Abstract
Meningiomas are among the most common primary tumors of the central nervous system (CNS) and originate from the arachnoid or meningothelial cells of the meninges. Surgery is the first option of treatment, but depending on the location and invasion patterns, complete removal of the tumor is not always feasible. Reports indicate many differences in meningiomas from male versus female patients; for example, incidence is higher in females, whereas males usually develop the malignant and more aggressive type. With this as motivation, we used shotgun proteomics to compare the proteomic profile of grade I meningioma biopsies of male and female patients. Our results listed several differentially abundant proteins between the two groups; some examples are S100-A4 and proteins involved in RNA splicing events. For males, we identified enriched pathways for cell-matrix organization and for females, pathways related to RNA transporting and processing. We believe our findings contribute to the understanding of the molecular differences between grade I meningiomas of female and male patients.
Similar content being viewed by others
Introduction
Meningioma is a high incidence tumor that typically emerges at the arachnoid cap or meningothelial cells of the meninges1, commonly from intracranial, intraspinal, or orbital locations, and are usually benign, slow-growing tumors2. Resonance imaging and molecular markers are frequently used for preliminary diagnosis; yet, surgical removal of the tumor is necessary for histological diagnostic confirmation and improved life quality1,2. When surgery for total removal of the tumor is not feasible, adjuvant radiation may be used1.
The World Health Organization (WHO) classifies meningiomas according to their histopathological characteristics, mitotic count, and brain invasion pattern in (i) grade I, also known as benign meningiomas (BMs, about 80% of termed cases); (ii) grade II, or atypical meningiomas (AMs, 17% of termed cases); and (iii) grade III, the malignant meningiomas (MMs, 3% of termed cases)3. Although this classification is valid in terms of prognosis, it lacks information about tumor aggressiveness and recurrence rates2. Controversially, most recurrent meningiomas correspond to BMs; their metabolic phenotype indicates an aggressive metabolism, resembling that of AM4.
Female patients present approximately double the incidence of meningiomas compared to men1. Interestingly, the main risk factor for meningiomas is related to hormonal changes as these tumors present hormones receptors (i.e., progesterone and estrogen)5,6. Moreover, association between meningiomas and breast cancer, mainly due to similar hormonal signaling and genetic predisposition, has also been reported5. The fact that women diagnosed with breast cancer are more likely to develop meningioma may also justify the higher incidence of this tumor in females7,8. In general, women usually develop the benign form while males develop the malignant type of meningioma, the aggressive grade III6. Besides gender, other risk factors associated with this disease include exposure to ionizing radiation9, family history5, and porting specific mutations such as the neurofibromatosis type 2 (NF2), characterized by a mutation on chromosome 22q1210 or in genes involved in the sonic hedgehog and phosphatidylinositol-3 kinase (PI3K)/AKT/mTOR pathways, i.e., AKT1, PIK3CA, and SMARCE1 1.
There are few proteomic studies on meningiomas. Sharma et al. characterized the serum proteome of patients with different degrees of meningiomas11 and identify differential regulation of important physiological pathways related to coagulation, lipid metabolism and cell growth. The results posed apolipoprotein E and A-I and hemopexin as potential predictors for meningiomas. The proteins vimentin, alpha-2-macroglobulin, apolipoprotein B and A-I, and antithrombin III presented differential abundancy according to the degree of meningioma and may function as predictive markers that complement the histological diagnosis. In another report, Papaioannou et al.12 evaluated the alteration of meningioma proteins of different degrees; 61 samples were analyzed and a total of 3,042 proteins were identified using a Q-Exactive Plus mass spectrometer (Thermo, San Jose). Dunn et al.13 also evaluated the proteomic and phosphoproteomic profile of meningiomas (grade I, II, and III) versus healthy meninges and identified 3,888 proteins and 3,074 phosphoproteins.
To date, no proteomic report has addressed the molecular mechanisms related to gender-specific tumorigenesis – much is still hypothesized14. With this as motivation, we employed shotgun proteomics to compare the proteomic profile of grade I meningioma biopsies of male versus female patients. Our results showed enriched pathways involved in cell-matrix organization for male samples and RNA splicing and processing for female patients, thus posing as a contribution to the understanding of the molecular differences between genders.
Results
Exploratory data analysis
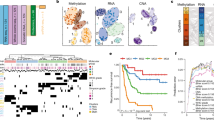
We performed a Clustergram (hierarchical clustering + heatmap), Principal Component Analysis (PCA), and a t-distribution Distributed Stochastic Neighbor Embedding (t-SNE) analyses on our dataset. This is accomplished by encoding data from the technical replicates of each biological replicate into a vector whose dimensions hold normalized quantitation values of the identified proteins. PCA and t-SNE are commonly used for dimensional reduction to provide a visual interpretation of the dataset. PCA projects vectors to a reduced set of orthogonal axis linked to maximum variance. t-SNE uses the t-distribution to group objects in the higher dimensional space and then relies on Kullback–Leibler divergence minimize the projections to a corresponding distribution at a lower dimension; t-SNE is know for producing more effective visualizations. The Clustergram was achieved using PatternLab’s Clustergram module with the Feature Stringency Selection parameter set to 0.9515. The PCA and t-SNE were generated using DiagnoProt 2.0; in brief this software clusters spectra and performs downstream analyses on these vectorized clusters and thus is search-engine unbiased16. These results are all provided in Supplementary File 1.
