Abstract
Lyme disease (LD) and relapsing fevers (RF) are vector-borne diseases caused by bacteria of the Borrelia genus. Here, we report on the widespread infection by a non-described Borrelia species in passerine-associated ticks in tropical rainforests of French Guiana, South America. This novel Borrelia species is common in two tick species, Amblyomma longirostre and A. geayi, which feed on a broad variety of neotropical mammal and bird species, including migratory species moving to North America. The novel Borrelia species is divergent from the LD and RF species, and is more closely related to the reptile- and echidna-associated Borrelia group that was recently described. Genome sequencing showed that this novel Borrelia sp. has a relatively small genome consisting of a 0.9-Mb-large chromosome and an additional 0.3 Mb dispersed on plasmids. It harbors an RF-like genomic organization but with a unique mixture of LD- and RF-specific genes, including genes used by RF Borrelia for the multiphasic antigen-switching system and a number of immune-reactive protein genes used for the diagnosis of LD. Overall, our data indicate that this novel Borrelia is an intermediate taxon between the LD and RF species that may impact a large host spectrum, including American mammals. The designation “Candidatus Borrelia mahuryensis” is proposed for this species.
Similar content being viewed by others
Introduction
Bacteria of the genus Borrelia (Spirochaetes) are the causative agents of major vector-borne diseases including Lyme disease (LD), the most important tick-borne disease in the northern hemisphere, and relapsing fevers (RF), which are transmitted by ticks and lice worldwide1,2. While RF epidemics occurred repeatedly in past centuries2, LD is an expanding infectious disease with more than 300,000 new human cases each year in the United States3,4. Most LD and RF Borrelia species are maintained in enzootic cycles and their presence in humans and domestic animals is incidental to their usual wildlife reservoir hosts1,2,5.
The LD and RF Borrelia species form two separate sister groups6,7, and are sometimes referred to as two sister genera by various authors8,9. LD and RF species were usually thought to be the only two groups within Borrelia. However, recent surveys have uncovered new Borrelia species and strains that do not fall into the LD or RF groups10,11,12,13,14,15,16,17,18,19,20,21. They form a third group, sometimes referred to as “reptile group” (REP), encompassing only two designated species, B. turcica and “Candidatus Borrelia tachyglossi” (B. tachyglossi hereafter), and a few strains not taxonomically described10,11,12,13,14,15,16,17,18,19,20,21. The only two genomes sequenced to date confirmed that these Borrelia are substantially different from the LD and RF Borrelia groups19. Members of the third Borrelia group have been detected worldwide, but not in Western Europe10,11,12,13,14,17,20,21. They have been detected in hard ticks (Amblyomma, Hyalomma, Bothriocroton, and Ixodes genera), which are associated mostly with reptiles, but also with echidna10,11,12,13,14,20,21. Recently, new strains of unknown Borrelia species, closely related to the third Borrelia group, were identified in ticks feeding on ground-dwelling birds (for immature stages) and mammals (for adult stages) in Brazil16, Argentina17, and the United States15,17,18. However, currently, we know little about these new American Borrelia strains: Only a small DNA fragment of the flagellin flaB gene has been used as an exclusive marker for their identification15,16,17,18. Unfortunately, this short gene fragment is inadequate for inferring their phylogenetic proximity with known Borrelia species since the inner tree topology remains poorly resolved at many nodes because of insufficient sequence polymorphism15,16,17,18.
In this study, we describe a novel species, belonging to the third Borrelia group, in passerine-associated ticks from tropical rainforests of French Guiana. This remote territory is a vast equatorial land located on the north-east coast of South America, mostly covered by dense rainforests and with a low density of human population. This novel Borrelia species is common in two tick species feeding on a variety of mammal and bird species. We further cultivated this bacterium and sequenced its complete genome, which showed that this novel Borrelia is intermediate between LD and RF Borrelia, sharing common features with both of them.
Results
A novel Borrelia species in passerine-associated ticks
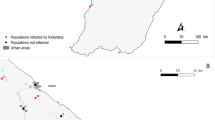
A total of 290 tick specimens were collected from 16 sites in French Guiana (from 2012 to 2018), mostly located along the coastline (Table S1 and Fig. 1). Most of the specimens were larvae (n = 274), with only few nymphs (n = 16), and no adults (Table S1). The 290 ticks were collected from 26 bird species, mainly the blue-backed manakin, Chiroxiphia pareola (n = 98 ticks; 34%), the wedge-billed woodcreeper, Glyphorynchus spirurus (n = 75; 26%), and the crimson-hooded manakin, Pipra aureola (n = 33; 11%). Morphological and genetic examination of tick specimens led to the identification of six tick species, all belonging to the Amblyomma genus: A. longirostre (n = 230), A. geayi (n = 52), A. varium (n = 4), A. cajennense (n = 2), A. calcaratum (n = 1), and A. humerale (n = 1). A. longirostre was found in 25 bird species, A. geayi in 11, and the four other Amblyomma species in one or two bird species each (Table S1 and Fig. 2).
Location of sampling sites in French Guiana. Localities are represented by dots, and numbers correspond to the sampling site number given in Table S1. A letter placed after number indicates a same sampling site sampled over several years as detailed in Table S1. Red and black dots indicate populations infected by Cand. Borrelia mahuryensis and populations not infected, respectively. Bar-charts represents the prevalence of Cand. Borrelia mahuryensis in the sites where infected ticks were collected. Pictures to the left represents a female Crimson-hooded Manakin (Pipra aureola) bearing ticks around its eyes and a nymph of the tick species Amblyomma longirostre.
