Abstract
Weill-Marchesani syndrome (WMS) is a rare disorder displaying short stature, brachydactyly and joint stiffness, and ocular features including microspherophakia and ectopia lentis. Brachydactyly and joint stiffness appear less commonly in patients with WMS4 caused by pathogenic ADAMTS17 variants. Here, we investigated a large family with WMS from Newfoundland, Canada. These patients displayed core WMS features, but with proportionate hands that were clinically equivocal for brachydactyly. Whole exome sequencing and autozygosity mapping unveiled a novel pathogenic missense ADAMTS17 variant (c.3068 G > A, p.C1023Y). Sanger sequencing demonstrated variant co-segregation with WMS, and absence in 150 population matched controls. Given ADAMTS17 involvement, we performed deep phenotyping of the patients’ hands. Anthropometrics applied to hand roentgenograms showed that metacarpophalangeal measurements of affected patients were smaller than expected for their age and sex, and when compared to their unaffected sibling. Furthermore, we found a possible sub-clinical phenotype involving markedly shortened metacarpophalangeal bones with intrafamilial variability. Transfection of the variant ADAMTS17 into HEK293T cells revealed significantly reduced secretion into the extracellular medium compared to wild-type. This work expands understanding of the molecular pathogenesis of ADAMTS17, clarifies the variable hand phenotype, and underscores a role for anthropometrics in characterizing sub-clinical brachydactyly in these patients.
Similar content being viewed by others
Introduction
Weill-Marchesani syndrome (WMS) is a rare heritable connective tissue disorder (prevalence 1 in 100,000)1 that manifests with short stature, brachydactyly, thick skin, and joint stiffness2. Patients may also have microspherophakia, severe myopia, ectopia lentis, glaucoma and cataracts. Occasionally, there is pulmonary or aortic valve stenosis. WMS is caused by highly penetrant pathogenic variants in fibrillin-1 (FBN1), that cause autosomal dominant WMS (WMS2, OMIM #608328), while pathogenic variants in three other genes, ADAMTS10 (WMS1, OMIM #277600), LTBP2 (WMS3, OMIM#614819) ADAMTS17 (WMS4, OMIM#613195) cause autosomal recessive WMS3,4,5. In addition to genetic heterogeneity, WMS also demonstrates subtle clinical heterogeneity, that together with its mechanistic basis, needs further clarification.
Fibrillin-1 forms tissue microfibrils that are key structural components of the extracellular matrix, also regulating TGFβ signaling and the biogenesis and homeostasis of elastic fibers, which support skin, ligaments, and blood vessels. Likewise, fibrillin microfibrils are the core structures of the ciliary zonule, which are compromised in WMS, and cause lens dislocations6,7. The other genes mutated in WMS encode ECM proteins that either localize to fibrillin-1 microfibrils in the ECM or can be involved in the formation of microfibrils. LTBP2 encodes latent TGFβ binding protein-2, which directly binds fibrillin-1, and is required for stable assembly of microfibril bundles within the ciliary zonule8,9. ADAMTS10 and ADAMTS17 encode secreted metalloproteases, which require fibrillin-1 microfibrils for ECM localization and regulate assembly of microfibrils. ADAMTS10 accelerates microfibril assembly through direct interactions with fibrillin-110, whereas ADAMTS17 binds fibrillin-111, and may instead play a role in biogenesis and maturation of microfibrils, or act as a chaperone that facilitates fibrillin-1 secretion12.
Family studies that characterize WMS patients are essential toward advancing our understanding of these molecular pathways within the extracellular matrix. Pathogenic ADAMTS17 variants were first described in 20095. Review of the literature indicates there are eight pathogenic ADAMTS17 variants reported in six studies, which collectively describe 18 WMS4 patients (Supplementary Table S1). Interestingly, among these reports, brachydactyly appears to be an uncommon manifestation. For example, Morales et al.5, and Khan et al.13, collectively identified four unrelated WMS families (i.e. nine affected patients) from Saudi Arabia. They reported four different pathogenic ADAMTS17 variants (p.E820GfsX23; p.Q254X; c.1721 + 1 G > A; p.Asp218ThrfsX41) in these families. Notably, affected individuals displayed core WMS features, but consistently had normal (i.e. proportionate) hands, without any evidence of brachydactyly or joint stiffness. This observation led the authors to hypothesize that pathogenic ADAMTS17 variants manifest as a ‘WMS-like’ syndrome (WMS4, OMIM #613195) typified by the above.
