 (PTGR2), prostaglandin E synthase 1 (PTGES) and 2 (PTGES2), and prostaglandin E receptor subtype EP4 (PTGER4) genes were also up-regulated in all samples stimulated with Cr-LAAO. Genes PTGES3, PTGR1, and PTGER1, 2, and 3 did not present equivalent expression in all samples analysed. Regarding genes involved in lipid body structure, we evaluated the expression of perilipines 1 to 5, but only PLIN2 and PLIN3 were up-regulated in all samples stimulated with Cr-LAAO. In addition, DGAT1 and DGAT2 enzymes that have major roles in LB formation were also up-regulated. To confirm the microarray gene expression profile, qRT-PCR was performed for PTGS1, PTGS2, PLAA, PTGES, PLA2G4A, and PLIN2 genes. The results showed a statistically significant increase in gene expression in the LPS- and Cr-LAAO-stimulated neutrophil samples compared to the negative control for all genes tested, except for PTGS1, concurring with the up-regulated genes observed in the microarray assay (Fig. 1).
Gene expression of COX pathway. The microarray and qRT-PCR were performed with 2 × 105 human neutrophils from 3 different donors, stimulated with Cr-LAAO (50 μg/mL), LPS (1 μg/mL; positive control) or RPMI (negative control) for 1 h at 37 °C and 5% CO2. This figure was created using images from Servier Medical Art Commons Attribution 3.0 Unported License (https://smart.servier.com) (A). Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License. The microarray fold change was represented in up regulation (red) and down regulation (green) for cells stimulated with Cr-LAAO in relation to control. The genes shown were for phospholipase A2-activating protein (PLAA), group IVA (PLA2G4A), IVB (PLA2G4B), IVC (PLA2G4C), IVD (PLA2G4D), IVE (PLA2G4E), IVF (PLA2G4F) and VI (PLA2G6) phospholipase A2, prostaglandin-endoperoxide synthase 1 (PTGS1) and 2 (PTGS2), prostaglandin E synthase 1 (PTGES), 2 (PTGES2) and 3 (PTGES3), prostaglandin reductase 1 (PTGR1) and 2 (PTGR2), prostaglandin E receptor 1 (PTGER1), 2 (PTGER2), 3 (PTGER3) and 4 (PTGER4), perilipin 1–5 (PLIN1, PLIN12, PLIN3, PLIN4 and PLIN5), diacylglycerol O-acyltransferase 1 and 2 (DGAT1 and DGAT2) (B). The genes analyzed by qRT-PCR were PLAA, PLA2G4A, PTGS1, PTGS2, PTGES and PLIN2. Hemoglobin subunit beta (HBB) was used as reference gene for normalization (C). Values are mean S.E.M. from 3 donors. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 compared to negative control (Data were presented with ANOVA followed by Dunnett post-test).
Cr-LAAO induces cPLA2-α and COX accumulation in human neutrophils
Figure 2 shows the western blot analysis of isolated neutrophils incubated with RPMI (negative control), LPS (positive control), or Cr-LAAO (50 µg/mL) for 1 h. COX-1 expression was evidenced in all conditions analysed, including the negative control, proving to be constitutive. With respect to COX-2 expression, we observed a larger protein expression in samples containing LAAO and LPS when compared to RPMI, demonstrating COX-2 accumulation. Furthermore, the phosphorylated form of cPLA2 (p-cPLA2-α) has been shown to be expressed in cells stimulated with Cr-LAAO and LPS.
Protein expression of cPLA2-α and COX in neutrophils. Western blot of cPLA2-α, p-cPLA2-α, COX-1, COX-2 and β-actin using human neutrophils (1 × 107) stimulated Cr-LAAO (50 μg/mL), LPS (1 μg/mL; positive control) or RPMI (negative control) pre-treated or not with CAY10650 (cPLA2-α inhibitor, 12 nM for 30 min) or AACOCF3 (cPLA2 inhibitor, 20 µM for 30 min) for 1 h at 37 °C and 5% CO2. This figure was created using images from Servier Medical Art Commons Attribution 3.0 Unported License (https://smart.servier.com) (A). Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License. Figure representative of one experiment of three independent experiments (B). Relative immunoreactivity analysis (fold of β-actin) of the western blots from COX-1, COX-2, PTGES, PLIN2, cPLA2-α and p-cPLA2-α proteins without inhibitor (C); cPLA2-α and p-cPLA2-α proteins from neutrophils pre-treated with CAY10650 or AACOCF3 (D). Values are mean S.E.M. from 3 donors. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 compared to negative control (Data were presented with ANOVA followed by Dunnett post-test).
To confirm the action of the cPLA2-α inhibitor (CAY10650), protein expression analysis for cPLA2 and p-cPLA2-α was performed on neutrophils pretreated with 12 nM of CAY10650. Results showed that there was expression of cPLA2-α in the native form but there was no expression of the phosphorylated cPLA2-α (p-cPLA2-α) in cells pretreated with both concentrations of the inhibitor. In comparison, western blotting was also performed for cPLA2 and p-cPLA2-α in neutrophils pretreated with AACOCF3 (cPLA2-α inhibitor) at 20 µM. Results showed that as well as with CAY10650, it was observed cPLA2-α expression in the native form this was not observed the p-cPLA2-α expression in neutrophils pretreated with the inhibitor (Fig. 2) confirming that both CAY10650 and AACOCF3 inhibit cPLA2-α.
In order to show the presence of COX-2 in the cell cytoplasm, immunofluorescence experiments were conducted. Cells stimulated with Cr-LAAO or LPS had a higher fluorescence intensity after COX-2 labelling when compared to negative control (RPMI), confirming the results obtained with protein expression (Fig. 3).