The network of ticks, passerines, and Cand. Borrelia mahuryensis. Each red node (with letters) and each black node (with numbers) designate a tick and passerine species, respectively. Solid edges indicate pairs of tick/passerine species for which Cand. Borrelia mahuryensis was detected (see Table S1 for details). (A) Amblyomma longirostre; (B) A. geayi; (C) A. varium; (D) A. cajennense; (E) A. calcaratum; (F) A. humerale; 1, Attila spadiceus; 2, Campylopterus largipennis; 3, Chiroxiphia pareola; 4, Cyanocompsa cyanoides; 5, Dendroplex picus; 6, Glyphorynchus spirurus; 7, Manacus manacus; 8, Mionectes macconnelli; 9, M. oleaginous; 10, Myrmeciza ferruginea; 11, Myrmotherula axillaris; 12, Percnostola rufifrons; 13, Picumnus exilis; 14, Pipra aureola; 15, P. erythrocephala; 16, Ramphocelus carbo; 17, Saltator maximus; 18, Thamnomanes caesius; 19, Thamnophilus doliatus; 20, T. punctatus; 21, Tolmomyias sulphurescens; 22, Turdus albicollis; 23, T. fumigatus; 24, T. leucomelas; 25, Xenops minutus; 26, Xiphorhynchus pardalotus.
Examination of the 290 Amblyomma spp. specimens through high-throughput 16 S rDNA sequencing showed that Borrelia was present in 20 specimens (6.9%), corresponding to 20 larvae. Infection was detected in 12 of the 230 A. longirostre specimens (5.2%) and eight of the 52 A. geayi (15.4%) specimens, but not in A. varium, A. cajennense, A. calcaratum, and A. humerale (Table S1, Figs. 1 and 2). Borrelia was detected in five sampling sites of the 16 examined, and in ticks collected from eight bird species: the woodcreeper G. spirurus (three sites), the crimson-hooded manakin P. aureola (two sites), the blue-backed manakin C. pareola (one site), the pale-breasted thrush Turdus leucomelas (one site), the silver-beaked tanager Ramphocelus carbo (one site), the straight-billed woodcreeper Dendroplex picus (one site), the black-headed antbird Percnostola rufifrons (one site), and the white-throated Xenops minutus (one site) (Table S1, Figs. 1 and 2). The prevalence of Borrelia in Amblyomma spp. varied from 0 to 28.5% depending on the sites and sampling dates (Table S1 and Fig. 1), but these variations were not significant (Fisher’s exact tests, all p > 0.10).
Specific polymerase chain reaction (PCR) surveys of the Borrelia 16 S rDNA (710 bp), flaB (540 bp), gyrB (1172 bp), groEL (600 bp), and glpQ (784 bp) genes further confirmed the presence of Borrelia in the 20 Amblyomma spp. specimens earlier found positive by high-throughput 16 S rDNA sequencing. On the basis of these five gene sequences, no nucleotide variation was observed in the Borrelia from the A. longirostre and A. geayi positive specimens, showing that only one Borrelia species was present. None of the 16 S rDNA, gyrB, groEL, and glpQ gene sequences observed in this study is 100% identical to other Borrelia sequences available on GenBank. However, based on partial flaB gene sequences (323 bp), this Borrelia strain is 100% identical to the Borrelia sp. clone 2 T (GenBank accession number: MN064675) recently found in A. longirostre from Brazil16. A maximum likelihood (ML) analysis based on these flaB gene sequences revealed a robust clustering of the new Borrelia of French Guiana with the Borrelia sp. clone 2 T of Brazil, along with several other Borrelia sp. found in A. maculatum of Texas18, suggesting that all these Borrelia belong to the same species (Fig. 3). ML phylogenetic analysis places this Borrelia cluster among members of the third Borrelia group, but the inner topology of the flaB tree remains too poorly resolved in most cases (as shown by the low support values of the inner branches) to infer the exact relatedness of this Borrelia cluster with other Borrelia species and groups (Fig. 3).
Phylogenetic relationship of Borrelia species and strains estimated using maximum likelihood analysis of flaB gene sequences (323 unambiguously aligned bp). In red, sequences of Cand. Borrelia mahuryensis obtained in this study. Branch numbers indicate percentage bootstrap support (1,000 replicates).
Cultivation of Borrelia
We obtained a viable Borrelia isolate (A-FGy1 hereafter) after 6 weeks of cultivation from a freshly molted tick specimen, obtained from an engorged A. longirostre larva collected from a wild passerine. The Borrelia A-FGy1 isolate has a typical Spirochaetes helical-shaped structure of ca. 0.2–0.3 μm diameter and 10–25 μm length (Movie S1). The growth rate was slow and the yield of culture was low. Genetic typing revealed no sequence variation in the flaB, gyrB, groEL, and glpQ genes between the Borrelia A-FGy1 isolated in cultivation and the Borrelia typed from tick DNA described above, showing that they belong to the same species. However, there was one single nucleotide polymorphism (SNP) in the 16 S rDNA sequence, suggesting that two closely related Borrelia strains are present in the study area. This SNP is upstream the 16 S rDNA V4 hypervariable region we sequenced through high-throughput sequencing, and was thus not detected in our primary bacterial bardocing investigation. However, Sanger sequencing of 16 S rDNA PCR product and Illumina complete genome sequencing consistently show that this SNP is well present in the Borrelia A-FGy1 isolate.