Subsequently, Radner et al.14 studied three families (i.e. four affected patients) with a complex phenotype involving both congenital ichthyosis and autosomal recessive WMS. They identified a contiguous 106.96 kb deletion involving CERS3 and ADAMTS17 (among other loci), in this unique 15q26.3 deletion syndrome. Notably, they described brachydactyly in three patients, and found joint stiffness in two patients. Unfortunately, given this causative deletion included multiple genes, it is difficult to correlate specific findings to ADAMTS17. Meanwhile, brachydactyly without joint stiffness was described by Shah et al.15, in the hands of one WMS patient with a pathogenic ADAMTS17 splice variant (c.873 + 1 G > T). Subsequently, Yi et al.16, studied three WMS siblings with brachydactyly and identified a nonsense variant (p.Lys351*) in ADAMTS17. Finally, Karoulias et al.12, reported a pathogenic ADAMTS17 missense variant (p.Thr343Ala) in a singleton WMS patient with no evidence of brachydactyly. There are inconsistencies however with how brachydactyly was ascertained among these studies (Supplementary Table S1). Diagnosis of brachydactyly can be either clinical, anthropometric or radiological; however, only one study defined their criteria for brachydactyly, and many appear to be based on clinical observation alone. Thus, to establish whether there is any utility in describing a ‘WMS-like’ syndrome, or more recently a specific designation of WMS-4 (OMIM #613195) specifically caused by ADAMTS17 mutations, a more detailed analysis of the hand phenotype in patients with ADAMTS17 variants is warranted.
The primary objective of our study was to solve the genetic etiology in a large multiplex family from Newfoundland, Canada with five affected siblings that displays autosomal recessive WMS. The family were first described in 1982, with core WMS features, but had broad appearing, proportionate hands, with stubby fingers, which were clinically equivocal for brachydactyly17. Subsequently, they were followed over a 40-year interval, during which no causative variant was found, prompting their enrollment in the FORGE Canada Consortium18,19, a large project designed to identify causative genetic variants in families with unexplained rare disease. Here, we report successful identification of a novel pathogenic ADAMTS17 variant in this family with WMS. We performed deep phenotyping of patients’ hands using anthropometry to refine hand analysis and identified a possible sub-clinical phenotype. Functional characterization of the impact of this variant on secretion of ADAMTS17 into the extracellular medium, provides further insights into the mechanistic impact of the variant and pathophysiology of this disorder.
Materials and Methods
Patient Ascertainment
Family members live in a small rural community on the island of Newfoundland, Canada. Patients were first seen at an ophthalmology clinic by GJJ and were the subject of a case report in 198317. Subsequently, they were referred to the Provincial Medical Genetics Program. Genetic testing during routine clinic visits did not identify any mutations in WMS genes known at the time (ADAMTS10 & FBN1). In 2007, family members gave written and informed consent to participate in an ocular genetics study. They consented to participate in the FORGE Canada Consortium study (now Care4Rare Canada Consortium) in 201218,19. The current study was approved by the Health Research Ethics Board of Newfoundland & Labrador (HREB#2011.060) and all experiments were performed in accordance with relevant guidelines and regulations.