Quality assessment of technical replicate reproducibility
The study includes six biological replicates for each condition; each biological replicate served for generating two technical replicates. We then used RawVegetable (Freely available at: http://www.patternlabforproteomics.org/rawvegetable/) to certify that the reproducibility of all 12 technical replicate pairs achieve a reproducibility RawVegetable k-score <0.1. The scatterplots of all analyses are provided as Supplementary File 2.
Unique proteins identified in female and male meningiomas
We performed our proteomic analysis following the PatternLab for proteomics protocol15. A summary of our identification numbers is reported in Table 1, and the complete report on Supplementary file 3.
We used PatternLab’s “Venn Diagram” to pinpoint proteins identified in two or more biological replicates of each biological condition; the results indicated 235 and 194 exclusive to the female and male groups, respectively (Fig. 1; complete information in Supplementary file 3). Table 2 shortlists proteins distinct to each gender that is related to cancer, cell growth or cell cycle, as per DAVID platform (complete information in Supplementary file 4).
Differentially abundant proteins identified in female and male meningiomas
We used PatternLab’s TFold analysis to assess the differences in protein abundances identified between the groups evaluated (Supplementary file 3). Figure 2 shows the graph corresponding to protein distribution under the two conditions studied. A total of 37 proteins were identified as differently abundant (blue dots), described in Table 3; of those, 11 were upregulated in the male group and 26 in the female group.
Differently abundant proteins identified in the female and male groups. The y- and x- axis are related to the fold change and p-value. Red dots are proteins that do not satisfy our fold-change cutoff and the established False Discovery Rate (FDR). Green dots are proteins whose abundancy fold change satisfy the criteria but not the FDR. The 37 blue dots represent proteins that satisfy both criteria.
Enriched pathways in female and male meningiomas
The proteins identified to only one gender or presenting differential abundancy were selected to search for enriched pathways using the Reactome software. Tables 4 and 5 list the enriched pathways together with their corresponding identified proteins for female and male meningioma samples, respectively. For the female group, most are related to RNA processing and transport, whereas for the male group, we highlight the extracellular matrix (ECM)-related pathways. The complete information of the Reactome analysis is available in Supplementary file 5.
Discussion
Protein alterations in male meningioma
Apolipoprotein L1
The human apolipoprotein L1 (APOL1) was uniquely identified in male grade I meningiomas (Supplementaty file 3). Apolipoproteins are glycoproteins that bind lipids to form and transport lipoproteins, acting in the homeostasis of lipid and lipoprotein metabolism in the liver17. APOL1 is a minor component of high density lipoprotein (HDL), but despite its main role in lipid transport and metabolism, recent studies have shown a role of APOL1 in apoptosis, innate immunity, and autophagy, all processes related to cancer, due to its similarity to Bcl-2 family proteins18.
Specifically, in meningiomas a great variety of apolipoproteins have been identified with differential abundance in a study that used serum quantitative proteomics to compare WHO grades I-III meningiomas11. Some examples are the APOE and A-I, considered potential predictors for meningiomas. APOB and A-I were differentially abundant in malignant grades of meningioma, posing as a potential biomarker for the disease. Also, apolipoproteins A-I, A-II, A-IV, B-100, C-II and E were altered in atypical and anaplastic meningiomas. In reference to other studies, APOE and APOA-I were also found with differential abundance in WHO grade I-III meningiomas19,20
Extracellular matrix (ECM) organization
Tumor cells show changes in cell-cell and cell-ECM adhesion processes, thus contributing to cancer progression from loss of contact with their original tissues21. Our data indicate that in male meningioma there are mostly changes in cell-ECM adhesion, while in females, there is an enrichment for RNA processing-related processes (Tables 5 and 6).
ECM is responsible for cell-cell communication, adhesion and cell proliferation and is highly modified by remodeling and degradation processes, with its regulation or dysregulation has direct effects on cell differentiation and adhesion22. The composition of ECM varies according to the needs of the tissue, which is remodeled according to biochemical signals23. The proteins decorin, integrin alpha-M, fibronectin, microfibrillar-associated protein 5, laminin subunit gamma-1, among others, participate in the organization of ECM and were identified exclusively or upregulated in male meningioma (Supplementary file 3). Integrins are responsible for anchoring cells to the ECM, and fibronectins connect integrins to other ECM proteins24. Our study identified integrin beta 3 precursor (ITGB3), alpha V integrin (ITGAV) and alpha M integrin (ITGAM) only in male meningioma samples (Table 2); these proteins are associated with meningiomas tumorigenesis 7,8. Integrins function as transmembrane receptors mediating ECM adhesion and other cellular processes, such as cell migration and angiogenesis 7. Reports indicate the ECM constitution to be altered in tumor cells, and an increase in fibronectin secretion to be noted; here, we identified this upregulation pattern in the male group24. In a recent study, proteins involved in ECM formation and were found differentially abundant in grade I meningiomas12.
Tumor cells are suggested to infiltrate the ECM and promote biochemical changes that increase metastatic spread25, and perhaps these events are related to the greater aggressiveness of male meningiomas.