Immunofluorescence of COX-2 in neutrophils. Immunofluorescence of COX-2 using human neutrophils (2 × 105) stimulated Cr-LAAO (50 μg/mL), LPS (1 μg/mL; positive control) or RPMI (negative control) for 1 h at 37 °C and 5% CO2. This figure was created using images from Servier Medical Art Commons Attribution 3.0 Unported License (https://smart.servier.com) (A). Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License. The images were collected using constant automatic gain among the samples to quantify the differences in absolute levels of fluorescence intensity different conditions in 100 × magnification oil immersion objective. Figure representative of one experiment of three independent experiments (B). Analysis of the mean fluorescence intensity of COX-2 immunofluorescence was performed using 10 cells in field of view of each condition collected impartially (C) and plotted at the fluorescence intensity per cell (D). Values are mean S.E.M. from 3 donors. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 compared to negative control (Data were presented with ANOVA followed by Dunnett post-test).
cPLA2-α activation are involved in increased lipid body formation
cPLA2-α (CAY10650) and diacylglycerol acyltransferase 1 (DGAT1) (A922500) inhibitors were used to evaluate the mechanisms of Cr-LAAO-induced LB biogenesis. LB quantification demonstrated an increase in neutrophils stimulated for 2 h with Cr-LAAO or LPS (positive control) when compared to RPMI (negative control). However, when the specific inhibitors were used, together or separately, the LB levels were reduced to a baseline similar to the negative control group, demonstrating that the cPLA2 pathway was activated and participated in LB biogenesis in these cells in a mechanism that also involves DGAT-1 dependent lipid remodelling. In addition, the LBs formation in the culture medium without fetal bovine serum (FBS) was evaluated. It was observed that even in the absence of FBS there is an increase in LBs formation in human neutrophils stimulated with Cr-LAAO, while the negative control showed baseline levels of LBs formation in human neutrophils both in presence or absence of FBS (Fig. 4).
Production of lipid bodies in neutrophils pretreated with DGAT and cPLA2 inhibitor. LBs quantification stained with neutrophils (2 × 105) pre-treated or not with inhibitors CAY10650 (cPLA2) and A922500 (DGAT) and stimulated Cr-LAAO (50 μg/mL), LPS (1 μg/mL; positive control) or RPMI (negative control) for 1 h at 37 °C and 5% CO2. This figure was created using images from Servier Medical Art Commons Attribution 3.0 Unported License (https://smart.servier.com) (A). Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License. Figure representative of one experiment of three independent experiments (B). The mean number of LBs from neutrophils stimulated in the absence of fetal bovine serum (FBS) (C) and in the presence of fetal bovine serum (D) was measured in 50 cells in the vision field of each condition impartially collected from 3 donors. Values are mean S.E.M. from 3 donors. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 compared to negative control, and #P < 0.05, ##P < 0.01, ###P < 0.001, ####P < 0.0001 compared to without pre-treatment with inhibitor (Data were presented with ANOVA followed by Tukey post-test).
Pharmacological treatment
cPLA2-α (CAY10650) and DGAT1 (A922500) inhibitors were used to characterize the mechanisms involved in the PGE2 release pathway. Cr-LAAO, similarly to LPS, stimulated PGE2 release compared to the negative control (RPMI) (Fig. 5). When neutrophils were treated with the DGAT inhibitor, we observed a reduction in PGE2 cell release after stimulation with Cr-LAAO and LPS, compared to cells without this treatment and with negative control cells. The treatment of neutrophils with a cPLA2-α inhibitor also showed reduced PGE2 in cells without treatment and in negative control cells. When both inhibitors were administered simultaneously, neutrophils incubated with Cr-LAAO or LPS showed reduced PGE2 release, demonstrating that Cr-LAAO and LPS used cPLA2 and lipid bodies to generate PGE2. We observed a greater effect of CAY10650 on inhibition of PGE2 release when used alone than when added in combination with the DAGT1 inhibitor (Fig. 5).
Prostaglandin E2 release by neutrophils pretreated with DGAT and cPLA2 inhibitor. PGE2 quantification in the supernatant of neutrophils (2 × 105) pre-treated or not with CAY10650 (cPLA2) and A922500 (DGAT) inhibitors and followed by stimulation with Cr-LAAO (50 μg/mL), LPS (1 μg/mL; positive control) or RPMI (negative control) for 1 h at 37 °C and 5% CO2. This figure was created using images from Servier Medical Art Commons Attribution 3.0 Unported License (https://smart.servier.com) (A). Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License. PGE2 concentrations were quantitated by specific EIA in supernatant collected after incubation with RPMI or LPS or Cr-LAAO. The results were expressed as pg/mL of PGE2 produced and represent the mean ± S.E.M of 3 donors (B). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 compared to negative control, and #P < 0.05, ##P < 0.01, ###P < 0.001, ####P < 0.0001 compared to without pre-treatment with inhibitor (Data were presented with ANOVA followed by Tukey post-test).
When considered as a whole, the data obtained in this study and previous results published in the literature, we proposed a mechanism of action of Cr-LAAO on human neutrophils for PGE2 production. Firstly, Cr-LAAO interacts with the cellular membrane by an unknown mechanism, leading to PKC activation, which stimulates the p38 MAPK phosphorylation, and cPLA2 activation. Activated cPLA2 cleaves membrane phospholipids to form arachidonic acid (AA). AA can be catalyzed by COX-1 or COX-2, forming PGH2, metabolized by PTGES to form PGE2 for release. Activation of cPLA2 and DGAT may lead to increased numbers of lipid bodies in neutrophils. Cr-LAAO can utilize lipid bodies content to release PGE2. Thus, in the presence of cPLA2 (CAY10650) and DGAT (A922500) inhibitors, there is a decrease in the level of PGE2 released (Fig. 6).
Suggestion of mechanism of activation of the cyclooxygenase pathway stimulated by Cr-LAAO. The representative scheme shows Cr-LAAO action on neutrophils. Firstly, Cr-LAAO interacts with the cellular membrane by an unknown mechanism (01), leading to PKC activation (02), which stimulates the p38 MAPK phosphorylation (03), and cPLA2 phosphorylation and activation (04). Activated cPLA2 cleaves membrane phospholipids to form arachidonic acid (AA) (05). AA can be catalyzed by COX-1 or COX-2 (06), forming PGH2, metabolized by PTGES (07) to form PGE2 (08) for release (09). Activation of cPLA2 (10) and DGAT (11) may lead to increased numbers of LBs in neutrophils because LB is rich in arachidonic acid (AA) for PGE2 synthesis. Cr-LAAO can utilize LBs content to release PGE2 (12). Thus, in the presence of cPLA2-α (CAY10650) (13) and DGAT (A922500) (14) inhibitors, there is a decrease in the PGE2 level released (15). This figure was created using images from Servier Medical Art Commons Attribution 3.0 Unported License (https://smart.servier.com). Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License.
Discussion
In leukocytes, cell membranes are permeable to hydrogen peroxide, which can activate leukocyte functions. In neutrophils, hydrogen peroxide regulates both the extension of pseudopodia and the orientation and direction of the cell32. Moreover, there is evidence demonstrating that hydrogen peroxide controls the recruitment of leukocytes to the inflammatory site33. Klyubin et al.34 were the first to propose that hydrogen peroxide acts as a chemotactic in leukocytes34.