Whole exome sequencing, variant filtering and homozygosity mapping
Whole blood was drawn and genomic DNA was extracted from peripheral leukocytes by standard protocols. Whole exome sequencing (Illumina Hiseq. 2000) was performed according to protocols at the Genome Quebec Innovation Center, Montreal, Canada18,19. Briefly, 3 µg of genomic DNA from each affected individual (Fig. 1: II-5, II-3) was used for library preparation, and exome capture and enrichment was accomplished using the SureSelect Human Exome Kit V.4 (Agilent Technologies, Inc., Santa Clara, CA). Alignment, variant calling, and annotation were performed as per previous FORGE projects20,21. Mapping of high quality paired-end reads (100 bp) against the UCSC Genome hg19 was performed using BWA (v. 0.5.9)22. The Genome Analysis Toolkit (GATK)23 was used for local realignment around indels (short insertions and deletions), and for coverage assessment. Samtools (v. 0.1.17) mpileup24 and ANNOVAR25 were then used for variant calling and annotation, respectively. A custom filtering protocol was applied to the whole exome data and sequence variants were excluded in the event that: (1) <3 reads supported the alternative variant; (2) variant allele frequency was >0.05 in ExAC database and if variants were seen in >30 individuals in our database (~1000 exomes sequenced from provincial residents previously in our center); (3) variants were homozygous in ExAC database; (4) variants were synonymous; (5) variants were within 5’ UTRs or intronic variants outside of splice-site boundaries.
Finally, hypothesizing autozygosity for our family, mapping was performed using in-house scripts written in Perl, to identify regions of homozygosity (ROH). An ROH was defined as a span of at least 25 SNPs where no more than two are called as heterozygous and the remainder were homozygous.
Validation of candidate variant, segregation analysis and population controls
To validate the candidate ADAMTS17 variant, primers were designed using Primer326, capturing exon 21 (forward primer: 5-′TCCCTTGACCTCA-3′; reverse primer: 5′-CACCGTCAGGGAG-3′). PCR conditions are available upon request. Sanger sequencing of PCR products was accomplished using an ABI 3130XL Genetic Analyzer, and chromatograms were interpreted using Sequencher 5.2.3.
In this manner, familial segregation was then tested in individuals I-2, II-3, II-4, II-5 and II-8 (Fig. 1). Finally, a cohort of 150 geographically matched population controls, from a previous study27 were screened for the ADAMTS17 variant.
Analysis of metacarpophalangeal lengths
Bilateral hand roentgenograms of an unaffected sibling (II-4) and two affected siblings (II-3, II-5) were obtained in 2016. To examine for dysmorphia, measurements of metacarpal bones, and proximal, middle and distal phalanges were taken. These measurements were compared against their expected proportions28, taking each person’s age and sex into account. In this study, we defined brachydactyly as hands in affected patients that were appreciably smaller compared to unaffected siblings.
Cloning
Mammalian expression plasmids for full-length human ADAMTS17 and its proteolytically inactive mutant ADAMTS17-EA were described previously11. ADAMTS17-EA was used for initial analysis of the effect of the mutation on secretion, since the wild-type ADAMTS17 is autocatalytically processed and therefore difficult to detect in conditioned medium. To introduce the variant of interest, a synthetic DNA fragment spanning nucleotides 2799–3352 of ADAMTS17 (Ref.Seq. XM_017021975) (Integrated DNA Technologies, Coralville, IA) was ligated with the gel-purified 8335 bp SrfI x AfeI fragment obtained from the ADAMTS17-EAplasmid, using the NEBuilder® HiFi DNA Assembly Kit (New England Biolabs, Ipswich, MA). Positive clones were screened via an additional ScaI restriction site introduced by the mutation and verified by DNA sequencing (Cleveland Clinic Lerner Research Institute Genomics Core).
Cell culture, transfection, and collection of cell lysate and conditioned medium
Human embryonic kidney cells (HEK293T, ATCC, Manassas, VA) which were maintained in DMEM supplemented with 10% FBS, 100 units/ml penicillin and 100 μg/ml streptomycin in a 5% CO2 atmosphere in a humidified incubator at 37 °C. HEK293T cells were seeded in 12-well plates (BD Bioscience, San Jose, CA) and transfected in triplicate with 1 μg plasmid DNA/well using Lipofectamine 3000 according to the manufacturer’s protocol. 16–24 h after transfection, the medium was removed and the cells were rinsed with 1 ml PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2KPO4, 1.8 mM KH2PO4) and incubated with 600 μl serum-free DMEM for 48 h. Conditioned medium was collected and cell debris were removed by centrifugation. The cell layer was rinsed with 1 ml PBS and cells were lysed in 0.1% NP40, 0.01% SDS, 0.05% Na-deoxycholate in PBS. Cell lysates were incubated for 5 min with rotation end-over-end and cleared by centrifugation (5 min, > 20,000 g, 4 °C).
