Abstract
Family studies have identified a heritable component to self-harm that is partially independent from comorbid psychiatric disorders. However, the genetic aetiology of broad sense (non-suicidal and suicidal) self-harm has not been characterised on the molecular level. In addition, controversy exists about the degree to which suicidal and non-suicidal self-harm share a common genetic aetiology. In the present study, we conduct genome-wide association studies (GWAS) on lifetime self-harm ideation and self-harm behaviour (i.e. any lifetime self-harm act regardless of suicidal intent) using data from the UK Biobank (n > 156,000). We also perform genome wide gene-based tests and characterize the SNP heritability and genetic correlations between these traits. Finally, we test whether polygenic risk scores (PRS) for self-harm ideation and self-harm behaviour predict suicide attempt, suicide thoughts and non-suicidal self-harm (NSSH) in an independent target sample of 8,703 Australian adults. Our GWAS results identified one genome-wide significant locus associated with each of the two phenotypes. SNP heritability (hsnp2) estimates were ~10%, and both traits were highly genetically correlated (LDSC rg > 0.8). Gene-based tests identified seven genes associated with self-harm ideation and four with self-harm behaviour. Furthermore, in the target sample, PRS for self-harm ideation were significantly associated with suicide thoughts and NSSH, and PRS for self-harm behaviour predicted suicide thoughts and suicide attempt. Follow up regressions identified a shared genetic aetiology between NSSH and suicide thoughts, and between suicide thoughts and suicide attempt. Evidence for shared genetic aetiology between NSSH and suicide attempt was not statistically significant.
Similar content being viewed by others
Introduction
Every year nearly one million people take their own lives1, making suicide a pressing issue of considerable social and economic burden. Moreover, self-harm behaviours are now recognized by the American Psychiatric Association as independent conditions for further study. Namely, non-suicidal self-injury and suicidal behaviour disorder were recently introduced in the section 3 of the Diagnostic and Statistical Manual of Mental Disorders (DSM-V)2. The lifetime prevalence estimate for suicide thoughts is ~10%, while suicide attempt and non-suicidal self-harm (NSSH) affect ~2.5 and ~5% of the population, respectively3,4,5. Higher rates have been reported amongst children and adolescents6. The key difference between suicidal and non-suicidal self-harm is that the former implies an intent to die as a consequence of the act. Non-suicidal self-harm acts include equally dangerous behaviours such as cutting, burning or poisoning, but are underlined by a different motivation such as seeking attention or the desire to feel pain.
Twin and family studies indicate that NSSH, suicide thoughts and suicide attempt are moderately heritable7,8. Multiple studies have documented that the presence of a psychiatric disorder considerably increases the risk for both suicidal and non-suicidal self-harm9,10,11 but a sizeable genetic component (33-51% of variance) of suicide risk is not explained by underlying psychiatric conditions12,13,14,15,16. Importantly, controversy exists about whether NSSH and suicide attempt are part of the same liability spectrum7,16,17,18,19,20. Notably, twin studies have identified a genetic correlation between suicide thoughts and NSSH7, but the extent to which shared genetic factors underlie NSSH, suicide thoughts and suicide attempt (i.e. a self-harm liability continuum hypothesis) still remains elusive. Investigating broad sense self-harm could improve our understanding of the aetiology and underpinnings of the liability to both suicidal and non-suicidal self-harm.
Although several genome-wide association studies (GWAS) on suicidality have been published to date21,22,23,24,25,26,27,28,29,30,31, robustly associated genetic variants are still elusive32. A number of studies assessing the genetic predictability of suicidality have suggested a shared aetiology between depression and suicidality, and suggestive evidence for suicidality-related associations32,33,34,35. Investigating the genetic architecture of self-harm regardless of suicidal intent, could be valuable to gain biological insights into the relationship between self-harm and suicide. In the present study, we explore the genetic aetiology of lifetime self-harm ideation and lifetime self-harm behaviour (regardless of suicidal intent) using a GWAS and PRS approach. An overview of the phenotypes and terminology used throughout this manuscript is available in Table 1.
Methods
Discovery sample
The discovery sample consisted of ~156,700 participants from the UK Biobank (as of February 2019) with self-harm data (lifetime history of self-harm ideation and self-harm behaviour)36. Self-harm was assessed as described previously37. DNA extraction and genotyping are described in ref. 38. Notably, genotyping was performed using two highly related arrays: the UK BiLEVE Axiom array and the UK Biobank Axion Array.