LAAO from snake venoms have been described to perform several biological functions, including proinflammatory activities27. According to previous studies, Cr-LAAO activates isolated human neutrophils and stimulates ROS production (hydrogen peroxide and superoxide anion), chemotaxis, phagocytosis, cytokine release such as IL-8, IL-6, TNF-α, and LTB4, as well as the release of PGE2, NETs, and MPO, in addition to activating p38MAPK and PI3K28,29. Moreover, Costa et al.35 recently reported the inflammatory responses induced by Cr-LAAO. The authors showed that this enzyme induced neutrophil recruitment and IL-6, IL-1β, LTB4, and PGE2 release by murine macrophages. Paloschi et al.30 showed that Cr-LAAO induces NADPH oxidase complex and PKC-α activation, which contributes to the ROS production (hydrogen peroxide and superoxide anion) observed earlier.
Studies from our laboratory showed that isolated human neutrophils stimulated with Cr-LAAO at several concentrations (6, 12.5, 25, 50, and 100 µg/mL) remained alive until 12 h of incubation, using MTT and trypan blue viability methods. Based on these results, we adopted a concentration equal to 50 μg/mL, an incubation period of 1 h, and a temperature of 37 °C in a humid atmosphere of 5% CO228,29. Moreover, this period of time was defined based on PGE2 production by neutrophils under Cr-LAAO action, as previously reported by Pontes et al.29. However, the pathways responsible for this effect were not identified.
Microarray technology allows the investigation of thousands of genes simultaneously, substantially increasing the analytical capacity of molecular processes36. A study employing microarray assays with the LAAO of Ophiophagus hannah (OH-LAAO) on human breast adenocarcinoma cells (MCF-7) showed the expression of 178 genes after treatment with the enzyme, of which 27 were expressed due to the cytotoxic action of LAAO in relation to apoptosis, autophagy, cell cycle, DNA replication, oxidative stress, proteolysis, and intracellular signalling37. In another study, Guo et al.38 used microarray analysis to screen differentially expressed genes related to molecules involved in the TGF-β signalling pathway in human hepatocellular carcinoma (HepG2) cells in response to the action of LAAO from Agkistrodon blomhoffii ussensis (Akbu-LAAO)38. Pontes et al.29 have previously demonstrated PGE2 release by human neutrophils stimulated with Cr-LAAO; therefore, we decided to perform a microarray analysis on 38 genes related to COX signalling pathway and LBs, which may be involved in PGE2 production and release. These selected genes corroborate previous data obtained by Pontes et al.29 and complement the cellular activation mechanism of this enzyme on human neutrophils.
The COX pathway is initially activated by PLA2s, which perform various functions in the maintenance of homeostasis through membrane phospholipid cleavage to produce lipid mediators39. Cytosolic PLA2 (cPLA2) is a type of PLA2 that is activated upon phosphorylation (p-cPLA2)39. PLA2G4A is one of the genes encoding cPLA2-α, the cytosolic form prominently expressed in neutrophils40. cPLA2-α, translocates from the cytoplasm to the intracellular membrane in response to calcium to stimulate the release of arachidonic acid from these membranes. cPLA2-α is phosphorylated by MAPKs, which increases its catalytic activity39,41. Once activated, cPLA2-α releases arachidonic acid from the membrane phospholipids, which triggers the activity of enzymes that oxygenate arachidonic acid (AA), generating various eicosanoids42. During this process, AA is formed and subsequently metabolised by COXs, which depending on the stimulus, can be driven by COX-1 (constitutively active) or COX-2 (induced), resulting in PGE2 as the end product9,10. Studies have shown that the expression of the PLAA gene is necessary for the production of AA, leading to an increase in the production of PGE2 through the activation of cPLA2, iPLA, and COX-2, in response to stimuli such as TNF-α and LPS43,44,45.
In the present study, we observed an association between PLAA and cPLA2 gene expression and the COX-2 accumulation for PGE2 release. Stimulation of neutrophils with Cr-LAAO resulted in the expression of cPLA2 and accumulation of its phosphorylated form (p-cPLA2), demonstrating enzyme activation. Gene and protein expression for COX-1 showed that there was an expression of this constitutive protein in all groups. However, with respect to COX-2, there was a greater expression in neutrophils stimulated with Cr-LAAO and LPS when compared to the negative control, demonstrating COX-2 accumulation in the cytosol. COX-2 presence in the cytoplasm was confirmed with immunofluorescence imaging, demonstrated with an increase in the fluorescence level of the labeled enzyme in Cr-LAAO-stimulated neutrophil cytosol. In addition, the expression of PTGES, the protein responsible for the conversion of PGH2 to PGE2, was evaluated, showing the presence of this protein in neutrophils stimulated with Cr-LAAO. This is the first study to elucidate the mechanism underlying the cyclooxygenase pathway in human neutrophils stimulated with Cr-LAAO, which can be important for the comprehension and management of the local aspects observed in snakebites.
Diverse enzymes that lead to eicosanoid biosynthesis may be associated with LBs under activation conditions, including cPLA2-α46,47. cPLA2-α can remodel phospholipids of the endoplasmic reticulum and increase LBs involving deacylation/reacylation reactions48,49,50. Gubern et al.51 demonstrated that cPLA2-α inhibition reduces the levels of LBs in CHO-K1 cells. The authors also showed that knocking down cPLA2-α expression with short interfering RNA is similar to pharmacological inhibition in terms of enzyme activity and LBs biogenesis51. The protein expression results showed that there was no expression of p-cPLA2-α in the cells pretreated with CAY10650. Similarly, this result was observed when AACOCF3, another effective cPLA2-α inhibitor52,53,54,55, was used. Meanwhile I take this opportunity to bring to the knowledge of that AACOCF3 stimulated the LBs formation in neutrophils as demonstrated by Bozza et al.56.
LB biogenesis during inflammation is regulated both by the stimulus that the cell is submitted to and by specific signalling pathways. Naïve leukocytes, including neutrophils, have few LBs in their cytoplasm. However, when cells are activated, there is a significant increase in the number and size of LBs31. Nose et al.57 demonstrated that perilipin 3 (PLIN3) plays a crucial role in the formation of LBs in neutrophils for the release of PGE2. In our study, we found a positive regulation of PLIN3 and PLIN2 in human neutrophils after stimulation with Cr-LAAO. Among the proteins that comprise the LBs, PLIN2 is one of the main structural proteins found in all cell types58, which was a relevant factor in this study to examine PLIN2. Chen et al.59 showed that the increase in PLIN2 mRNA level is directly linked to increased mRNA levels of proinflammatory cytokines, such as TNF-α and IL-6. This finding corroborates the results obtained by Pontes et al.28,29 which showed that Cr-LAAO induces TNF-α and IL-6 release by neutrophils. Gene expression results obtained in the current study showed that there was an increase in PLIN2 expression in neutrophils stimulated with Cr-LAAO, supporting previous findings.