Western blotting
Equal volumes of conditioned medium or cell lysate from each transfection were subjected to 7.5% SDS-PAGE under reducing conditions. Proteins were transferred onto PVDF membranes (Immobilon F, EMD Millipore, Billerica, MA) for 1.5 h at 70 V at 4 °C in 25 mM Tris, 192 mM glycine, 20% methanol buffer. The membrane was blocked with 5% (w/v) milk in TBS (10 mM Tris-HCl, pH 7.2, 150 mM NaCl) for 1 h at room temperature and anti-myc (clone 9E10, Invitrogen, 1:500) was diluted in 5% (w/v) milk in TBS + 0.1% Tween 20 (TBST) and incubated overnight at 4 °C. Membranes were washed three times with TBST for 5 min at RT and incubated with fluorophore-labeled IRDye goat-anti-mouse secondary antibody (1:10,000) (LI-COR Biosciences, Lincoln, NE) in 5% milk in TBST + 0.01% SDS for 1 h at room temperature. Membranes were washed three times with TBST for 5 min, once with TBS for 5 min at room temperature, and scanned wet on an Odyssey CLx scanner (LI-COR Biosciences, Lincoln, NE). The fluorescent signal was quantified using the Image Studio software.
Results
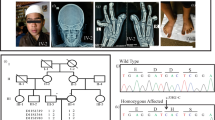
Five out of eight siblings were affected with WMS in this family (Fig. 1). The pattern of WMS is autosomal recessive, and both parents live in a small rural community, sharing the same surname prior to marriage, but are unaware of any consanguinity. Their 40-year clinical histories are summarized in Table 1. Affected individuals have characteristic WMS features such as short stature (mean height 156.5 cm), early onset myopia (5/5 siblings), microspherophakia (4/5 siblings) and ectopia lentis (4/5 siblings). They all had high IOPs diagnosed from early ages (10–26 years). Cataracts and glaucoma developed in 2/5. Longitudinal clinical histories for each patient are provided (Supplementary Results). Joint stiffness was an uncommon manifestation and only 1/5 siblings (sibling II-8) presented with limited range of motion in the wrists and shoulders and had a winged scapula.
We next analyzed the hand phenotype in these patients, given the incomplete clinical penetrance of brachydactyly in previous descriptions of individuals with WMS4. The hands of our patients appeared wide with stubby fingers, but clinically, were proportionate and equivocal for brachydactyly17. Roentgenograms demonstrated shortened metacarpophalangeal bones. For example, sibling II-6 had bilateral shortening of 4th and 5th metacarpals, while sibling II-8 had bilateral shortening of 1st, 4th and 5th metacarpals (Table 1). To perform a deeper phenotyping, new roentgenograms were obtained from available affected siblings (II-3, II-5) and their unaffected sibling (II-4) (Fig. 2). Anthropometrics of the hands, adjusted for age and sex, were applied to characterize differences in metacarpophalangeal measurements28 (Fig. 2B). In the unaffected sibling (II-4), metacarpophalangeal measurements predominantly varied between 1 and −1 standard deviations from expected values. However, in the affected siblings (II-3 and II-5), these values predominantly varied from −1 to −3 standard deviations below their expected values. Moreover, in affected sibling II-5 there was marked unilateral shortening of the 1st metacarpal on the left hand (−7 standard deviations below expected). Likewise, we observed a unilaterally shortened 1st proximal phalanx (−5.3 standard deviations below expected) on the right hand of affected sibling II-3.
Roentgenogram and anthropometric analysis of family members demonstrates intrafamilial variability of hand features. Bilateral roentgenograms (A) of unaffected sibling II-4 (left) compared with affected sibling II-5 (center) and II-3 (right). There is bilateral 5th finger clinodactyly in II-3 and, overall, the hands of affected siblings are shorter and stubbier than their unaffected sibling. Row B shows age and sex corrected anthropometrics of metacarpophalangeal bones. Unaffected sibling II-4 is compared against affected sibling II-5 (top panel), and II-3 (middle panel), or both siblings (bottom panel). There was marked shortening (−7 S.D) of left 1st metacarpal in II-5 (light orange line) and shortened (−5.3 S.D) right 1st proximal phalanx in II-3 (dark blue line). Overall, measurements of the affected siblings (orange and blue lines) are shorter (−1 to 3 S.D) as compared to their unaffected sibling (1 to −1 S.D; red lines).