Genomic features of the Borrelia A-FGy1 isolate
The sequencing of the Borrelia A-FGy1 isolate produced 230,143,188 paired-end reads of 150 bases, which were assembled into 42 contigs with a mean coverage depth of 1,849×(318–7,854×). The final Borrelia A-FGy1 genome is 1,236,294 bp in size and consists of a 918,483-bp linear chromosome and 41 contigs of putative plasmids (987–42,459 bp) (Table S2). Overall, the Borrelia A-FGy1 genome contains 1,123 genes (812 on linear chromosome and 311 on putative plasmids) including 1,055 predicted protein-coding genes, 37 RNA genes, and 31 pseudogenes. The linear chromosome of the Borrelia A-FGy1 isolate had greatest average nucleotide identity (ANI) with members of the third Borrelia group, B. turcica (90.2%) and B. tachyglossi (82.3%).
Phylogenetic analysis based on 590 single-copy orthologous genes (197,675 AA) present in the linear chromosome of 18 other Borrelia species showed that the Borrelia A-FGy1 isolate clusters within a robust clade with the two known species of the third Borrelia group, B. turcica and B. tachyglossi (Fig. 4). The closest relative of this novel Borrelia species is B. turcica, which was primarily reported from reptiles and reptile-associated ticks. The third Borrelia group, including the Borrelia A-FGy1 isolate, clearly forms an independent Borrelia lineage. The third Borrelia group, however, is more closely related to the RF group than to the LD group (Fig. 4).
Phylogenetic relationship of 19 Borrelia genomes, including the Cand. Borrelia mahuryensis A-FGy1 genome (in red). The phylogenetic tree was inferred using maximum likelihood analysis of a concatenated alignment of 590 single-copy orthologous genes (197,675 AA). The numbers on each node represent the support of 1,000 bootstrap replicates.
The Borrelia A-FGy1 genome is roughly similar to the genomes of the two other sequenced members of the third Borrelia group, B. turcica and B. tachyglossi. The Borrelia A-FGy1 linear chromosome shows an extensive synteny with chromosomes of other Borrelia species and, compared with LD, RF, and the third group of Borrelia chromosomes, it exhibits a comparable size and organization (Fig. S1). The single exception concerns a specific 8.5-kb inversion at the 5′ end of the B. turcica chromosome but that is absent in the Borrelia A-FGy1 chromosome as in other Borrelia chromosomes (Fig. S1). Likewise, some Borrelia A-FGy1 plasmid contigs were similar in organization to other plasmids common to all Borrelia groups: Indeed, the Borrelia p6A-FGy1 plasmid contig was largely collinear with linear plasmids of B. tachyglossi (lp25), B. turcica (lp35), RF species (e.g., B. miyamotoi [lpB] and B. hermsii [lp53]), and with the circular LD plasmid cp26 (Fig. S2A). Only one Borrelia A-FGy1 plasmid contig (p7A-FGy1) was partly collinear with the large linear plasmids (megaplasmids) known exclusively from Borrelia species of the third and RF groups. A few plasmid contigs (e.g., p11A-FGy1), however, were highly syntenic with LD, but not RF, Borrelia plasmids (Fig. S2B). Another Borrelia A-FGy1 plasmid contig, p9A-FGy1, was very similar to the circular plasmid cp33 of Borrelia turcica that is absent in all other Borrelia species. Several large Borrelia A-FGy1 plasmid contigs (e.g., p1A-FGy1, p2A-FGy1, p3A-FGy1, p4A-FGy1 and p5A-FGy1) did not share any common genetic architecture with any previously known Borrelia plasmids. However, genes located on these plasmids were homologous to either LD or RF Borrelia genes.
Based on gene orthologs, the pan-genome (including the linear chromosome and all plasmids) of third Borrelia group members (B. turcica and B. tachyglossi along with Borrelia A-FGy1) shared 806 genes while the core genome of the genus Borrelia contained 638 genes, including 590 on the chromosome and 148 on plasmids (Fig. 5). Overall, the third Borrelia group genomes shared 8 genes (kduD, involved in D-galacturonate catabolic process, cof, involved in thiamine biosynthetic process, and 6 hypothetical protein genes) that were not present in the LD and RF Borrelia genomes (Table S3). Borrelia A-FGy1 however contained 168 unique genes (including a variable large protein vlp gene, two iron-sulfur cluster carrier protein genes, one IS200/IS605 family transposase and many hypothetical protein genes), mostly located on plasmids and not present in other Borrelia genomes (Table S3). Some genes only present in members of the third Borrelia group were absent in Borrelia A-FGy1: this includes a pair of genes involved in maltose metabolism (glvA and glvC) which are inserted within the rRNA operon of B. turcica and B. tachyglossi but that are absent in Borrelia A-FGy1. The Borrelia A-FGy1 chromosome also harbors a conserved RF-like gene architecture of the rRNA operon: there is only one copy each of the 23 S rRNA and 5 S rRNA genes (duplicated in LD Borrelia), and a horizontally acquired set of three purine salvaging pathway genes (purA, purB, and htp) that are inserted between the 23 S rRNA and 16 S rRNA genes (present in RF but absent in LD Borrelia species). The Borrelia A-FGy1 genome also contains 19 unique orthologs with B. tachyglossi, B. turcica and all RF Borrelia that are absent in all LD species, including genes involved in important cellular functions such as glycerophospholipid metabolism (glpT, glpQ) and DNA repair (RecF, RecR) (Table S3). Finally, the Borrelia A-FGy1 genome also contains genes commonly found in LD Borrelia genomes but that are absent from all RF Borrelia genomes: these genes include the ATP-dependent DNA helicase PcrA gene, a tRNAMet(CAT) gene and several hypothetical protein genes (Table S3).