Lipid bodies are intracellular organelles that mainly store triacylglycerols (TGs) and sterol esters (SEs) as a bioenergy source. Harris et al.60 showed that DGAT1 and DGAT2 are responsible for the synthesis of almost all TGs in adipocytes, because in the absence of DGAT, adipocytes lack TGs and LBs, but in macrophages, they are not absolutely necessary for the formation of LBs. DGAT1 is proposed to possess dual topology, contributing to triacylglyceride (TAG) synthesis on both sides of the endoplasmic reticulum membrane and esterifying only the pre-formed fatty acids. Studies suggest that DGAT2 translocates to LBs and is associated with other structural proteins to synthesis TGs from endogenous and exogenous fatty acids56,61. In this study, microarray data demonstrated for the first time the signalling dependent up-regulation of DGAT1 and DGAT2 in human neutrophils. In addition, an increase in the amount of LBs present in the neutrophil cytoplasm stimulated with Cr-LAAO was verified. Treatment with a DGAT1 inhibitor (A922500) diminished the biogenesis of LBs to baseline numbers in neutrophils stimulated with Cr-LAAO. This data is in agreement with previous results that demonstrate that TAG is the main component in LB and consistent remodeling of AA pools from human neutrophil57.
Previous studies have shown that AA-rich LBs rapidly associate with phagosomes, suggesting that AA derived from LBs functions as an activator of the NADPH oxidase complex in phagosomes62,63. According to Paloschi et al.30, NADPH oxidase is activated by Cr-LAAO in human neutrophils. Additionally, as reported by Pontes et al.29, these cells phagocytose more zymosan particles under Cr-LAAO stimulation. Therefore, the present data support that LB biogenesis could be mediated by the activation of DGAT and cPLA2 in neutrophils stimulated with Cr-LAAO, and it is possible that the increase in LBs may participate in the activation of NADPH oxidase, as shown previously by Paloschi et al.30.
Other studies conducted with snake venoms and isolated toxins have demonstrated the production of LBs in macrophages. De Carvalho et al.64 investigated the venom capacity of Crotalus durissus ruruima and an isolated phospholipase A2 (CBr) to activate macrophages. The researchers focused on lipid droplet formation and the synthesis of lipid mediators involved in these effects, demonstrating PLIN2 recruitment, as well as the expression and presence of PGE2 in LBs as sites for prostatenoid synthesis. The isolated phospholipase A2 from Crotalus durissus terrificus venom, which has an anti-inflammatory effect, caused increased LB and PLIN2 recruitment in macrophages65. The increase in LB formation in macrophages from J774 cell line has also been observed upon stimulation with two basic myotoxic phospholipase A2: BaTX-I, a catalytically inactive Lys-49 variant, and BaTX-II, a catalytically active Asp-49, and of one acidic myotoxic PLA2, the BaPLA2, a catalytically active Asp-49, all of them isolated from Bothrops atrox snake venom66. MT-II, a Lys49-PLA2 from Bothrops asper venom devoid of catalytic activity, induces the formation of LBs and the synthesis of PGE2, as well as its localisation in LBs67. In addition, a study on the function of MT-III, a sPLA2 from B. asper, in the biogenesis of LBs in murine macrophages, demonstrated the expression of PLIN2, which depends on the PKC, PI3K, p38MAPK, ERK1/2, cPLA2, and iPLA2 signalling pathways, but not in the PTK, COX-1, or COX-2 pathways68. In human neutrophils, the relation between cPLA2 and LB biogenesis is unknown. In the present study we showed that Cr-LAAO could activate cPLA2 in neutrophils to induce an increase in the LBs formation in the cell. This accumulation of LBs may be directly related to the release of PGE2, a product of COX-2 activation, which has been shown to accumulate in the cytosol of activated neutrophils, together with PLIN2 recruitment. In addition to this finding, we have seen in our previous studies the presence of important signalling proteins for this process, such as PKC, PI3K and p38MAPK28,29,30.
Collectively, the data of the present study demonstrate for first time that Cr-LAAO induces the regulation of lipid signalling and metabolism in human neutrophils, leading to increased LB biogenesis and PGE2 production. Moreover, Paloschi et al.30 and Pontes et al.29 demonstrated that Cr-LAAO induces the activation of PKC and p38-MAPK, respectively; and here, we demonstrated that Cr-LAAO induces the cPLA2 activation which present an important role in the mechanisms of LBs biogenesis in human neutrophils (Fig. 6). The inhibition of LB formation by DGAT-1 inhibitor also lead to decrease the PGE2 production suggesting that LB participate in the increase of the PGE2 synthesis in Cr-LAAO-stimulated neutrophils. However, the data shown here do not rule out the possibility that LB formation and PGE2 release occur simultaneously and independently of each other. Although the mechanism through which Cr-LAAO interacts with the cellular membrane for neutrophil activation is still not fully understood, Cr-LAAO regulates lipid metabolism and signalling in human neutrophils through the expression and activation of enzymes and structural proteins, including cPLA2, DGAT-1, COX-2, PGES and PLIN2 and PLIN3, that participates in the amplification of the inflammatory process by triggering intracellular signalling cascade that culminate in lipid body formation and increased PGE2 synthesis.
Methods
Chemicals and reagents
Crystallized Calloselasma rhodostoma venom, lipopolysaccharides from Escherichia coli O111:B4 (LPS), Histopaque 1077, 3,30-diaminobenzidine tablets, hydrogen peroxide 30%, ethylene glycol-bis (β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), HEPES, Triton X-100, leupeptin, aprotinin, phenylmethylsulfonyl fluoride (PMSF), sodium orthovanadate, protease and phosphatase inhibitor cocktail, ethylenediaminetetraacetic acid disodium salt dihydrate (Na2 EDTA), bicinchoninic acid protein assay Kit (BCA), oil Red O, AACOCF3 (arachidonyl trifluoromethyl ketone), anti-mouse Fab specific-FITC and anti-β-actin were purchased from Sigma Chem. Co. (Misssouri, USA). CAY10650, A922500, prostaglandin E2 ELISA Kit, anti-COX-1 and anti-COX-2 were purchased from Cayman Chemical (Michigan, USA). Anti-cPLA2-α, anti-PTGES and anti-p-cPLA2-α were purchased from Santa Cruz Biotechnology (Texas, USA). PureLink kit, SuperScript III Reverse Transcriptase, GeneChip WT PLUS Reagent Kit and GeneChip Clariom S Array Human were purchased from Thermo Fisher Scientific (Massachusetts, USA). iTaq Universal SYBR Green Supermix were purchased from Bio-Rad (California, USA). All salts and reagents used obtained from Merck Millipore (Darmstadt, Germany) with low endotoxin or endotoxin-free grades.