Neutrophil degranulation
Proteins involved in neutrophil degranulation, such as ARSA, GCA, FUCA1, PRTN3, ELANE, MNDA, and LCN2, were identified uniquely or with differential abundance in male meningiomas (Supplementary file 3). Neutrophils are the first defense line cell population to reach the site of inflammation and are capable of binding to tumor cells, providing greater angiogenesis, cell matrix remodeling and tumor progression26. They secrete three types of granulocytes that modulate cell function, called primary, secondary and tertiary27. Primary factors mainly secrete myeloperoxidases, proteolytic proteins, and bactericides; secondary and tertiary factors secrete proteins that interact and degrade the cellular matrix, such as metalloproteinase-9 (MMP-9)26,27.
Papaioannou and collaborators reported the enrichment of the neutrophil degranulation pathway in aggressive meningiomas, based on the time of recurrence12. Templeton and collaborators showed that a neutrophil/lymphocyte ratio of more than 4 in the peripheral blood of patients with different types and stages of cancer is correlated with a poor prognosis and survival28. However, in the tumor microenvironment, it is not known if the presence of neutrophils is related to the worsening of prognosis. Shen and collaborators evaluated the solid tumor neutrophil population of 3,946 patients with different cancers and concluded that increased intratumoral neutrophil levels are associated with decreased patient survival29.
CCN family member 3 (NOV)
CCN family member 3 (NOV) was identified uniquely in male meningiomas (Supplementary file 3). CCN family proteins are involved in cell adhesion, proliferation, and differentiation30. Under normal biological conditions, NOV is related to neuronal and muscular differentiation31. However, given pathological stimuli, it is suggested that the protein is involved in tumorigenic events31. Thibout and collaborators showed that NOV levels are significantly higher in malignant adrenocortical tumors when compared to benign tumors, revealing the participation of NOV in the aggressiveness and worse prognosis of the disease32. The role of NOV in tumorigenesis was investigated by Dankner and collaborators; they analyzed samples from 1,500 patients with primary prostate cancer and concluded that NOV protein abundance correlates with bone metastasis events33. These data corroborate the findings of Chen and collaborators, that demonstrated that NOV silencing decreases tumor growth34. Positively regulated NOV is related to a poor prognosis of cervical cancer35 and metastatic primary musculoskeletal tumors36.
However, the effects of NOV are indicated to be beneficial in cases of glioblastomas, as in vitro assays have shown that the protein has antiproliferative effects and prevents the S/G2 transition in the cell cycle37. Fukunaga-Kalabis and collaborators. indicated that NOV prevented melanoma cell invasion and that reduced NOV levels may facilitate the invasive character of melanoma cells38. In addition, the protein had antiproliferative effects in gliomas39 and Wilms tumors40,41. These findings suggest NOV may decrease cell proliferation and increase apoptotic events. NOV interacts with different proteins and participates in different cellular events, depending on the type of cell in which it is located, and this may be the reason for its dual role in tumorigenesis41.
NOV interacts with the Notch-1 transmembrane receptor and thus promote downstream effects on the Notch signaling pathway42 that is linked to embryonic cell development, coordinating cell differentiation, cell proliferation, and apoptosis. Liao and collaborators showed that Notch-1 silencing inhibits cell growth and promotes apoptosis in HT29 cells, a model colorectal carcinoma cell culture43. However, data from Sin and collaborators showed that NOV reduces cell growth by regulating actin cytoskeleton reorganization and increasing intercellular adhesion in breast cancer cells44. These studies show different mechanisms of NOV protein actions, revealing a close relationship with tumor development.
S100-A4
The S100 protein family is composed of proteins that play key roles in regulating cell events, such as cell cycle progression, and are described at multiple stages of tumorigenesis45,46. One of the CCN3 interaction partners is the calcium-binding protein S100-A447, which in our study was identified with a higher abundancy in male meningioma (Table 4). S100-A4 is involved in cell cycle control, angiogenesis, motility, and cell adhesion, and therefore, related to tumor progression and metastasis48. Albeit S100-A4 having a role in tumorigenesis and metastasis49, this protein is also found in normal human cells, such as macrophages, fibroblasts, granulocytes, and T lymphocytes49. The interaction between S100-A4 and p53 promotes p53 degradation50, an important tumor suppressor. Loss of protein function prevents the cell cycle from progressing moving on to the next phase, which may result in the development of tumors51,52,53, such as brain tumors54, breast55, colon56 and lung57 carcinomas. S100-A4 poses as a strong biomarker candidate to indicate early tumor detection and also offers possible evidence of metastatic events of these tissues, being identified in the breast58, brain59, and liver60 cancer metastasis.
Metastasis events related to the action of S100-A4 linked to its intra and extracellular functions61. In the extracellular context, the protein may be related to cytokine recruitment62 and inflammation, increased secretion of growth factors in the tumor environment, and increased angiogenesis63,64. In addition, the tumor cells themselves can secrete S100A4 and stimulate angiogenesis, favoring nutrient and oxygen exchange, as well as the disposal of metabolites and carbon dioxide49. In the intracellular context, S100-A4 covalently binds to actins, non-muscular myosin IIA, and tropomyosin and together alter cell migration and adhesion48. The reduction in S100A4 levels is reported to correlate with the decrease in epithelial-mesenchymal transition (EMT) events65. EMT is a process in which an epithelial cell assumes a mesenchymal phenotype, and then presents greater migratory capacity and invasiveness, and also presents apoptosis resistance. Non-muscular myosin IIA protein regulates EMT events, and may be related to increased EMT events, thus potentially contributing to metastatic events65,66.