Briefly, the self-harm behaviour item was “Have you deliberately harmed yourself, whether or not you meant to end your life?” (No = 150,008, yes = 6,872). The self-harm ideation item was: “Have you contemplated harming yourself (for example by cutting, biting, hitting yourself or taking an overdose)?” (No = 133,524, yes = 23,192). Notably, both these phenotypes make no distinction between suicidal and non-suicidal self-harm. As such, they include both suicidal and non-suicidal self-harm. A liability threshold model (tetrachoric correlation; psych package in R) estimated the traits to be highly correlated ρ = 0.89 (95% c.i. 0.88–0.89; N reporting both ideation and behaviour = 6446).
GWAS
We conducted two GWAS, on lifetime self-harm ideation and lifetime self-harm behaviour. Association analyses were performed using BOLT_LMM39, based on a linear mixed model and allele dosages (of the effect allele based on imputed data) accounting for the first 20 genetic ancestry principal components, standard covariates (age, age2, sex, sex*age as fixed effect predictors), and correcting for cryptic relatedness and population stratification using a genetic relatedness matrix as the random effects variance covariance structure. Approximately 48,000 individuals of non-European ancestry were excluded from the analyses. A stringent, but standard quality-control (QC) protocol40 was applied: variants with low minor allele count (MAC < 25), low quality imputation (INFO < 0.8) or with a deviation from the Hardy-Weinberg equilibrium (HWEp < 1e-10) were excluded from further analyses.
Gene-based test analyses
Gene-based association analysis was conducted for self-harm ideation or self-harm behaviour using MAGMA41 as implemented on the FUMA web platform42. Briefly, SNPs were mapped to ~20,000 protein coding genes based on their genomic location. Then, the independent SNP association statistics were combined to yield gene-based mean χ2 statistics. Genome-wide significance level was defined as 2.652e-6 (Bonferroni corrected alpha <0.05).
PRS target sample
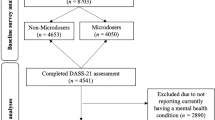
The target sample consisted of individuals from two cohorts of the Queensland Twin Registry. Individuals were recruited and participated in structured telephone or paper interviews assessing psychiatric disorders, substance abuse and living conditions. Detailed information on the cohorts has been published previously43,44. Items from the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) assessing self-harm behaviours were included in both cohorts, a detailed description of the items and their application is available in34. Briefly, participants were first asked whether they experienced any suicidal thoughts, then suicide attempt, and finally acts of self-harm not related to suicide attempts. The items used to determine lifetime prevalence of self-harm behaviours were: “Have you ever thought about taking your own life?”; “Have you ever tried to take your own life?” and “(Other than when you tried to take your own life) Did you ever hurt yourself on purpose, for example, by cutting or burning yourself?”.
The genotyping and quality control (QC) protocol for the target sample have been described previously34,45. Briefly, standard protocols for DNA collection and extraction were used. Genotyping was carried out using commercial Illumina SNP arrays. Platform specific QC was performed including: Hardy Weinberg equilibrium deviation, individual SNP call rates, minor allele frequency threshold, and removal of population outliers (i.e. non-European ancestry as determined by principal component analysis). Genotype data were imputed using the haplotype reference consortium (HRC) reference panel.
SNP heritability and genetic correlation
The amount of variance in risk explained by SNP effect sizes (i.e. the SNP heritability or hsnp2) was calculated using LD-score regression as previously described46. The software ldsc v 1.0.0 was used to calculate both the hsnp2 and the genetic correlation between both GWAS summary statistics. This method relies on the relationship between the non-centrality parameter (NCP or χ2) of GWAS results and LD scores (the sum of LD-r2 of a SNP against all other SNPs on the same population) expected under a polygenic assumption (the bigger the LD score, the more likely to tag a causal variant)47. The fact that the expected value of the NCP for a given SNP is a function of the genetic covariance (hg2) of the trait and the LDscore of that SNP, allow us to estimate the SNP based heritability46 (hsnp2) and co-heritability (rg)48 of a set of traits given their summary statistics and known LD patterns. As the population prevalence of broad sense self-harm behaviours on the UK has not been reported, we assumed the population prevalence to be equal to the discovery sample prevalence when transforming to the liability scale using ldsc and thus estimates of hsnp2 should be referred to with caution. Genetic correlations between the traits under study and other traits and diseases (~760 traits) were explored using the LD-Hub web platform46,49. We used a stringent definition of a significant genetic correlation (FDR < 0.01 using a Benjamini-Hochberg multiple testing correction, for a list of traits see Supplementary Data 1).