Whole-genome comparison of the pan-genome of the 19 Borrelia species and strains used in Fig. 4. Presence of a gene in a genome is indicated in black. The core genome of 638 genes and the genes unique to LD, RF, and third groups are clustered together in the heat map.
The Borrelia A-FGy1 genome harbors a number of previously described genes encoding immune-reactive Borrelia proteins, including the Flagellin (flaB) and Borrelia membrane protein A (BmpA) genes located on the linear chromosome and an outer membrane protein OspC homolog gene located on plasmid. Three Borrelia plasmid contigs contain variable large protein (vlp) and variable small protein (vsp) genes homologous to those used in the multiphasic antigen variation system of RF Borrelia to evade detection by the host immune system. Three Borrelia A-FGy1 plasmid contigs contained vlp genes: Borrelia p6A-FGy1, p33A-FGy1, and p35A-FGy1, which contained one vlp gene each. One vsp gene was identified on the Borrelia p6A-FGy1 plasmid contig and shared high homology to B. turcica vsp35, B. hermsii vsp24, B. turicatae vspB, and OspC from LD Borrelia species.
Proposal of candidate name
On account of these distinct and coherent microbiological, phylogenetic, and evolutionary traits described above, we propose the designation “Candidatus Borrelia mahuryensis” for this novel bacterium associated with the passerine-associated ticks A. longirostre and A. geayi. The specific name refers to Mount Mahury, French Guiana, which was the first sampling site where we detected the presence of this bacterium.
Discussion
We found that a novel Borrelia species, Cand. Borrelia mahuryensis, divergent from the LD and RF Borrelia species, is common in passerine-associated ticks in tropical rainforests of French Guiana. We obtained a pure culture isolate of Cand. Borrelia mahuryensis (A-FGy1 isolate) that is morphologically similar to those of other Borrelia species. This novel Borrelia species is more closely related, although distinct, to the two known species of the third Borrelia group, B. turcica and B. tachyglossi. At the five loci extensively examined, only one SNP was observed between all our Cand. Borrelia mahuryensis DNA samples, showing that at least two very closely related strains are circulating in French Guiana. The recent detection of Borrelia isolates in Brazil17 sharing the same flaB sequence with Cand. Borrelia mahuryensis, along with the presence of closely related strains in Texas18, suggests that Cand. Borrelia mahuryensis may have a broad geographic distribution across American countries, with a substantial intraspecific variation. Finally, examination of the Cand. Borrelia mahuryensis A-FGy1 genome confirmed its difference from other Borrelia species, but revealed the presence of shared features with either LD or RF or both Borrelia species.
The repeated detection of Cand. Borrelia mahuryensis from 2012 to 2018 confirms that infection persists durably in French Guiana through its circulation in at least two tick species, A. longirostre and A. geayi. Infected tick specimens were collected from eight passerine species, suggesting that these vertebrates can be natural hosts for Cand. Borrelia mahuryensis. Interestingly, the related Borrelia strains detected in Brazil were also detected in A. longirostre16 collected from ground-dwelling birds, and those detected in the United States were detected in A. maculatum15,18, a species that also parasitizes birds among other hosts22,23. All of this evidence supports the hypothesis of birds as natural hosts for Cand. Borrelia mahuryensis. However, A. longirostre, A. geayi, and A. maculatum are also known to be associated with a variety of vertebrate species, with the immature stages usually feeding on passerine birds and the adults feeding instead on arboreal mammals such as new world porcupines and sloths (for A. longirostre and A. geayi)24,25 or large mammals such as cattle (for A. maculatum)22,23. Natural hosts of Cand. Borrelia mahuryensis can thus be either passerine birds, or mammals, or both. The observation that tick larvae are commonly infected suggests the existence of two non-exclusive scenarios: (i) passerine birds are the hosts, with tick larvae acquiring Borrelia through feeding on infected passerine birds; (ii) mammals are the hosts, with tick females acquiring Borrelia through feeding on infected mammals and further transmitting the infection transovarially to their larvae. Transovarial transmission, however, is not a common feature in the Borrelia genus since it has been demonstrated often for RF, but rarely for LD, species26,27,28,29. Interestingly, the isolation of Cand. Borrelia mahuryensis in culture from a freshly molted tick demonstrates that infection is maintained through molting to subsequent developmental stages in the tick host. Thus, through this transstadial transmission, infected ticks have the potential to infect animals on which they subsequently feed, including humans on whom A. longirostre and A. maculatum can occasionally feed22,23,30,31. Furthermore, while both A. longirostre and A. geayi are native to South and Central America24,32,33,34, migratory birds also regularly introduce A. longirostre to North America32,35,36,37,38,39. Bird migration (for A. longirostre, A. geayi, and A. maculatum) and cattle transportation (for A. maculatum) may be two important factors affecting the distribution of Cand. Borrelia mahuryensis over long distances and across geographical barriers, thereby explaining its wide geographic distribution.
The genome of Cand. Borrelia mahuryensis is more similar to those of the Borrelia third group, B. tachyglossi and B. turcica, which were also recently sequenced19. Members of this group shared similar gene content, including specific genes not present in LD or RF Borrelia. When compared with LD or RF Borrelia species, the genome of Cand. Borrelia mahuryensis shows more similarities with RF Borrelia species and harbors genes not found in LD Borrelia. However, it also harbors LD-specific genes and plasmids, showing that Cand. Borrelia mahuryensis exhibits intermediate features between the RF and LD groups. The Cand. Borrelia mahuryensis genome encodes several immunogenic vlp and vlp proteins (used in the multiphasic antigen variation system of RF Borrelia to evade detection by the host immune system40) as well as a number of immune-reactive proteins, including flab and BmpA, which are used for the diagnosis of LD41. Altogether, these genomic features suggest that Cand. Borrelia mahuryensis and other members of the third Borrelia group form a continuum of Borrelia species between the LD and RF groups, affecting our ability to clearly distinguish between these two groups.