The alterations in the proteomic profile of male meningiomas when compared to the female group may be related to the higher cancer aggressiveness and poor prognosis of male patients.
Protein alterations in female meningioma
RNA splicing and transport
We identified RNA splicing and transport-related proteins as uniquely identified or differentially abundant in the female group (Supplementary file 3). RNA splicing is a very important mechanism for increasing proteome diversity, and its proper control is required to maintain cellular processes; several studies suggest that aberrant splicing of some genes may be related to cancer67. We have identified differentially abundant protein interactions related to RNA processing, such as DDX42, BCAS2, DDX3×, PRPF6, PRPF31 (Supplementary file 3). In a study using quantitative proteomics to compare WHO grades I, II and III meningiomas, proteins involved in RNA splicing/processing were found differentially abundant in malignant grades II-III meningiomas, when compared to benign grade I12.
Reports indicate a close relationship between the occurrence of meningiomas and stimulation by hormones, especially estrogens68. Estrogen receptors (ERs) are associated with cases of meningiomas and breast cancer, but there are more studies in cases of breast tumors69. In general, ER-alpha is described as a transcription factor that increases cell growth and proliferation, while ER-beta performs antiproliferative functions70. Dago and collaborators performed RNA sequencing to evaluate the difference in cell transcripts expressing only ER-alpha or ER-alpha and ER-beta in response to estradiol hormone, which is a natural estrogen71; the results indicated that both forms induced RNA splicing in response to estradiol, and that ER-beta positive cells exhibited about twice as many mRNA splicing events as receptor negative cells.
As aforementioned, meningiomas of female patients have more progesterone receptors (PRs) than those of male patients72. Other results indicate that the presence of PR is higher in benign tumors and that PR status is inversely related to mitotic intensity and degree of meningiomas73. The same was reported for breast cancer, where decreased PR is related to a worse prognosis. Loss of PR can be related to several factors, such as PR promoter hypermethylation and PR pre-mRNA alternative splicing events, which can generate receptor variants with different domains, and thus modify how the cells respond to progesterone, contributing to the growth and abnormal proliferation of these cells73.
The proliferative cell nuclear antigen (PCNA)
The protein called proliferative cell nuclear antigen (PCNA) has multiple functions, including DNA repair and replication, chromatin remodeling and cell cycle regulation74. In our study, the protein was identified as differentially abundant in female compared to male meningioma. Studies have found that ER-alpha is associated with PCNA, and Norton-Schultz and collaborators showed that PCNA helps maintain the basal expression of estrogen-responsive genes75. In breast cancer, ER-alpha has been shown to affecting the cell cycle by suppressing p53 / p21 activity, and by increasing the levels of PCNA and KI-67 antigen (Ki-67)76. It has also been found that stimulation of ER-alpha by 17-β-estradiol increases MCF-7 cell proliferation by increasing PCNA and Ki-67 levels76. This information provides important clues as to how stimulation by hormones can influence the development of meningioma, especially the female, which is more frequent.
Conclusions
Our study compared the proteomes of biopsies derived from meningiomas of male and female patients. Most of the proteins were identified in both genders (82%); those uniquely identified in female meningiomas pointed to enriched pathways related to RNA splicing and transport-related pathways; and have been described as enriched for breast cancer and related to hormone exposure. The enriched pathways in the male group were related to extracellular matrix organization which are linked to cancer aggressiveness and metastasis. We recall that proteins uniquely identified in one condition does not mean a complete absence in the other one but only that they were not identified by our approach; regardless, this is still suggestive of differential abundancy77. We also point out that exclusively identified proteins to a single biological condition does not mean that they are fully absent in the other; their absence could be due to the stochastic nature of data-dependent acquisition or with an abundancy lower than the detection limits of our approach.
Material and Methods
Materials
Qubit Protein Assay Kit (Cat. no Q33212) and RapiGest SF acid-labile surfactant (Cat. no 186001861) were purchased from Invitrogen (Carlsbad, CA) and Waters Corp. (Milford, MA), respectively. Sequence grade modified trypsin (V511A) was purchased from Promega. All other laboratory reagents were acquired from Sigma-Aldrich (St. Louis, MO), unless specified otherwise.
Patients
This study was approved by the ethics committee of Oswaldo Cruz Foundation and the Federal University of Clinical Hospital of Curitiba under the numbers 63056316.8.0000.5248 and 63056316.8.3001.0096, respectively. A written informed consent was acquired from each patient. As such, all methods were carried out in accordance with relevant guidelines and regulations for this manuscript. The tumor fragments were collected by neurosurgeons belonging to the clinical staff of the Clinical Hospital of the Federal University of Paraná. The collected samples were stored in sterile 15 mL capped tubes, which were transported on dry ice, following all necessary biosecurity standards for such procedure. All collected material was aliquoted using sterile material and then stored at -80 °C. All patients were diagnosed with type I meningioma, as shown in the Table 6.