Genetic analysis was pursued as part of the FORGE Canada Consortium. Whole exome sequencing and homozygosity mapping of genomic DNA samples from siblings II-3 and II-5 revealed a homozygous missense variant in ADAMTS17 (NM_139057.3: c.3068 G > A: p.C1023Y). GnomAD reports that p.C1023Y is rare, with a minor allele frequency (MAF) of 0.0016% (3 out of 187,624 alleles; no homozygotes). Sanger sequencing for p.C1023Y in 150 population matched controls from Newfoundland was negative. Familial co-segregation analysis corroborated the expected pattern of transmission for a fully penetrant, autosomal recessive disorder (Fig. 1). Several in silico tools predicted highly deleterious consequences for this substitution. For example, SIFT predicted deleterious effects (score = 0), PolyPhen-2 predicted damaging (score = 1.0), MutationTaster predicted disease-causing (score: 0.99), CADD score was 32, and GERP score was 5.23, suggesting a region of high evolutionary conservation. Positionally, p.C1023Y substitution resides in a thrombospondin type 1 repeat, which spans amino acids 976–1028 of the ADAMTS17 polypeptide (Fig. 3).
Distribution of reported pathogenic ADAMTS17 variants in WMS families worldwide. 1. Exon structure of ADAMTS17. Asterisks (red) show approximate position of reported pathogenic variants. Introns are not drawn to scale. 2. Shows the relative position of protein domains coinciding with exonic structure. 3. Describes whether a study reported brachydactyly (Y) or proportionate hands (N).
Thus, having identified a novel pathogenic ADAMTS17 variant, we proceeded to characterize the functional consequences of p.C1023Y (Fig. 4). HEK293T cells were transfected with a plasmid where the p.C1023Y variant was introduced in ADAMTS17 and its active site mutant form (p.E390A) to analyze secretion of ADAMTS17 into the extracellular medium. Fluorescence intensities of ADAMTS17 p.C1023Y mutants were compared against wild type constructs or active site mutant constructs in either extracellular medium or cell lysates. The p. E390A active site mutants are useful as they do not undergo autoproteolysis, and thus are more readily detected in the medium of transfected cells11. A significant reduction of ADAMTS17 p.C1023Y mutants was seen in the extracellular medium in both wild type (p = 0.0012) and p.E390A constructs (p = 0.00014). In cell lysates, there were no significant differences in wild type or p. E390A constructs harboring either wild type residue or the p.C1023Y substitution. Thus, ADAMTS17 expression was not significantly reduced inside HEK293T cells nor did it accumulate inside the cell, whereas secretion into extracellular medium was significantly impaired in p.C1023Y mutants.
Impaired secretion of the ADAMTS17 p.C1023Y mutant in transfected HEK293T cells. (A) Western blot comparing levels of mutant and wild-type ADAMTS17 (detected by anti-myc, green, upper panel) in the medium and lysate. There is a noticeable reduction in intensities in the medium of p.C1023Y expressing cells (“Y” allele in red) compared to wild type (“C” allele) in both ADAMTS17 and ADAMTS17-E390A constructs. No difference in signal intensity was seen in the corresponding cell lysates. Z, zymogen; M. mature enzyme. The lower panel shows the corresponding GAPDH western blot (red). (B) Quantification of ADAMTS17 fluorescence intensity in the medium normalized to ADAMTS17 in the corresponding lysate. Note the reduced intensities for ADAMTS17 levels in the medium in the p.C1023Y mutants (“Y” allele in red) (p = 0.0012 ADAMTS17 and p = 0.00014 in ADAMTS17E390A).