Isolation and biochemical characterization of Cr-LAAO
Cr-LAAO was isolated according to Pontes et al.28. In brief, Calloselasma rhodostoma venom (30 mg) was dissolved in 1.0 mL of 0.02 M Tris–HCl buffer, pH 8.0, centrifuged at 755 g for 10 min at room temperature and the clear supernatant applied on a 70 cm × 0.9 cm Superdex G-75 column, which was previously equilibrated and then eluted with the same buffer. The fraction I showing LAAO activity was lyophilized, diluted with 0.02 M Tris–HCl buffer, pH 8.0 and then applied on a 4.0 × 0.6 cm Q-Sepharose Fast Flow column (GE Healthcare), previously equilibrated with the same buffer, using a crescent concentration NaCl gradient (0–100%). The activity of l-amino acid oxidase was performed according to Paloschi et al.30 using 50 µg/mL of Cr-LAAO added to the mixture containing horseradish peroxidase (50 µg/mL), 100 mM l-leucine, 10 mM 3′3′-diaminobenzidine in 100 mM Tris–HCl buffer (pH 7.8), incubated at 37 °C for 30 min. The reaction was stopped using a solution of 10% citric acid and the absorbance was measured on a spectrophotometer (Synergy HT, Biotek) at 490 nm.
Neutrophil isolation
Peripheral blood neutrophils were obtained from self reportedly healthy donators (18–40 years old). Informed consents were obtained at the time of the blood draw. All experiments were performed in accordance with relevant guidelines and regulations. All participants gave informed consent prior to their inclusion in the study, and the Brazilian IRB (Institutional Review Board) of the Center of Tropical Medicine Research (CEPEM, Rondônia, Brazil—Approval Number 1.739.023) approved it. In brief, blood was collected in vacuum tubes containing heparin and diluted in phosphate buffered saline (PBS, 14 mM NaCl, 2 mM NaH2PO4·H2O, 7 mM Na2HPO4·12H2O), pH 7.4, after local asepsis. For the separation of leukocytes, Histopaque 1077 was added to the tubes and then the diluted blood was carefully added over the reagent. After centrifugation at 400×g for 30 min, neutrophils were collected from the bottom of the tube, along with the erythrocytes and were transferred to another tube, according to Pontes et al.28,29. Lysis of red blood cells was performed using lysis buffer (0.15 M NH4Cl, 0.01 M KHCO3, 0.0001 M Na2 EDTA), homogenized and subjected to a temperature of − 20 °C for 5 min, and then centrifuged. Neutrophils were washed with PBS and an aliquot of isolated neutrophils was used for determining the total number of neutrophils in a Neubauer’s chamber after cell staining (1:20, v/v) with Turk solution (violet crystal 0.2% in acetic acid 30%). The purity of the isolated cell population was determined by Panotic staining of cytospin preparations and by flow cytometry analysis (FACScan). The average purity achieved by our isolation technique was 99% of the neutrophils28.
Neutrophil activation
Neutrophils isolated according to item above from 3 different donors, were suspended in a RPMI assay medium [RPMI-1640 medium supplemented with 100 mg/mL of gentamicin, 2 mM of l-glutamine and 2% of fetal bovine serum (FBS)]. Then, they were plated and incubated with RPMI (negative control), LPS (1 µg/mL; positive control) or Cr-LAAO (50 µg/mL) at 37 °C, in a humidified atmosphere (5% CO2) for 1 h.
Gene expression
Human neutrophil mRNA (5 × 106) isolated and stimulated for 1 h was extracted for microarray and RT-qPCR (Reverse transcription-quantitative polymerase chain reaction) using the PureLink kit (Thermo Fisher Scientific) according to manufacturer’s instructions. The cDNA for RT-qPCR was generated from 1 μg of total RNA using the SuperScript III Reverse Transcriptase kit (Thermo Fisher Scientific) and random primers according to the manufacturer’s instructions. Part of the extracted RNA was treated for microarray expression using the GeneChip WT PLUS Reagent Kit (Applied Biosystems, Thermo Fisher Scientific) for purification, reverse transcription, fragmentation and labeling of the samples, for application in a GeneChip Clariom S Array Human (Applied Biosystems, Thermo Fisher Scientific) according to the manufacturer's instructions. The microarray genes selected for this study are in Table S1. RT-qPCR was performed using the iTaq Universal SYBR Green Supermix kit (Bio-Rad) on Rotor-Gene Q (QIAGEN), with primers pre-designed for mRNA gene expression (DNA Express Biotechnology) analysis (Table S2). The relative fold change quantification of each gene was calculated by the 2ΔΔCt method69 using the reference gene hemoglobin subunit beta (HBB) for normalization.
Western blot
For this assay, 1 × 107 isolated and stimulated human neutrophils for 1 h according to items above pre-treated with CAY10650 (cPLA2-α inhibitor, 12 nM for 30 min70) or AACOCF3 (cPLA2 inhibitor, 20 µM for 30 min71) or the same vehicle used to dissolve the inhibitors in RPMI (control). For β-actin, cPLA2-α, p-cPLA2-α, COX-1, COX-2 and PTGES determinations, total protein extracts were prepared, resolved by 10% SDS/PAGE and transferred onto a PVDF membrane (Hybond, Amersham Pharmacia Biotech). Immunoblotting was performed using monoclonal antibodies to the referent proteins (Fig. S1). Blots were developed with 3,30-diaminobenzidine tablets and hydrogen peroxide (Sigma-Fast)29. The relative immunoreactivity bands of three independent experiments were quantified by densitometry using Image Studio Lite Ver 5.2 (LI-COR, Lincoln, Nebraska, EUA). The mean densitometry values of tested proteins were divided by the mean densitometry values of respective β-actin values to show the relative expression of each protein as a ration mean of the protein/β-actin30.