Polygenic risk scores and prediction analysis
To calculate the genetic predisposition (risk) of our target sample to the traits of interest, each variant’s effect size was obtained from the GWAS summary statistics. Our PRS estimation pipeline excluded indels, strand ambiguous- and low (R2 < 0.6) imputation quality-variants. The most significant independent SNPs were selected using a conservative clumping procedure (PLINK1.9; p1 = 1, p2 = 1, r2 = 0.1, kb=10000)50 to correct for inflation arising from linkage disequilibrium (LD). Eight different PRS were calculated for each individual using different p-value thresholds (p < 5 × 10−8, p < 1 × 10−5, p < 0.001, p < 0.01, p < 0.05, p < 0.1, p < 0.5, p < 1) as criteria for SNP inclusion on the PRS calculation. PRS were calculated using a dosage assumption, therefore multiplying the effect size of a given SNP by the imputed number of copies (using dosage probabilities) of the effect allele present in an individual. Finally, the SNP dosage effects were summed across all loci per individual.
To assess the association between the genetic liability to self-harm behaviour and self-harm ideation (i.e. the PRS) with actual self-harm phenotypes in the target sample, we employed a linear mixed model regression framework. Briefly, the PRS was added to the model as a predictor variable while accounting for sex, age, age2, sex*age, the first five genetic principal components and imputation run, an in-house set of variables that capture array and cohort differences34,51, as fixed effects. Correcting for relatedness is crucial when examining family cohorts. Given varying degrees of relatedness in our sample, we employed a linear mixed model using genetic restricted maximal likelihood (GREML) with a random effects variance covariance structure defined by the sample’s genetic relatedness matrix obtained from GCTA 1.91.752,53. This method has been previously used to deal with related individuals in related target samples of PRS studies51,54. A partial R2 was used to estimate the variance explained by the PRS using the formula:
where β represents the PRS fixed effect estimate, \({\sigma }_{pheno}\) the standard deviation of the phenotype and \({\sigma }_{PRS}\) the standard deviation of the PRS respectively. Statistical significance threshold was defined accounting for multiple testing using a matrix spectral decomposition approach55,56 to estimate the number of effective variables being tested. The final significance threshold was defined at α < =0.0064
As sensitivity analysis, we tested whether the liability to major depressive disorder was driving polygenic prediction. To this end, MDD-PRS were calculated using SBayesR57 based on available summary statistics leaving out the Australian sample58. The self-harm behaviour (and self-harm ideation) phenotype associations were reproduced with MDD-PRS as a covariate to identify the variance explained by self-harm behaviour (or self-harm ideation) over and above the effect of MDD liability.
Further, we were interested on whether the prediction of our PRS on the traits (Suicide attempt, suicide thoughts and NSSH) was due to shared or independent genetic factors. To this end, the significant associations between the PRS for self-harm ideation and suicide thoughts were reproduced including NSSH as a covariate. Likewise, the association between PRS for self-harm ideation and NSSH was reproduced with suicide thoughts as a covariate. The same approach was used for the self-harm behaviour PRS but using either suicide thoughts or suicide attempt (the phenotypes with some evidence of association) as a covariate. If the significant PRS-phenotype associations were driven through the same genetic components, adding one of the phenotypes as a covariate should implicitly capture the shared genetic predisposition, thus removing the observed association. Any residual prediction would imply that independent genetic factors, captured by our GWAS, are underlying each phenotype.
Results
Sample demographics and self-harm behaviours prevalence
The demographic composition for both the discovery and target samples are given in Tables 2 and 3 respectively. In the discovery sample, where genetic correlates of broad sense self-harm ideation and self-harm behaviour were assessed, males and females presented a similar age range, but females showed a higher prevalence of both self-harm ideation and self-harm behaviour. Prevalence of NSSH, suicide thoughts and suicide attempt on the target sample were 3.4, 26.4 and 3.8% respectively. Prevalence of suicide thoughts was slightly lower in the female subgroup while suicide attempt was higher (Table 3).