To conclude, the description of Cand. Borrelia mahuryensis shows that the third Borrelia group is both widespread and biologically diverse. Most of the known members of this group were found in association with reptile hosts10,13,14,20,21, with the exception of one echidna host11,12. However, the description of Cand. Borrelia mahuryensis now suggests that members of the third Borrelia group have a large host spectrum that may also include a variety of birds and mammals as natural hosts. It is perhaps not unexpected to find other members of the third Borrelia group infecting a wide diversity of vertebrates. They have RF-like genomes with several housekeeping and macronutrient metabolism genes only present in the RF Borrelia species. Paralogous vlp and vsp proteins playing key roles in the RF multiphasic antigenic variation system and pathogenicity are also conserved, but Cand. Borrelia mahuryensis also have LD-specific orthologs. Future studies should give pivotal clues about the biology of Cand. Borrelia mahuryensis, as recently done through in vitro experiments with other members of the third Borrelia group: Indeed, variable levels of B. turcica resistance to vertebrate serum suggest that tortoises are reservoir host species while birds or humans are not42. Additional studies of Cand. Borrelia mahuryensis are needed to determine its transmission cycle and to establish whether these bacteria are pathogenic for birds and mammals, including humans.
Material and methods
Study area and tick sampling
Birds were captured using mist nets during the dry season (2012–2018) at 16 sites within forest patches (Table S1 and Fig. 1). Before releasing the birds, ticks were collected with fine forceps and immediately stored in 75% ethanol until examination and molecular screening (n = 290). An additional batch of ticks (n = 19) were collected from the Rémire-Montjoly site (site 14B in Table S1) and were taken alive to the laboratory for Borrelia cultivation (see below). All ticks were morphologically identified to species level using morphological and genetic diagnostic criteria25. The global connectivity between tick and bird species was visualized using the network analysis software package Gephi43.
Molecular screening and typing
Tick whole-body DNA was extracted using an extraction kit according to the manufacturer’s instructions (Qiagen). The presence of Borrelia was further examined through DNA barcoding involving the production of PCR amplicons from a 251-bp portion of the V4 variable region of the bacterial 16 S rDNA using a Multiplex PCR Kit (Qiagen) and universal primers (16 SV4F: 5′-GTGCCAGCMGCCGCGGTAA-3′ and 16SV4R: 5′-GGACTACHVGGGTWTCTAATCC-3′)44 as previously described45. Amplified bacterial 16S rDNA products were purified and sequenced using an Illumina MiSeq platform (GenSeq, Montpellier University) and 250-bp end sequence reads were obtained. All bioinformatic analyses were conducted using the pipeline Frogs46 as previously described45. Sequences with 97% similarity were clustered together and identified as an operational taxonomic unit (OTU). Each representative OTU sequence was aligned and taxonomically assigned using the Silva database (https://www.arb-silva.de/).
Independent PCR assays for Borrelia species identification were performed through the amplification of flaB, gyrB, groEL, glpQ, and 16 S rRNA gene fragments using specific primers (Table S4). All PCR products were visualized via electrophoresis in a 1.5% agarose gel. Positive PCR products were purified and sequenced in both directions using Sanger method (Eurofins). Sequence chromatograms were manually cleaned with CHROMAS LITE (http://www.technelysium.com.au/chromas_lite.html), and alignments were performed using CLUSTALW47, implemented in the MEGA V7 software48.
Borrelia cultivation
Engorged tick specimens (n = 19) were taken alive to the laboratory for blood meal digestion. Ticks were kept in humidified chamber (80–90% relative humidity) until molting (only three remained after molting: one female and two nymphs). Freshly molted ticks (n = 3) were further surface-sterilized with a bleach solution, rinsed with PBS before being individually cut into two parts. The two parts of the ticks were transferred together into a 6 ml-tube to isolate borreliae that was further cultured at 34 °C in BSK-II modified medium (6% rabbit serum, 6% gelatin, 30 µg rifampicin) in anaerobic conditions49 for several weeks and regularly examined under dark-field microscopy. Presence of Borrelia in remnants of the ticks was investigated through a specific PCR assay targeting the flab gene (using primers listed in Table S4).
Genome sequencing and analyses
One Borrelia-positive culture was used to prepare DNA-seq libraries using the Illumina Nextera DNA Flex sample preparation kit. Library validity was assessed by quantification using a Fragment Analyzer and a Qubit (Invitrogen). DNA-seq experiments were performed on an Illumina MiniSeq via a platform (MGX, Montpellier) using Illumina MiniSeq Mid Output Reagent Cartridge with a paired-end read length of 2×150 bp. A total of 6 GB of data were obtained. The quality of Illumina reads was analyzed with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and reads were further cleaned and trimmed using Trimmomatic50. The remaining reads were assembled into contigs and then into scaffolds with the SPAdes v3.8 assembler51. The annotation was performed via the NCBI prokaryotic genome annotation pipeline52. Dot plot analyses were done using YASS53. Whole-genome alignments were performed using Mauve54.