Immunofluorescence
For immunofluorescence microscopy, 2 × 105 isolated and stimulated human neutrophils as mentioned above for 1 h were seeded on 70% alcohol-washed coverslips and treated with Poly-l-Lysine (Sigma Aldrich) and placed in 24-well plates. The cells were fixed with 4% paraformaldehyde at room temperature for 15 min. Next, cells were permeabilized with acetone PA for 5 min at room temperature and the cells were incubated with the anti-COX-2 primary antibody (Cayman Chemical) overnight, followed by incubation with FITC-conjugated Fab anti-mouse secondary antibody (Sigma Aldrich) 1 h and staining with Alexa Fluor 647 phalloidin (Invitrogen), according to the manufacturer's instructions. After DAPI staining, the coverslips were mounted with Fluoromout G (Sigma Aldrich) and analyzed under a Nikon Eclipse 80i microscope with a 100 × magnification oil immersion objective. The images were collected using constant automatic gain among the samples to quantify the differences in absolute levels of fluorescence intensity different conditions. Ten fields of view of each condition were collected impartially. The acquired images were subsequently analyzed using ImageJ software (National Institutes of Health) to quantify the absolute total fluorescence intensity. The calculated fluorescence intensity of the fields of view was plotted as mean normalized intensity for the total number of cells72.
Lipid bodies quantification
The 0.3% Oil Red O (ORO) solution was prepared in isopropanol P.A., diluted in distilled water (3:2) and filtered through a paper filter to avoid precipitates. Isolated and stimulated neutrophils (2 × 105) for 2 h in the presence or absence of inhibitors CAY10650 (cPLA2-α inhibitor, 12 nM for 30 min70) and A922500 (DGAT1 inhibitor; 5 µM for 30 min73), were seeded on 70% alcohol-washed coverslips, treated with Poly-l-Lysine (Sigma Aldrich) and placed in 24-well plates. The supernatant was removed and the cells were fixed with 4% paraformaldehyde at room temperature for 15 min. The coverslips were stained with ORO (300 μL/well) for 2 min, washed with 30% isopropanol and subsequently with distilled water. The coverslips were mounted with Fluoroshild with DAPI (Sigma Aldrich) and analyzed under a Nikon Eclipse 80i microscope with a 100 × magnification oil immersion objective. LBs were quantified from fifty consecutive cells and the results were expressed of mean normalized for the total number of cells74.
Prostaglandin E2 (PGE2) assay
PGE2 concentrations were measured in the supernatant of neutrophils (2 × 105 cells) suspended in assay RPMI. Briefly, isolated and stimulated neutrophils were incubated with assay medium (negative control), LPS (1 μg/mL; positive control) or Cr-LAAO (50 μg/mL) diluted in assay medium for 1 h at 37 °C in a humidified atmosphere (5%CO2) in the presence or absence of inhibitors CAY10650 (cPLA2-α inhibitor, 12 nM for 30 min66) and A922500 (DGAT1 inhibitor; 5 µM for 30 min73). PGE2 concentrations in the supernatant were determined by a specific enzymatic immunoassay (EIA) previously described by Pontes et al.29 using a commercial kit (Cayman Chemicals).
Statistical analysis
The graphs were plotted using GraphPad Prism Ver 7.04 (GraphPad Software Incorporated). The means and standard error of the mean (S.E.M.) of all data were obtained and compared by one-way ANOVA followed by Dunnett or Tukey post-test with significance probability levels less than 0.05. Individual comparisons using t-test was also checked.
References
Granger, D. N. & Kubes, P. The microcirculation and inflammation: modulation of leukocyte-endothelial cell adhesion. J. Leukoc. Biol. 55, 662–675 (1994).
Witko-Sarsat, V., Rieu, P., Descamps-Latscha, B., Lesavre, P. & Halbwachs-Mecarelli, L. Neutrophils: molecules, functions and pathophysiological aspects. Lab. Investig. 80, 617–653 (2000).
Kolaczkowska, E. & Kubes, P. Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 13, 159–175 (2013).
Morita, I. et al. Different intracellular locations for prostaglandin endoperoxide H Synthase-1 and -2. J. Biol. Chem. 270, 10902–10908 (1995).
Harris, S. G., Padilla, J., Koumas, L., Ray, D. & Phipps, R. P. Prostaglandins as modulators of immunity. Trends Immunol. 23, 144–150 (2002).
Kawahara, K., Hohjoh, H., Inazumi, T., Tsuchiya, S. & Sugimoto, Y. Prostaglandin E2-induced inflammation: Relevance of prostaglandin E receptors. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1851, 414–421 (2015).
Rucker, D. & Dhamoon, A. S. Physiology, Thromboxane A2. StatPearls (2020).
Hurley, B. P. & McCormick, B. A. Multiple roles of phospholipase A2 during lung infection and inflammation. Infect. Immun. 76, 2259–2272 (2008).
Bapna, M. & Chauhan, L. The ambidextrous cyclooxygenase: An enduring target. Inflamm. Allergy Drug Targets 13, 387–392 (2015).
Gupta, R. A. & DuBois, R. N. Colorectal cancer prevention and treatment by inhibition of cyclooxygenase-2. Nat. Rev. Cancer 1, 11–21 (2001).
Bozza, P. T., Yu, W. & Weller, P. F. Mechanisms of formation and function of eosinophil lipid bodies: Inducible intracellular sites involved in arachidonic acid metabolism. Mem. Inst. Oswaldo Cruz 92(Suppl 2), 135–140 (1997).
Bozza, P. T., Magalhães, K. G. & Weller, P. F. Leukocyte lipid bodies: Biogenesis and functions in inflammation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1791, 540–551 (2009).
Farese, R. V. & Walther, T. C. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell 139, 855–860 (2009).
Service, R. F. ExxonMobil fuels venter’s efforts to run vehicles on algae-based oil. Science 325(379), 1–379 (2009).
Zhang, S. O. et al. Genetic and dietary regulation of lipid droplet expansion in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 107, 4640–4645 (2010).
Murphy, D. J. The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma 249, 541–585 (2012).
Irshad, Z., Dimitri, F., Christian, M. & Zammit, V. A. Diacylglycerol acyltransferase 2 links glucose utilization to fatty acid oxidation in the brown adipocytes. J. Lipid Res. 58, 15–30 (2017).
Dvorak, A. M., Weller, P. F., Harvey, S., Morgan, E. S. & Dvorak, H. F. Ultrastructural localization of prostaglandin endoperoxide synthase (cyclooxygenase) to isolated, purified fractions of guinea pig peritoneal macrophage and line 10 hepatocarcinoma cell lipid Bodies. Int. Arch. Allergy Immunol. 101, 136–142 (1993).
Bozza, P. T. et al. Eosinophil lipid bodies: Specific, inducible intracellular sites for enhanced eicosanoid formation. J. Exp. Med. 186, 909–920 (1997).
Beller, M. et al. Characterization of the drosophila lipid droplet subproteome. Mol. Cell. Proteomics 5, 1082–1094 (2006).