Sample preparation
The twelve tissue samples of grade I meningiomas were pulverized in liquid nitrogen, as previously described78. Then, protein extraction was made in a solution of 0.1% of RapiGest (w/v) in 50 mM triethylammonium bicarbonate (TEAB). Subsequently, the extracted proteins were centrifuged at 18,000 × g at 4 °C, for 15 minutes and supernatant was collected. The protein content was quantified by a fluorimetric assay using the Qubit 2.0 platform, according to the manufacturer’s instructions. Next, 180 µg of total protein from each sample was reduced with 10 mM of dithiothreitol (DTT) at 60 °C for 30 minutes. Then, all samples were cooled to room temperature and incubated in the dark with 25 mM of iodoacetamide (IAA) for 30 minutes. The samples were subsequently digested for 20 hours with high sequence grade modified trypsin at a 1:50 (Enzyme/Substrate) ratio at 37 °C. Following digestion, all reactions were acidified with 10% (v/v) trifluoroacetic acid (0.5% v/v final concentration) to stop proteolysis and precipitate RapiGest. The samples were centrifuged for 15 minutes at 18,000 × g at 20 °C to remove insoluble materials. Then, peptides were desalted with a C18 spin column, according to the manufacturer’s instructions (Harvard Apparatus).
Mass spectrometry analysis
We used a nanoLC Easy1000 coupled online with a Q-Exactive plus mass spectrometer to generate two proteomic profiles of each biological replicate using the same materials and methods as we previously described78.
Reproducibility assessment
The study consisted of 12 biological samples; six from male and six from female grade I meningioma biopsies. For each biological sample, two technical replicates were generated, producing a total of 24 Q-Exactive Plus runs. All technical replicates were assessed for reproducibility; those achieving RawVegetable’s reproducibility k-score below 0.1 were repeated. PatternLab’s bioinformatic analysis merges the information from the technical replicates to reduce under sampling.
Peptide spectrum matching (PSM)
Our bioinformatic analysis was guided by the steps described in the PatternLab for proteomics protocol15; the software version we used was PaternLab for proteomics 4.1.1.17 that is freely available at http://www.patternlabforproteomics.org. The H. Sapiens Swiss-Prot database79 was downloaded on February 19th, 2019; a reversed version of each sequence plus those from 127 common mass spectrometry contaminants was included. The search considered semi-tryptic and fully-tryptic peptide candidates, allowing a maximum of 2 lost cleavage sites. Oxidation of methionine and carbamidomethylation of cysteine were considered as variable and fixed modifications, respectively.
Validation of PSMs
The Search Engine Processor (SEPro), built into PatternLab, was used for converging to a list of identifications with less than 1% of false discovery rate (FDR) at the protein level, as previously described80. Briefly, the identifications were grouped by charge state (2 + and ≥3 +), and then by tryptic status, resulting in four distinct subgroups. For each group, the XCorr, DeltaCN, DeltaPPM, and Peaks Matched values were used to generate a Bayesian discriminator. The identifications were sorted in non-decreasing order according to the discriminator score. A cutoff score was established to accept a false-discovery rate (FDR) of 1% at the peptide level based on the number of labeled decoys. This procedure was independently performed on each data subset, resulting in an FDR that was independent of charge state or tryptic status. Additionally, a minimum sequence length of six amino-acid residues was required. Results were post-processed to only accept PSMs with less than 10 ppm from the global identification average. One-peptide identifications (i.e., proteins identified with only one mass spectrum) with the peptide having an XCorr of less than 2 were discarded. This last filter led to FDRs, now at the protein level, to be lower than 1% for all search results.
Relative quantitation of proteins
Quantitation was performed according to PatternLab’s Normalized Ion Abundance Factors (NIAF) as a relative quantitation strategy. We recall that NIAF is the equivalent to NSAF81, but applied to extracted ion chromatogram (XIC)82. The PatternLab TFold module83 was used to pinpoint differentially abundant proteins between the female and male groups. The proteins log fold change was estimated by obtaining the log of the averaged corresponding peptide folds. Our differential proteomic comparison only considered proteins identified with two or more unique peptides (i.e., peptides that map to a single sequence in the database), a q-value ≤ 0.1 and an absolute peptide fold change cutoff > 3. Only proteins present in at least two technical replicates (from six biological replicates for each gender) were considered for the TFold analysis. In summary, the procedure briefly described in our bioinformatics protocol15. Finally, we used the Reactome84, DAVID85 tools to help interpret the data.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE86 partner repository with the dataset identifier PXD015979.
References
Wang, N. & Osswald, M. Meningiomas: Overview and New Directions in Therapy. Semin. Neurol. 38, 112–120 (2018).
Shaikh, N., Dixit, K. & Raizer, J. Recent advances in managing/understanding meningioma. F1000Research 7, 490 (2018).
Louis, D. N. et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. (Berl.) 131, 803–820 (2016).
Monleón, D. et al. Metabolic aggressiveness in benign meningiomas with chromosomal instabilities. Cancer Res. 70, 8426–8434 (2010).