Pangenomic analysis (presence/absence of genes in the 19 complete genomes) was conducted with prokka55 and roary56. Roary was run with default parameters that are well tailored to infer of the most accurate number of genes56. Paralogs were then split according to the authors methodology. Briefly, if multiple genes from the same sample were found in an orthologs cluster, the neighborhood −5 genes up and downstream - of putative paralogs are used to infer information about synteny and split the cluster accordingly. This can be problematic for mis-assembled contigs or for multigenic family repeated in tandem, but allow genes with similar sequence to be placed in different cluster if they lie in different genomic position.
Molecular phylogenetic analyses
For analyses of single gene sequences, the GBLOCKS program57 with default parameters was used to remove poorly aligned positions and to obtain unambiguous sequence alignments. Closely related organisms obtained from GenBank were also included in the analyses. The evolutionary models that best fit the sequence data were determined using the Akaike information criterion with the program MEGA v748. Tree-based phylogenetic analyses were performed using maximum likelihood (ML) analyses. ML heuristic searches using a starting tree obtained by neighbor joining were conducted in MEGA v748. Clade robustness was assessed by bootstrap analysis using 1,000 replicates.
A phylogenomic approach was followed using ITEP58. All other complete Borrelia genomes were obtained from GenBank. Multiple orthologs were aligned with MAFFT59 v7. The concatenated multiple alignment was cleaned with trimal. The phylogenetic tree was computed by RaxML v8.2.460 using an ML approach with a GAMMA-LG model and 1,000 bootstrap replicates.
Ethics approval
All animals were handled in strict accordance with good animal practice as defined by the French code of practice for the care and use of animals for scientific purposes, established by articles R214-87 to R214-137 of the French rural code. All captures were performed by competent people without causing avoidable pain, suffering, distress, or lasting harm to the birds. Bird sampling was done under permission granted by several organizations: the Direction de l’Environnement, de l’Aménagement et du Logement (DEAL) from Guyane, the Direction Régionale de l’Office National des Forêts (ONF) de Guyane, the Conservatoire du Littoral (CEL) de Guyane, the Collectivité Territoriale de Guyane (CTG), the Centre National d’Etudes Spatiales (CNES), the Centre Spatial Guyannais (CSG). The use of the genetic resources was declared to the French Ministry of the Environment (reference TREL1902817S/156), in compliance with the Access and Benefit Sharing procedure implemented by the Loi pour la Reconquête de la Biodiversité.
Data availability
The genome of Cand. Borrelia mahuryensis A-FGy1 has been deposited at GenBank under the accession number #SAMN12690807.
References
Ogden, N. H., Artsob, H., Margos, G. & Tsao, J. Non-ricketsial tick-borne bacteria and the diseases they cause. in Biology of Ticks 278–312 (Edited by Sonenshine, D. E., New York, NY: Oxford University Press, 2014).
Talagrand-Reboul, E., Boyer, P. H., Bergström, S., Vial, L. & Boulanger, N. Relapsing fevers: neglected tick-borne siseases. Front Cell Infect Microbiol 8, 98 (2018).
Nelson, C. A. et al. Incidence of clinician-diagnosed Lyme disease, United States, 2005–2010. Emerging Infect. Dis. 21, 1625–1631 (2015).
Hinckley, A. F. et al. Lyme disease testing by large commercial laboratories in the United States. Clin. Infect. Dis. 59, 676–681 (2014).
Lopez, J. E., Krishnavahjala, A., Garcia, M. N. & Bermudez, S. Tick-borne relapsing fever spirochetes in the Americas. Vet Sci 3 (2016).
Ras, N. M. et al. Phylogenesis of relapsing fever Borrelia spp. Int. J. Syst. Bacteriol. 46, 859–865 (1996).
Takano, A. et al. Multilocus sequence typing implicates rodents as the main reservoir host of human-pathogenic Borrelia garinii in Japan. J Clin Microbiol 49, 2035–2039 (2011).
Adeolu, M. & Gupta, R. S. A phylogenomic and molecular marker based proposal for the division of the genus Borrelia into two genera: the emended genus Borrelia containing only the members of the relapsing fever Borrelia, and the genus Borreliella gen. nov. containing the members of the Lyme disease Borrelia (Borrelia burgdorferi sensu lato complex). Antonie Van Leeuwenhoek 105, 1049–1072 (2014).
Barbour, A. G., Adeolu, M. & Gupta, R. S. Division of the genus Borrelia into two genera (corresponding to Lyme disease and relapsing fever groups) reflects their genetic and phenotypic distinctiveness and will lead to a better understanding of these two groups of microbes (Margos et al. (2016). There is inadequate evidence to support the division of the genus Borrelia. Int. J. Syst. Evol. Microbiol. 67, 2058–2067, https://doi.org/10.1099/ijsem.0.001717) (2017). International Journal of Systematic and Evolutionary Microbiology.
Güner, E. S. et al. Borrelia turcica sp. nov., isolated from the hard tick Hyalomma aegyptium in Turkey. Int. J. Syst. Evol. Microbiol. 54, 1649–1652 (2004).
Loh, S.-M. et al. Novel Borrelia species detected in echidna ticks, Bothriocroton concolor, in Australia. Parasit Vectors 9, 339 (2016).
Loh, S.-M., Gillett, A., Ryan, U., Irwin, P. & Oskam, C. Molecular characterization of ‘Candidatus Borrelia tachyglossi’ (family Spirochaetaceae) in echidna ticks, Bothriocroton concolor. Int. J. Syst. Evol. Microbiol. 67, 1075–1080 (2017).