Cermelli, S., Guo, Y., Gross, S. P. & Welte, M. A. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 16, 1783–1795 (2006).
Hodges, B. D. M. & Wu, C. C. Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J. Lipid Res. 51, 262–273 (2010).
Du, X.-Y. & Clemetson, K. J. Snake venom l-amino acid oxidases. Toxicon 40, 659–665 (2002).
Zuliani, J. et al. Snake venom l-amino acid oxidases: Some consideration about their functional characterization. Protein Pept. Lett. 16, 908–912 (2009).
Guo, C., Liu, S., Yao, Y., Zhang, Q. & Sun, M.-Z. Past decade study of snake venom l-amino acid oxidase. Toxicon 60, 302–311 (2012).
Izidoro, L. F. M. et al. Snake venom l-amino acid oxidases: Trends in pharmacology and biochemistry. Biomed Res. Int. 2014, 1–19 (2014).
Paloschi, M. V., Pontes, A. S., Soares, A. M. & Zuliani, J. P. An update on potential molecular mechanisms underlying the actions of snake venom l-amino acid oxidases (LAAOs). Curr. Med. Chem. 25, 2520–2530 (2018).
Pontes, A. S. et al. Effect of l-amino acid oxidase from Calloselasma rhodosthoma snake venom on human neutrophils. Toxicon 80, 27–37 (2014).
Pontes, A. S. et al. p38 MAPK is involved in human neutrophil chemotaxis induced by l-amino acid oxidase from Calloselasma rhodosthoma. Toxicon 119, 106–116 (2016).
Paloschi, M. V. et al. Role of l-amino acid oxidase isolated from Calloselasma rhodostoma venom on neutrophil NADPH oxidase complex activation. Toxicon 145, 48–55 (2018).
Bozza, P. T. & Bandeira-Melo, C. Mechanisms of leukocyte lipid body formation and function in inflammation. Mem. Inst. Oswaldo Cruz 100, 113–120 (2005).
Hattori, H., Subramanian, K. K., Sakai, J. & Luo, H. R. Reactive oxygen species as signaling molecules in neutrophil chemotaxis. Commun. Integr. Biol. 3, 278–281 (2010).
Niethammer, P., Grabher, C., Look, A. T. & Mitchison, T. J. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature 459, 996–999 (2009).
Klyubin, I. V., Kirpichnikova, K. M. & Gamaley, I. A. Hydrogen peroxide-induced chemotaxis of mouse peritoneal neutrophils. Eur. J. Cell Biol. 70, 347–351 (1996).
Costa, T. R. et al. CR-LAAO, an l-amino acid oxidase from Calloselasma rhodostoma venom, as a potential tool for developing novel immunotherapeutic strategies against cancer. Sci. Rep. 7, 42673 (2017).
Mocellin, S. & Rossi, C. R. Principles of Gene Microarray Data Analysis. In Microarray Technology and Cancer Gene Profiling 19–30 (Springer, New York, 2007). https://doi.org/10.1007/978-0-387-39978-2_3.
Fung, S. Y., Lee, M. L. & Tan, N. H. Molecular mechanism of cell death induced by king cobra (Ophiophagus hannah) venom l-amino acid oxidase. Toxicon 96, 38–45 (2015).
Guo, C. et al. Akbu-LAAO exhibits potent anti-tumor activity to HepG2 cells partially through produced H2O2 via TGF-β signal pathway. Sci. Rep. 5, 1–14 (2015).
Sun, G. Y. et al. Phospholipases A2 and inflammatory responses in the central nervous system. NeuroMol. Med. 12, 133–148 (2010).
Ghosh, M., Tucker, D., Burchett, S. & Leslie, C. Properties of the Group IV phospholipase A2 family. Prog. Lipid Res. 45, 487–510 (2006).
Leslie, C. C. Cytosolic phospholipase A2: Physiological function and role in disease. J. Lipid Res. 56, 1386–1402 (2015).
Yonker, L. M. et al. Neutrophil-derived cytosolic PLA2α contributes to bacterial-induced neutrophil transepithelial migration. J. Immunol. 199, 2873–2884 (2017).
Ribardo, D. A., Crowe, S. E., Kuhl, K. R., Peterson, J. W. & Chopra, A. K. Prostaglandin levels in stimulated macrophages are controlled by phospholipase A2-activating protein and by activation of phospholipase C and D. J. Biol. Chem. 276, 5467–5475 (2001).
Chopra, A. K. et al. Molecular characterization of cDNA for phospholipase A2-activating protein. Biochim. Biophys. Acta 1444, 125–130 (1999).
Zhang, F. et al. Alteration in the activation state of new inflammation-associated targets by phospholipase A2-activating protein (PLAA). Cell. Signal. 20, 844–861 (2008).
Wooten, R. E. et al. Novel translocation responses of cytosolic phospholipase A2alpha fluorescent proteins. Biochim. Biophys. Acta 1783, 1544–1550 (2008).
Khatchadourian, A., Bourque, S. D., Richard, V. R., Titorenko, V. I. & Maysinger, D. Dynamics and regulation of lipid droplet formation in lipopolysaccharide (LPS)-stimulated microglia. Biochim. Biophys. Acta 1821, 607–617 (2012).
Pol, A., Gross, S. P. & Parton, R. G. Review: Biogenesis of the multifunctional lipid droplet: Lipids, proteins, and sites. J. Cell Biol. 204, 635–646 (2014).
Lagace, T. A. & Ridgway, N. D. The role of phospholipids in the biological activity and structure of the endoplasmic reticulum. Biochim. Biophys. Acta 1833, 2499–2510 (2013).
Wilfling, F., Haas, J. T., Walther, T. C. & Farese, R. V. Lipid droplet biogenesis. Curr. Opin. Cell Biol. 29, 39–45 (2014).
Gubern, A. et al. Group IVA phospholipase A2 is necessary for the biogenesis of lipid droplets. J. Biol. Chem. 283, 27369–27382 (2008).
Street, I. P. et al. Slow- and tight-binding inhibitors of the 85-kDa human phospholipase A2. Biochemistry 32, 5935–5940 (1993).
Platelets, I. et al. Arachidonyl trifluoromethyl ketone, potent a inhibitor of 85-kDa phospholipase 4, blocks production of arachidonate and acid by calcium. Biochemistry 269, 15619–15624 (1994).
Triggiani, M. et al. Migration of human inflammatory cells into the lung results in the remodeling of arachidonic acid into a triglyceride pool. J. Exp. Med. 182, 1181–1190 (1995).