Discussion
Here, we investigated the molecular underpinnings for WMS in a large family from Newfoundland & Labrador and identified a novel homozygous pathogenic missense ADAMTS17 variant (NM_139057.3: c.3068 G > A: p.C1023Y). Having been followed since 1983, this family provides us with a unique and longitudinal view of their WMS history (Supplementary Results). Patients demonstrated core WMS features such as short stature, early onset myopia, microspherophakia and ectopia lentis, but visually their hands were not strongly suggestive of brachydactyly (Table 1). Interestingly, hand roentgenograms revealed shortened metacarpophalangeal bones with intrafamilial variability. Anthropometric analysis allowed us to delineate this sub-clinical hand phenotype in our WMS4 patients (Fig. 2). The analysis showed that hands of the affected siblings are objectively smaller than their unaffected sibling. For example, in the unaffected sibling (II-4), metacarpophalangeal measurements predominantly ranged from 1.0 to −1.0 standard deviations around expected values. This contrasted with the affected siblings (II-3, II-5), whose metacarpophalangeal measurements predominantly ranged between −1.0 and −3.0 standard deviations below expected values. Therefore, siblings II-3 and II-5 have a mild brachydactyly. Notably, our deep phenotyping in II-3 and II-5 also quantified a marked unilateral shortening of metacarpophalangeal bones (i.e. 1st proximal phalanx and 1st metacarpal, respectively) in these two family members. Thus, our findings further underscore the clinical heterogeneity and intrafamilial variability of the brachydactyly phenotype in WMS and emphasize that the overall clinical hand appearance can mask significant anomalies in individual metacarpals, which can be elicited by antropometry.
An interesting correlation with published literature is that brachydactyly is uncommonly found among patients with pathogenic ADAMTS17 variants. For example, after excluding the 15q26.3 deletion syndrome, 10 out of 14 WMS patients with reported pathogenic variants had reportedly normal hands (Supplementary Table S1). However, many of these studies relied solely on a clinical approach to reporting brachydactyly. In our study, WMS patients had hands that were proportionate and clinically equivocal, but roentgenograms revealed variably dysmorphic features, which we quantified with anthropometrics (Fig. 2). Our findings suggest the possibility of a sub-clinical phenotype, which may have been overlooked among previously reported studies. We therefore emphasize the importance of applying roentgenogram and anthropometric analysis when addressing the question of the ADAMTS17 hand phenotype.
Ultimately, it remains plausible that the hand phenotype in ADAMTS17 mutations demonstrates a spectrum of WMS severity. For example, some patients will develop normal hands, others might have sub-clinical dysmorphia, while others display more obvious brachydactyly. Future studies might consider whether a unique function of ADAMTS17 within the extracellular matrix, or potential functional overlap with ADAMTS10 leading to a degree of compensation might explain the variability in the hand phenotype compared to the other causative genes; since ADAMTS10 is universally expressed in cells in mouse paws29. Present evidence does not suggest any genotype-phenotype correlation when comparing the position of ADAMTS17 variants and the presence of brachydactyly (Fig. 3), recognizing the limitations given the low number of reported cases. Importantly, the p.C1023Y variant had a severe impact on the secretion of ADAMTS17, meaning that it cannot inform about a possible domain-specific effect for genotype-phenotype correlations.
Currently, there are nine pathogenic variants in ADAMTS17 that cause WMS4. There are two frameshift mutations (p.E820GfsX23; p.Asp218ThrfsX41), two splice site mutations (c.1721 + 1 G > A; c.873 + 1 G > T), two nonsense mutations (p.Q254*; p.Lys351*), a contiguous gene deletion on 15q26.3, and now two missense variants (p.Thr343Ala; p.C1023Y), which occur in multiple regions of ADAMTS17 (Supplementary Table S1, Fig. 3). Most previous studies show altered mRNA isoforms caused by high impact mutations with subsequent predictions of truncated ADAMTS17 proteins. Studies of a missense ADAMTS17 variants (p.Thr343Ala; p.C1023Y) by Karoulias et al.12, and our current study respectively, further delineate the functional consequences of pathogenic ADAMTS17 variants. The p.Thr343Ala variant lies in a catalytic domain and results in decreased secretion of ADAMTS17 into the extracellular matrix, whereas the p.C1023Y variant resides in the thrombospondin-1 repeat domain of ADAMTS17 and results in comparable deficiency in secretion. Our analysis of ADAMTS17 p.C1023Y mutants in HEK293T cells revealed significantly reduced levels of ADAMTS17 in cellular medium as compared with wild-type ADAMTS17 (p = 0.0012) and ADAMTS17E390A (p = 0.00014) constructs (Fig. 4). Together, these studies of the ADAMTS17 missense variants demonstrate that ADAMTS17 secretion can be reduced or eliminated as a consequence of missense mutations, suggesting that the consequences of these pathogenic variants is a functional loss of ADAMTS17 in the extracellular matrix or at the cell surface. Their effect is thus independent of the location of the mutations in ADAMTS17 protein, and is unlikely to reflect impairment of domain-specific interactions or functions.