Wiemels, J., Wrensch, M. & Claus, E. B. Epidemiology and etiology of meningioma. J. Neurooncol. 99, 307–314 (2010).
Sun, T., Plutynski, A., Ward, S. & Rubin, J. B. An integrative view on sex differences in brain tumors. Cell. Mol. Life Sci. 72, 3323–3342 (2015).
Farrag, A. et al. Intracranial meningioma as primary presentation for an undiagnosed collision metastatic breast cancer: Case report and literature review. Mol. Clin. Oncol. 8, 661–664 (2018).
Lin, J.-W. et al. Breast carcinoma metastasis to intracranial meningioma. J. Clin. Neurosci. 16, 1636–1639 (2009).
Umansky, F., Shoshan, Y., Rosenthal, G., Fraifeld, S. & Spektor, S. Radiation-induced meningioma. Neurosurg. Focus FOC 24, E7 (2008).
Goutagny, S. et al. Long-term follow-up of 287 meningiomas in neurofibromatosis type 2 patients: clinical, radiological, and molecular features. Neuro-Oncol. 14, 1090–1096 (2012).
Sharma, S., Ray, S., Moiyadi, A., Sridhar, E. & Srivastava, S. Quantitative Proteomic Analysis of Meningiomas for the Identification of Surrogate Protein Markers. Sci. Rep. 4, 7140 (2014).
Papaioannou, M.-D. et al. Proteomic analysis of meningiomas reveals clinically distinct molecular patterns. Neuro-Oncol. 21, 1028–1038 (2019).
Dunn, J. et al. Proteomic analysis discovers the differential expression of novel proteins and phosphoproteins in meningioma including NEK9, HK2 and SET and deregulation of RNA metabolism. EBioMedicine 40, 77–91 (2019).
Ostrom, Q. T. et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro-Oncol. 20, iv1–iv86 (2018).
Carvalho, P. C. et al. Integrated analysis of shotgun proteomic data with PatternLab for proteomics 4.0. Nat. Protoc. 11, 102–117 (2015).
Silva, A. R. F. et al. DiagnoProt: a tool for discovery of new molecules by mass spectrometry. Bioinformatics 33, 1883–1885 (2017).
Jiang, J., Xu, N., Zhang, X. & Wu, C. Lipids changes in liver cancer. J. Zhejiang Univ. Sci. B 8, 398–409 (2007).
Hu, C.-A. A., Klopfer, E. I. & Ray, P. E. Human apolipoprotein L1 (ApoL1) in cancer and chronic kidney disease. FEBS Lett. 586, 947–955 (2012).
Okamoto, H. et al. Comparative Proteomic Profiles of Meningioma Subtypes. Cancer Res. 66, 10199–10204 (2006).
Cui, G. Q. et al. Proteomic analysis of meningiomas. Acta Neurol. Belg. 114, 187–194 (2014).
Oh, E.-S., Seiki, M., Gotte, M. & Chung, J. Cell adhesion in cancer. Int. J. Cell Biol. 2012, 965618–965618 (2012).
Gérard, C. & Goldbeter, A. The balance between cell cycle arrest and cell proliferation: control by the extracellular matrix and by contact inhibition. Interface Focus 4, 20130075–20130075 (2014).
Walker, C., Mojares, E. & Del Río Hernández, A. Role of Extracellular Matrix in Development and Cancer Progression. Int. J. Mol. Sci. 19, 3028 (2018).
Malik, R., Lelkes, P. I. & Cukierman, E. Biomechanical and biochemical remodeling of stromal extracellular matrix in cancer. Trends Biotechnol. 33, 230–236 (2015).
Eble, J. A. & Niland, S. The extracellular matrix in tumor progression and metastasis. Clin. Exp. Metastasis 36, 171–198 (2019).
Mollinedo, F. Neutrophil Degranulation, Plasticity, and Cancer Metastasis. Trends Immunol. 40 (2019).
Borregaard, N., Sørensen, O. E. & Theilgaard-Mönch, K. Neutrophil granules: a library of innate immunity proteins. Trends Immunol. 28, 340–345 (2007).
Templeton, A. J. et al. Prognostic Role of Neutrophil-to-Lymphocyte Ratio in Solid Tumors: A Systematic Review and Meta-Analysis. JNCI J. Natl. Cancer Inst. 106 (2014).
Shen, M. et al. Tumor-Associated Neutrophils as a New Prognostic Factor in Cancer: A Systematic Review and Meta-Analysis. PLOS ONE 9, e98259 (2014).
Kim, H., Son, S. & Shin, I. Role of the CCN protein family in cancer. BMB Rep. 51, 486–492 (2018).
Li, C. L., Martinez, V., He, B., Lombet, A. & Perbal, B. A role for CCN3 (NOV) in calcium signalling. Mol. Pathol. MP 55, 250–261 (2002).
Thibout, H. et al. Characterization of Human NOV in Biological Fluids: An Enzyme Immunoassay for the Quantification of Human NOV in Sera from Patients with Diseases of the Adrenal Gland and of the Nervous System. J. Clin. Endocrinol. Metab. 88, 327–336 (2003).