Tripathi, T., Abdi, M. & Alizadeh, H. Role of phospholipase A2 (PLA2) inhibitors in attenuating apoptosis of the corneal epithelial cells and mitigation of Acanthamoeba keratitis. Exp. Eye Res. 113, 182–191 (2013).
Bozza, P. T. & Weller, P. F. Arachidonyl trifluoromethyl ketone induces lipid body formation in leukocytes. Prostaglandins Leukot. Essent. Fat. Acids 64, 227–230 (2001).
Nose, F. et al. Crucial role of perilipin-3 (TIP47) in formation of lipid droplets and PGE2 production in HL-60-derived neutrophils. PLoS ONE 8, e71542 (2013).
Robenek, H. et al. Lipid droplets gain pat family proteins by interaction with specialized plasma membrane domains. J. Biol. Chem. 280, 26330–26338 (2005).
Chen, F. L. et al. Adipophilin affects the expression of TNF-α, MCP-1, and IL-6 in THP-1 macrophages. Mol. Cell. Biochem. 337, 193–199 (2010).
Harris, C. A. et al. DGAT enzymes are required for triacylglycerol synthesis and lipid droplets in adipocytes. J. Lipid Res. 52, 657–667 (2011).
Bhatt-Wessel, B., Jordan, T. W., Miller, J. H. & Peng, L. Role of DGAT enzymes in triacylglycerol metabolism. Arch. Biochem. Biophys. 655, 1–11 (2018).
Berthier, S. et al. Changing the conformation state of cytochrome b 558 initiates NADPH oxidase activation. J. Biol. Chem. 278, 25499–25508 (2003).
van Manen, H.-J., Kraan, Y. M., Roos, D. & Otto, C. Single-cell Raman and fluorescence microscopy reveal the association of lipid bodies with phagosomes in leukocytes. Proc. Natl. Acad. Sci. 102, 10159–10164 (2005).
de Carvalho, A. E. Z. et al. Crotalus durissus ruruima snake venom and a phospholipase A2 isolated from this venom elicit macrophages to form lipid droplets and synthesize inflammatory lipid mediators. J. Immunol. Res. 2019, 2745286 (2019).
Giannotti, K. C. et al. A snake venom group IIA PLA2 with immunomodulatory activity induces formation of lipid droplets containing 15-d-PGJ2 in macrophages. Sci. Rep. 7, 4098 (2017).
Furtado, J. L. et al. Activation of J77A1 macrophages by three phospholipases A2 isolated from Bothrops atrox snake venom. Biomed. Res. Int. 2014, 683123 (2014).
Giannotti, K. C. et al. A Lys49 phospholipase A2, isolated from Bothrops asper snake venom, induces lipid droplet formation in macrophages which depends on distinct signaling pathways and the C-terminal region. Biomed. Res. Int. 2013, 807982 (2013).
Leiguez, E. et al. A group IIA-secreted phospholipase A2 from snake venom induces lipid body formation in macrophages: The roles of intracellular phospholipases A2 and distinct signaling pathways. J. Leukoc. Biol. 90, 155–166 (2011).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 (2008).
Drews, A. et al. 1-(5-Carboxyindol-1-yl)propan-2-one inhibitors of human cytosolic phospholipase A2 α with reduced lipophilicity: Synthesis, biological activity, metabolic stability, solubility, bioavailability, and topical in vivo activity. J. Med. Chem. 53, 5165–5178 (2010).
Fonteh, A. N. Differential effects of arachidonoyl trifluoromethyl ketone on arachidonic acid release and lipid mediator biosynthesis by human neutrophils. Eur. J. Biochem. 269, 3760–3770 (2002).
Allen, L.-A.H. Immunofluorescence and confocal microscopy of neutrophils. Neutrophil Methods Protoc https://doi.org/10.1007/978-1-62703-845-4_16 (2014).
Zhao, G. et al. Validation of diacyl glycerolacyltransferase I as a novel target for the treatment of obesity and dyslipidemia using a potent and selective small molecule inhibitor. J. Med. Chem. 51, 380–383 (2008).
Melo, R. C. N. et al. Lipid bodies in inflammatory cells. J. Histochem. Cytochem. 59, 540–556 (2011).
Acknowledgements
Mauro Paloschi thanks his family and friends for their support and encouragement. The authors express their gratitude to the Dr. Gustavo Menezes from UFMG for the discussion and help with immunofluorescence experiments; Dr. Andreimar M. Soares for provide the Cr-LAAO from Calloselasma rhodostoma venom from CEBIO—FIOCRUZ-RO; the team of the Laboratório de Imunofarmacologia do Instituto Oswaldo Cruz—FIOCRUZ for the contribution with this work, the team of the Laboratório de Imunologia Celular Aplicada à Saúde da Fiocruz-RO who rest and collaborate with this project at the program Mobility Academic from FIOCRUZ. The authors express their gratitude to Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Fundação de Amparo à Pesquisa do Estado de Rondônia (FAPERO) for the financial support. The authors thank the Program for Technological Development in Tools for Health-PDTIS-FIOCRUZ, Laboratório de Imunofarmacologia, Instituto Oswaldo Cruz—Instituto Oswaldo Cruz (IOC), and Empresa Brasileira de Pesquisa Agropecuária—Rondônia (EMBRAPA-RO) for their facilities use. This study was supported by Grants (482562/2010-2 and 479316-2013-6) from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). Juliana Pavan Zuliani was a recipient of productivity Grant 306672/2014-6 and 306197/2017-0 from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Mauro Valentino Paloschi was the beneficiary of CAPES by doctorate fellowship. "This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001".
Author information
Authors and Affiliations
Contributions
J.P.Z. and M.V.P. designed the study; M.V.P., J.A.L., C.N.B., M.D.S.S., J.R.E., A.S.P., S.S.S., C.M.A.R., N.M.N., A.A.F.F., W.L.P. and K.P.F. performed the experiments; G.E.M.F. performed and supervised the microarray procedures; J.P.Z., M.V.P., P.T.B. and G.E.M.F. collected and analyzed the data; J.P.Z., P.T.B. and G.E.M.F. provided reagents; J.P.Z., P.T.B. and M.V.P. wrote the manuscript. All the authors discussed the results and implications and commented on the manuscript at all stages.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Paloschi, M.V., Lopes, J.A., Boeno, C.N. et al. Cytosolic phospholipase A2-α participates in lipid body formation and PGE2 release in human neutrophils stimulated with an l-amino acid oxidase from Calloselasma rhodostoma venom. Sci Rep 10, 10976 (2020). https://doi.org/10.1038/s41598-020-67345-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-67345-3
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.