In conclusion, this study of a large multiplex family with WMS identified a novel pathogenic missense variant (p.C1023Y) in ADAMTS17. This variant caused severely reduced secretion of ADAMTS17 into the extracellular medium, suggesting functional loss of ADAMTS17 rather than impairment of a domain-specific function or interaction. Deep phenotyping of our patients’ hands provides novel observations regarding the spectrum of the WMS hand phenotype, highlighting a possible sub-clinical phenotype with intrafamilial variability. This work expands our understanding of the molecular pathogenesis of ADAMTS17, clarifies the resultant hand phenotype, and underscores a role for anthropometrics in characterizing brachydactyly in these patients.
References
Tsilou, E. & MacDonald, I. M. Weill-Marchesani Syndrome. In GeneReviews® (eds. Adam, M. P. et al.) (University of Washington, Seattle, 1993).
Le Goff, C. et al. Mutations in the TGFβ Binding-Protein-Like Domain 5 of FBN1 Are Responsible for Acromicric and Geleophysic Dysplasias. Am. J. Hum. Genet. 89, 7–14 (2011).
Dagoneau, N. et al. ADAMTS10 Mutations in Autosomal Recessive Weill-Marchesani Syndrome. Am. J. Hum. Genet. 75, 801–806 (2004).
Haji-Seyed-Javadi, R. et al. LTBP2 mutations cause Weill-Marchesani and Weill-Marchesani-like syndrome and affect disruptions in the extracellular matrix. Hum. Mutat. 33, 1182–1187 (2012).
Morales, J. et al. Homozygous mutations in ADAMTS10 and ADAMTS17 cause lenticular myopia, ectopia lentis, glaucoma, spherophakia, and short stature. Am. J. Hum. Genet. 85, 558–568 (2009).
Cain, S. A. et al. Proteomic analysis of fibrillin-rich microfibrils. Proteomics 6, 111–122 (2006).
Jones, W., Rodriguez, J. & Bassnett, S. Targeted deletion of fibrillin-1 in the mouse eye results in ectopia lentis and other ocular phenotypes associated with Marfan syndrome. Dis Model Mech 12 (2019).
Fujikawa, Y. et al. Latent TGF-β binding protein 2 and 4 have essential overlapping functions in microfibril development. Sci. Rep. 7, 43714 (2017).
Hirani, R., Hanssen, E. & Gibson, M. A. LTBP-2 specifically interacts with the amino-terminal region of fibrillin-1 and competes with LTBP-1 for binding to this microfibrillar protein. Matrix Biol. 26, 213–223 (2007).
Kutz, W. E. et al. ADAMTS10 protein interacts with fibrillin-1 and promotes its deposition in extracellular matrix of cultured fibroblasts. J. Biol. Chem. 286, 17156–17167 (2011).
Hubmacher, D. et al. Unusual life cycle and impact on microfibril assembly of ADAMTS17, a secreted metalloprotease mutated in genetic eye disease. Sci. Rep. 7, 41871 (2017).
Karoulias, S. Z. et al. A novel ADAMTS17 variant that causes Weill-Marchesani syndrome 4 alters fibrillin-1 and collagen type I deposition in the extracellular matrix. Matrix Biol., https://doi.org/10.1016/j.matbio.2019.11.001 (2019).