GWAS of broad sense self-harm behaviours
Two GWAS assessing self-harm ideation and self-harm behaviour were performed. After QC, the GWAS for self-harm ideation identified one genome-wide significant locus on chromosome five. The GWAS for self-harm behaviour presented one genome-wide significant hit on chromosome nine (Table 4 and Fig. 1). A gene-based association test identified seven significantly associated genes with self-harm ideation: SYT14, RPP14, FAM172A, SEMA3D, DCC, DDX27 and ZNFX1. For the GWAS on self-harm behaviour, four genes LINGO2, DCC, FBXO27 and WRB showed an association surpassing genome-wide significance (Fig. 2).
GWAS results and genetic correlations of broad sense self-harm thoughts and behaviours. Miami plot (left panel) depicts the genome-wide association results for the phenotypes studied. The x-axis represents the genomic position, while the y axis represents the significance of the association between each SNP and the phenotype; the top panel represents significance as –log10 (pvalue), while the bottom panel uses log10 (pvalue), in both cases, the farther from the x axis (middle) line, the more significant the association between the phenotype and the variant. On the right side, a heat map depicts the genetic correlations (rg) between published trait GWAS and our GWAS for self-harm ideation or self-harm behaviour. Only traits with a Benjamini-Hochberg fdr <0.01 for at least one phenotype and generated using studies independent from the UK-Biobank are depicted here (All the results, including UK-B traits, are available in Supplementary Data 1).
Gene based association. Manhattan plots depicting gene-based test results of the GWAS. The x-axis represents the genes genomic position, and the y axis the significance (−log10(p-value)) of the association between the genes and the studied phenotype. The phenotypes for the top and bottom panels are self-harm ideation and self-harm behaviour respectively.
Heritability and genetic correlation of broad sense self-harm behaviours and thoughts
The SNP heritability (hsnp2) on the liability scale for both traits was estimated to be 11.1% (SE = 1.7%) for self-harm behaviour and 10.1% (SE = 1.0%) for self-harm ideation. Further, the traits were highly genetically correlated with each other (rg = 0.85, p = 7.8e−53). High genetic correlations with psychiatric disorders such as anxiety, depression and schizophrenia, symptoms such as insomnia, and personality traits such as irritability, miserableness, mood swings, and risk-taking, among others, were identified for both traits (Fig. 1). Furthermore, a negative correlation with subjective well-being and age at first birth (i.e. the age in which a person has their first child) was observed (Fig. 1 and Supplementary Fig. 1). As expected from their high genetic correlation, the genetic correlations of self-harm ideation and self-harm behaviour across a range of available traits were highly similar (R > 0.8, Supplementary Fig. 2).
Polygenic risk score prediction
We calculated PRS on an independent sample of ~8,700 Australian adults. A summary of the PRS variables and results is available on Supplementary Tables S1 and S2. The PRS for self-harm ideation significantly predicted suicide thoughts (maximum variance explained 0.45%, p = 4.5e−6) and NSSH (maximum variance explained 0.27%, p = 3.6e−4) (Fig. 3). Notably, the associations were significant for PRS including variants with p value cut-offs <0.001 (or less stringent cut-offs) for suicide thoughts, and p < 0.01 (or less stringent cut-offs) for NSSH. The PRS for self-harm behaviour predicted suicide attempt (maximum variance explained = 0.20%) and suicide thoughts (maximum variance explained = 0.13%) (Fig. 3). Although the PRS for self-harm behaviour did not predict NSSH in our sample, the PRS for self-harm behaviour was predictive of broad sense self-harm (regardless of suicidal intent; maximum variance explained >0.30% p < 0.001). This association was also diminished when correcting for NSSH and further diminished when correcting for suicide attempt (Supplementary Fig. 3).
Polygenic prediction of self-harm behaviours. Bar plots represent the amount of variance explained by the polygenic risk scores on the self-harm phenotypes. The red colour (left side) shows the associations of the PRS for self-harm ideation whereas the blue colour (right side) depicts the associations of the PRS for self-harm behaviour. For each phenotype studied, the amount of variance explained by a PRS including variants with an increasingly liberal p-value threshold (from left to right) is shown. The bars are ordered based on the p-value cut-off used to construct the PRS (increasingly liberal p-values). The height of each bar represents the amount of variance explained. The p-value for the association between the PRS and the phenotype is shown with a colour scale. *Represents p < 0.05; **represents significant after multiple testing correction).