Panetta, J. L. et al. Reptile-associated Borrelia species in the goanna tick (Bothriocroton undatum) from Sydney, Australia. Parasit Vectors 10, 616 (2017).
Takano, A. et al. Isolation and characterization of a novel Borrelia group of tick-borne borreliae from imported reptiles and their associated ticks. Environ. Microbiol. 12, 134–146 (2010).
Mitchell, E. A. et al. Frequency and distribution of rickettsiae, borreliae, and ehrlichiae detected in human-parasitizing ticks, Texas, USA. Emerg Infect Dis 22, 312–315 (2016).
Pacheco, A. et al. Hemoparasites in ticks of wild birds of Serra dos Órgãos National Park, state of Rio de Janeiro, Brazil. Rev Bras Parasitol Vet 28, 238–244 (2019).
Cicuttin, G. L., De Salvo, M. N., Venzal, J. M. & Nava, S. Borrelia spp. in ticks and birds from a protected urban area in Buenos Aires city, Argentina. Ticks and Tick-borne Diseases 10, 101282 (2019).
Lee, J. K. et al. Detection of a Borrelia species in questing Gulf Coast ticks, Amblyomma maculatum. Ticks Tick Borne Dis 5, 449–452 (2014).
Gofton, A. W. et al. Genome-wide analysis of Borrelia turcica and ‘Candidatus Borrelia tachyglossi’ shows relapsing fever-like genomes with unique genomic links to Lyme disease Borrelia. Infect. Genet. Evol. 66, 72–81 (2018).
Kaenkan, W. et al. Reptile-associated Borrelia spp. In Amblyomma ticks, Thailand. Ticks and Tick-borne Diseases 11, 101315 (2020).
Trinachartvanit, W. et al. Borrelia sp. phylogenetically different from Lyme disease- and relapsing fever-related Borrelia spp. in Amblyomma varanense from Python reticulatus. Parasit Vectors 9, 359 (2016).
Teel, P. D., Ketchum, H. R., Mock, D. E., Wright, R. E. & Strey, O. F. The Gulf Coast tick: a review of the life history, ecology, distribution, and emergence as an arthropod of medical and veterinary importance. J. Med. Entomol. 47, 707–722 (2010).
Wilson, N. & Durden, L. A. Ectoparasites of terrestrial vertebrates inhabiting the Georgia Barrier Islands, USA: an inventory and preliminary biogeographical analysis. Journal of Biogeography 30, 1207–1220 (2003).
Floch, H. & Fauran, P. Ixodides de la Guyane et des Antilles Françaises. Publ Inst Pasteur Guyane Fr Inini 19, 1–94 (1958).
Binetruy, F., Chevillon, C., de Thoisy, B., Garnier, S. & Duron, O. Survey of ticks in French Guiana. Ticks and Tick-borne Diseases 10, 77–85 (2019).
Richter, D., Debski, A., Hubalek, Z. & Matuschka, F.-R. Absence of Lyme disease spirochetes in larval Ixodes ricinus ticks. Vector Borne Zoonotic Dis. 12, 21–27 (2012).
Rollend, L., Fish, D. & Childs, J. E. Transovarial transmission of Borrelia spirochetes by Ixodes scapularis: a summary of the literature and recent observations. Ticks Tick Borne Dis 4, 46–51 (2013).
Scoles, G. A., Papero, M., Beati, L. & Fish, D. A relapsing fever group spirochete transmitted by Ixodes scapularis ticks. Vector Borne Zoonotic Dis. 1, 21–34 (2001).
van Duijvendijk, G. et al. Larvae of Ixodes ricinus transmit Borrelia afzelii and B. miyamotoi to vertebrate hosts. Parasit Vectors 9, 97 (2016).
Guglielmone, A. A. et al. Ticks (Ixodidae) on humans in South America. Experimental and Applied Acarology 40, 83–100 (2006).
Nava, S., Velazco, P. M. & Guglielmone, A. A. First record of Amblyomma longirostre (Koch, 1844) (Acari: Ixodidae) from Peru, with a review of this tick’s host relationships. Systematic and Applied Acarology 15, 21–30 (2010).
Jones, E. K., Clifford, C. M., Keirans, J. E. & Kohls, G. M. Ticks of Venezuela (Acarina: Ixodoidea) with a key to the species of Amblyomma in the Western Hemisphere. Brigham Young University Science Bulletin, Biological Series 17 (1972).
Ogrzewalska, M., Uezu, A. & Labruna, M. B. Ticks (Acari: Ixodidae) infesting wild birds in the eastern Amazon, northern Brazil, with notes on rickettsial infection in ticks. Parasitol. Res. 106, 809–816 (2010).
Ogrzewalska, M. et al. Ticks (Acari: Ixodidae) infesting birds in an Atlantic rain forest region of Brazil. J. Med. Entomol. 46, 1225–1229 (2009).
Mukherjee, N. et al. Importation of exotic ticks and tick-borne spotted fever group rickettsiae into the United States by migrating songbirds. Ticks Tick Borne Dis 5, 127–134 (2014).
Noden, B. H., Arnold, D. & Grantham, R. First report of adult Amblyomma longirostre (Acari: Ixodidae) in Oklahoma. Systematic and Applied Acarology 20, 468–470 (2015).
Durden, L. A. & Kollars, J. T. M. An annotated list of the ticks (Acari: Ixodoidea) of Tennessee, with records of four exotic species for the United States. Bulletin of the Society of Vector Ecology 17, 125–131 (1992).
Hamer, S. A. et al. Wild birds and urban ecology of ticks and tick-borne pathogens, Chicago, Illinois, USA, 2005–2010. Emerging Infect. Dis. 18, 1589–1595 (2012).