Dankner, M. et al. CCN3/Nephroblastoma Overexpressed Is a Functional Mediator of Prostate Cancer Bone Metastasis That Is Associated with Poor Patient Prognosis. Am. J. Pathol. 189, 1451–1461 (2019).
Chen, P.-C., Lin, T.-H., Cheng, H.-C. & Tang, C.-H. CCN3 increases cell motility and ICAM-1 expression in prostate cancer cells. Carcinogenesis 33, 937–945 (2012).
Zhang, T. et al. The Clinical and Prognostic Significance of CCN3 Expression in Patients with Cervical Cancer. Adv. Clin. Exp. Med. Off. Organ Wroclaw Med. Univ. 22, 839–45 (2013).
Manara, M. C. et al. The expression of ccn3(nov) gene in musculoskeletal tumors. Am. J. Pathol. 160, 849–859 (2002).
Bleau, A. M. et al. Antiproliferative activity of CCN3: Involvement of the C-terminal module and post-translational regulation. J. Cell. Biochem. 101, 1475–1491 (2007).
Fukunaga-Kalabis, M. et al. Downregulation of CCN3 expression as a potential mechanism for melanoma progression. Oncogene 27, 2552 (2007).
Gupta, N. et al. Inhibition of glioma cell growth and tumorigenic potential by CCN3 (NOV). Mol. Pathol. MP 54, 293–299 (2001).
Liu, S. et al. CCN3 (NOV) regulates proliferation, adhesion, migration and invasion in clear cell renal cell carcinoma. Oncol. Lett. 3, 1099–1104 (2012).
Bleau, A.-M., Planque, N. & Perbal, B. CCN proteins and cancer: two to tango. Front. Biosci. J. Virtual Libr. 10, 998–1009 (2005).
Suresh, S. et al. The matricellular protein CCN3 regulates NOTCH1 signalling in chronic myeloid leukaemia. J. Pathol. 231, 378–387 (2013).
Liao, W. et al. Antitumor activity of Notch-1 inhibition in human colorectal carcinoma cells. Oncol. Rep. 39, 1063–1071 (2018).
Sin, W.-C. et al. Matricellular protein CCN3 (NOV) regulates actin cytoskeleton reorganization. J. Biol. Chem. 284, 29935–29944 (2009).
Chen, H., Xu, C., Jin, Q. & Liu, Z. S100 protein family in human cancer. Am. J. Cancer Res. 4, 89–115 (2014).
Sedaghat, F. & Notopoulos, A. S100 protein family and its application in clinical practice. Hippokratia 12, 198–204 (2008).
Bleau, A.-M., Planque, N. & Perbal, B. CCN proteins and cancer: two to tango. Front. Biosci. J. Virtual Libr. 10, 998–1009 (2005).
Fei, F., Qu, J., Zhang, M., Li, Y. & Zhang, S. S100A4 in cancer progression and metastasis: A systematic review. Oncotarget 8, 73219–73239 (2017).
Boye, K. & Maelandsmo, G. M. S100A4 and metastasis: a small actor playing many roles. Am. J. Pathol. 176, 528–535 (2010).
Orre, L. M. et al. S100A4 interacts with p53 in the nucleus and promotes p53 degradation. Oncogene 32, 5531 (2013).
Cooper, G. M. The cell: a molecular approach. (ASM Press; Sinauer Associates (2000).
Bertram, J. S. The molecular biology of cancer. Mol. Aspects Med. 21, 167–223 (2000).
Orre, L. M. et al. S100A4 interacts with p53 in the nucleus and promotes p53 degradation. Oncogene 32, 5531 (2013).
Bögler, O., Su Huang, H.-J., Kleihues, P. & Cavenee, W. K. The p53 gene and its role in human brain tumors. Glia 15, 308–327 (1995).
Gasco, M., Shami, S. & Crook, T. The p53 pathway in breast cancer. Breast Cancer Res. 4, 70 (2002).
Garima, P. S., Pandey, L. K., Saxena, A. K. & Patel, N. The Role of p53 Gene in Cervical. Carcinogenesis. J. Obstet. Gynaecol. India 66, 383–388 (2016).
Gibbons, D. L., Byers, L. A. & Kurie, J. M. Smoking, p53 mutation, and lung cancer. Mol. Cancer Res. MCR 12, 3–13 (2014).
Rudland, P. S. et al. Prognostic Significance of the Metastasis-inducing Protein S100A4 (p9Ka) in Human Breast Cancer. Cancer Res. 60, 1595 (2000).
Zakaria, R. et al. Metastasis-inducing proteins are widely expressed in human brain metastases and associated with intracranial progression and radiation response. Br. J. Cancer 114, 1101–1108 (2016).
Taylor, S., Herrington, S., Prime, W., Rudland, P. S. & Barraclough, R. S100A4 (p9Ka) protein in colon carcinoma and liver metastases: association with carcinoma cells and T-lymphocytes. Br. J. Cancer 86, 409–416 (2002).
Xia, C., Braunstein, Z., Toomey, A. C., Zhong, J. & Rao, X. S100 Proteins As an Important Regulator of Macrophage Inflammation. Front. Immunol. 8, 1908 (2018).
Li, Z.-H., Dulyaninova, N. G., House, R. P., Almo, S. C. & Bresnick, A. R. S100A4 regulates macrophage chemotaxis. Mol. Biol. Cell 21, 2598–2610 (2010).