Khan, A. O., Aldahmesh, M. A., Al-Ghadeer, H., Mohamed, J. Y. & Alkuraya, F. S. Familial spherophakia with short stature caused by a novel homozygous ADAMTS17 mutation. Ophthalmic Genet. 33, 235–239 (2012).
Radner, F. P. W. et al. Mutations in CERS3 cause autosomal recessive congenital ichthyosis in humans. PLoS Genet. 9, e1003536 (2013).
Shah, M. H., Bhat, V., Shetty, J. S. & Kumar, A. Whole exome sequencing identifies a novel splice-site mutation in ADAMTS17 in an Indian family with Weill-Marchesani syndrome. Mol. Vis. 20, 790–796 (2014).
Yi, H. et al. A novel nonsense mutation in ADAMTS17 caused autosomal recessive inheritance Weill–Marchesani syndrome from a Chinese family. J. Hum. Genet. 64, 681–687 (2019).
Johnson, G. J. & Bosanquet, R. C. Spherophakia in a Newfoundland family: 8 years’ experience. Can. J. Ophthalmol. 18, 159–164 (1983).
Fahiminiya, S. et al. Whole exome sequencing unravels disease-causing genes in consanguineous families in Qatar. Clin. Genet. 86, 134–141 (2014).
McDonald-McGinn, D. M. et al. Hemizygous mutations in SNAP29 unmask autosomal recessive conditions and contribute to atypical findings in patients with 22q11.2DS. J. Med. Genet. 50, 80–90 (2013).
Dyment, D. A. et al. Mutations in PIK3R1 cause SHORT syndrome. Am. J. Hum. Genet. 93, 158–166 (2013).
Smith, A. C. et al. Mutations in the enzyme glutathione peroxidase 4 cause Sedaghatian-type spondylometaphyseal dysplasia. J. Med. Genet. 51, 470–474 (2014).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
McKenna, A. et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Untergasser, A. et al. Primer3–new capabilities and interfaces. Nucleic Acids Res. 40, e115 (2012).
Green, R. C. et al. Very high incidence of familial colorectal cancer in Newfoundland: a comparison with Ontario and 13 other population-based studies. Fam. Cancer 6, 53–62 (2007).
Poznanski, A. K. The hand in radiologic diagnosis: With gamuts and pattern profiles. (Saunders, 1984).
Wang, L. W. et al. Adamts10 inactivation in mice leads to persistence of ocular microfibrils subsequent to reduced fibrillin-2 cleavage. Matrix Biol. 77, 117–128 (2019).
Acknowledgements
This work was funded by the Government of Canada through Genome Canada, the Canadian Institutes of Health Research and the Ontario Genomics Institute (OGI-049). Additional funding was provided by Genome Quebec, Genome British Columbia, and the McLaughlin Centre, as well as the Research & Development Corporation of Newfoundland & Labrador (RDCNL). SF received a postdoctoral Fellowship from the Network of Applied Medical Genetics. SSA was supported by NIH award EY024943.
Author information
Authors and Affiliations
Consortia
Contributions
D.R.E. validated genetic variants in cases and population controls, reviewed clinical histories, interpreted bioinformatic data and prepared the manuscript. J.S.G. conceived of the study, assisted in manuscript revision and collected patient records and consent. J.M. and S.F. aided in the collection and analysis of high throughput sequencing data. B.A.F. assisted in the collection of radiological scans and interpretation of the clinical data. M.A.D. assisted in the measurement and interpretation of radiological scans. G.J.J. and J.H.W. performed ophthalmic examination of individuals and assisted with clinical interpretation of disease. D.H. and S.A. performed biochemical assays of the candidate ADAMTS17 variant. M.O.W. participated in the study design, supervised the collection of molecular and genetic data, assisted in the analysis of genetic data and helped draft the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Evans, D.R., Green, J.S., Fahiminiya, S. et al. A novel pathogenic missense ADAMTS17 variant that impairs secretion causes Weill-Marchesani Syndrome with variably dysmorphic hand features. Sci Rep 10, 10827 (2020). https://doi.org/10.1038/s41598-020-66978-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-66978-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.