Scott, J. D. et al. Birds disperse ixodid (Acari: Ixodidae) and Borrelia burgdorferi-infected ticks in Canada. J. Med. Entomol. 38, 493–500 (2001).
Barbour, A. G., Dai, Q., Restrepo, B. I., Stoenner, H. G. & Frank, S. A. Pathogen escape from host immunity by a genome program for antigenic variation. Proc. Natl. Acad. Sci. USA 103, 18290–18295 (2006).
Cook, M. J. & Puri, B. K. Commercial test kits for detection of Lyme borreliosis: a meta-analysis of test accuracy. Int J Gen Med 9, 427–440 (2016).
Hepner, S. et al. First investigations on serum resistance and sensitivity of Borrelia turcica. Ticks and Tick-borne Diseases 10, 1157–1161 (2019).
Bastian, M., Heymann, S. & Jacomy, M. Gephi. An open source software for exploring and manipulating networks. International AAAI Conference on Weblogs and Social Media 2 (2009).
Galan, M. et al. 16S rRNA amplicon sequencing for epidemiological surveys of bacteria in wildlife. mSystems 1 (2016).
Binetruy, F., Dupraz, M., Buysse, M. & Duron, O. Surface sterilization methods impact measures of internal microbial diversity in ticks. Parasites & Vectors 12, 268 (2019).
Escudié, F. et al. FROGS: Find, Rapidly, OTUs with Galaxy Solution. Bioinformatics 34, 1287–1294 (2018).
Thompson, J. D., Gibson, T. J. & Higgins, D. G. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2, Unit 2, 3 (2002).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Barbour, A. G. Isolation and cultivation of Lyme disease spirochetes. Yale J Biol Med 57, 521–525 (1984).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Bankevich, A. et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19, 455–477 (2012).
Tatusova, T. et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44, 6614–6624 (2016).
Noé, L. & Kucherov, G. YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Res. 33, W540–543 (2005).
Darling, A. C. E., Mau, B., Blattner, F. R. & Perna, N. T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14, 1394–1403 (2004).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Page, A. J. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015).
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17, 540–552 (2000).
Benedict, M. N., Henriksen, J. R., Metcalf, W. W., Whitaker, R. J. & Price, N. D. ITEP: An integrated toolkit for exploration of microbial pan-genomes. BMC Genomics 15, 8 (2014).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Acknowledgements
This work has benefited from an “Investissement d’Avenir” grant managed by Agence Nationale de la Recherche (CEBA, ref. ANR-10-LABX-25-01, with the MicroBIOMES Strategic project 2016–2018 and the MiTick Annual project 2016) and recurrent funding from CNRS and IRD, the ERA-Net Net-Biome 2010, the Agence Nationale de la Recherche, the Conseil Régional de Guyane, the Conseil Régional de Bourgogne, and the Observatoire des Sciences de l’Univers Theta de Franche-Comté-Bourgogne. FB benefits from a PhD fellowship financed by the CEBA Laboratoire d’Excellence and Montpellier University. We are grateful to Groupe d'Étude et de Protection des Oiseaux en Guyane (GEPOG), Vincent Pelletier, Tanguy Deville, Maxime Loubon, Frank Théron, Gilles Leblond, Denis Roussel, Aurélie Khimoun, Antoine Perrin, and Renaud Scheifler for their precious help in tick sampling. We are grateful to Rolland Ruffine and to members of Institut Pasteur de Guyane, in particular to Agathe Chavy and Benoit de Thoisy, for their help. We are also grateful to the technicians of the Centre National de Référence Borrelia (Strasbourg, France) for their help in DNA isolation. Data used in this work were (partly) produced through the MGX core facilities supported by France Génomique National infrastructure and through GenSeq technical facilities of the « Institut des Sciences de l’Evolution de Montpellier » with the support of LabEx CeMEB. The authors acknowledge the IRD i-Trop HPC (South Green Platform) at IRD Montpellier for providing HPC resources that have contributed to the research results reported within this paper. We also acknowledge useful discussions with members of the working group Tiques et Maladies à Tiques (TMT) from the Réseau Ecologique des Interactions Durables (REID).
Author information
Authors and Affiliations
Contributions
F.B. and O.D. designed the study and wrote the manuscript. F.B., S.G., B.F. and O.D. collected the samples. F.B., M.B. and R.B. performed the molecular typing. N.B. and E.T.R. performed Borrelia cultivation and DNA isolation. F.B., E.L., V.N. and O.D. performed the phylogenetic and genomic analyses. All authors agreed on the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41598_2020_66828_MOESM2_ESM.mp4
Movie S1. Microscopy movie of the culture of Cand. Borrelia mahuryensis A-FGy1. Three motile bacteria are apparent in this low-density culture medium.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Binetruy, F., Garnier, S., Boulanger, N. et al. A novel Borrelia species, intermediate between Lyme disease and relapsing fever groups, in neotropical passerine-associated ticks. Sci Rep 10, 10596 (2020). https://doi.org/10.1038/s41598-020-66828-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-66828-7
This article is cited by
-
Molecular pathogen screening of louse flies (Diptera: Hippoboscidae) from domestic and wild ruminants in Austria
Parasites & Vectors (2023)
-
Characterisation and comparative genomics of three new Varanus-associated Borrelia spp. from Indonesia and Australia
Parasites & Vectors (2023)
-
Comparative genomics of the Western Hemisphere soft tick-borne relapsing fever borreliae highlights extensive plasmid diversity
BMC Genomics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.