Wu, Y., Zhang, J. & Qin, Y. S100A4 promotes the development of lipopolysaccharide-induced mouse endometritis†. Biol. Reprod. 99, 960–967 (2018).
Ambartsumian, N., Klingelhöfer, J. & Grigorian, M. The Multifaceted S100A4 Protein in Cancer and Inflammation: From Basics to Medical Applications. in. Methods in molecular biology (Clifton, N.J.) 1929, 339–365 (2019).
Stein, U. et al. The Metastasis-Associated Gene S100A4 Is a Novel Target of β-catenin/T-cell Factor Signaling in Colon Cancer. Gastroenterology 131, 1486–1500 (2006).
Kalluri, R. & Weinberg, R. A. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428 (2009).
Wang, B.-D. & Lee, N. H. Aberrant RNA Splicing in Cancer and Drug Resistance. Cancers 10, 458 (2018).
Custer, B., Longstreth, W. T. Jr, Phillips, L. E., Koepsell, T. D. & Van Belle, G. Hormonal exposures and the risk of intracranial meningioma in women: a population-based case-control study. BMC Cancer 6, 152–152 (2006).
Srebrow, A. & Kornblihtt, A. R. The connection between splicing and cancer. J. Cell Sci. 119, 2635 (2006).
Claus, E. B. et al. Epidemiology of Intracranial Meningioma. Neurosurgery 57, 1088–1095 (2005).
Dago, D. N. et al. Estrogen receptor beta impacts hormone-induced alternative mRNA splicing in breast cancer cells. BMC Genomics 16, 367–367 (2015).
Hsu, D. W., Efird, J. T. & Hedley-Whyte, E. T. Progesterone and estrogen receptors in meningiomas: prognostic considerations. J. Neurosurg. 86 (1997).
Cork, D. M. W., Lennard, T. W. J. & Tyson-Capper, A. J. Alternative splicing and the progesterone receptor in breast cancer. Breast Cancer Res. BCR 10, 207–207 (2008).
Strzalka, W. & Ziemienowicz, A. Proliferating cell nuclear antigen (PCNA): a key factor in DNA replication and cell cycle regulation. Ann. Bot. 107, 1127–1140 (2011).
Schultz-Norton, J. R. et al. Interaction of estrogen receptor alpha with proliferating cell nuclear antigen. Nucleic Acids Res. 35, 5028–5038 (2007).
Liao, X. et al. Estrogen receptor α mediates proliferation of breast cancer MCF–7 cells via a p21/PCNA/E2F1-dependent pathway. FEBS J. 281, 927–942 (2014).
Carvalho, P. C. et al. Analyzing marginal cases in differential shotgun proteomics. Bioinforma. Oxf. Engl. 27, 275–276 (2011).
Wippel, H. H. et al. Comparing intestinal versus diffuse gastric cancer using a PEFF-oriented proteomic pipeline. J. Proteomics (2017) https://doi.org/10.1016/j.jprot.2017.10.005.
UniProt Consortium. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 41, D43–47 (2013).
Carvalho, P. C. et al. Search engine processor: Filtering and organizing peptide spectrum matches. Proteomics 12, 944–949 (2012).
Zybailov, B. et al. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5, 2339–2347 (2006).
Neilson, K. A. et al. Less label, more free: Approaches in label-free quantitative mass spectrometry. PROTEOMICS 11, 535–553 (2011).
Carvalho, P. C., Yates, J. R. III & Barbosa, V. C. Improving the TFold test for differential shotgun proteomics. Bioinforma. Oxf. Engl. 28, 1652–1654 (2012).
Fabregat, A. et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 44, D481–D487 (2016).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Vizcaíno, J. A. et al. The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 (2012).
Acknowledgements
The authors would like to thank the medical residents of the Federal University of Clinical Hospital of Curitiba for assisting with the sample’s collections and the financial support agencies: CAPES, CNPq, Cancer Foundation, and PAPES.
Author information
Authors and Affiliations
Contributions
L.A.B.B. and J.S.G.F. proposed the study. D.C.A.V., L.A.B.B., S.L.S. and G.A.R.P. are medical doctors and responsible for acquiring tumor biopsies and patient care. M.D.M.S., H.H.W., J.M.S. prepared the proteomic samples. R.M.S. and F.C.S.N. generated the mass spectrometry data. P.C.C., M.D.M.S., H.H.W., J.M.S. and J.S.G.F. analyzed the data and wrote the manuscript draft. All authors revised and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Silva, J.M., Wippel, H.H., Santos, M.D.M. et al. Proteomics pinpoints alterations in grade I meningiomas of male versus female patients. Sci Rep 10, 10335 (2020). https://doi.org/10.1038/s41598-020-67113-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-67113-3
This article is cited by
-
Proteomic interrogation of the meninges reveals the molecular identities of structural components and regional distinctions along the CNS axis
Fluids and Barriers of the CNS (2023)
-
Male sex and presence of preoperative symptoms are associated with early recurrence of WHO grade I meningiomas after surgical resection: analysis from the nationwide Brain Tumor Registry of Japan
Neurosurgical Review (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.