Sensitivity analyses
Given the high genetic correlation identified with depression, we performed a sensitivity analysis to assess whether depression specific genetic factors were driving the polygenic prediction. To this end, PRS for MDD were calculated (see methods), and the associations described above were reproduced but including MDD-PRS as a covariate. The overall variance explained was reduced. Nonetheless, there was still evidence for an association between self-harm behaviour PRS and suicide attempt or suicide thoughts; and between self-harm ideation PRS and suicidal thoughts and non-suicidal self-harm (Supplementary Fig. 4).
We also performed secondary analyses to test whether the associations between PRS and phenotypes were driven by shared genetic factors (see methods). The PRS for self-harm ideation significantly predicted suicide thoughts after including NSSH as a covariate; it also predicted NSSH after including suicide thoughts as a covariate, albeit with a smaller proportion of variance explained (~0.33% and ~0.15 respectively; Fig. 4). The PRS for self-harm behaviour continued to significantly predict suicidal attempts after including suicide thoughts as a covariate, but the association with suicide thoughts disappeared when suicidal attempt was included as a covariate in the model (Fig. 4).
Assessing the shared genetic aetiology of self-harm behaviours. The PRS-phenotype associations were repeated but accounting for the other significantly associated phenotype as a covariate (see methods). Panel (a) shows the result of PRS for self-harm ideation while accounting for suicide thoughts, (b) PRS for self-harm ideation while accounting for NSSH, (c) PRS for self-harm behaviour while accounting for suicide attempt and (d) PRS for self-harm behaviour while accounting for suicide thoughts. *p < 0.05, **significant after multiple testing correction.
Discussion
We explored the genetic architecture of broad sense self-harm by performing GWAS of self-harm ideation and self-harm behaviour in a population-based sample. The identification of only one genome-wide significant locus for each phenotype suggests that better powered genetic studies of self-harm are needed, and indicate that these traits are highly polygenic. Given the self-reported nature of the phenotypes, it is possible that recall bias and differences in subjective understanding of the mental health items could increase the noise to signal ratio in the discovery GWAS. Self-harm is a complex behaviour that encompasses subtypes with varying severities and recurrence rates59, which are not captured by the single item used to ascertain self-harm on the UKB.
We also identified eleven genome wide significant genes using gene-based association tests. Seven were associated with self-harm ideation and four with self-harm behaviour. The gene with the strongest association with self-harm ideation, and the only gene associated with both phenotypes, was DCC. DCC is a gene involved in prefrontal cortex innervation and development. This observation could be consistent with reports of structural abnormalities on the brains of suicidal subjects60,61. Consistent with our results, DCC has been independently linked to suicidal severity on the UK-B62 and there is evidence of elevated DCC expression in the prefrontal cortex of post-mortem brains of subjects that died by suicide63.
Regarding genes associated with self-harm ideation, FAM172A has been previously associated with differential methylation linked to childhood stress in girls64, and is located in a locus recently associated with insomnia65, a phenotype known to be associated with self-harm and suicidality66,67. Notably, previous studies suggest that the gene SEMA3D, known to be associated with schizophrenia68,69, could also be associated with suicidality24. A link between SYT14 and bipolar disorder has been reported70. DDX27 has been associated with intelligence71,72, and a study reporting a relationship between lower IQ and suicide attempt has been published73. Variants near DDX27 and ZNFX1 have been nominally linked to proneness to anger74. No obvious relationship between RPP14 and any psychiatric or behavioural phenotype has been reported in GWAS databases or the literature. Notably, we identified suggestive associations of a cluster of protocadherin genes (PCDH) on chromosome 5. PCDHAC1 is enriched in serotonergic cells in mice75 and PCDH-family differential methylation has been recently associated with early-onset major depression76 and previously associated with schizophrenia, bipolar disorder77 and autism78.
Three genes: LINGO2, FBXO27 and WRB were associated with self-harm behaviour. LINGO2 and its paralog LINGO1 have been linked to neurodegenerative and psychiatric disorders79,80. Notably, previous results suggest that differential methylation on the promoter of FBXO27 might be linked with childhood physical aggression81, which is known to be highly associated with suicidal behaviours82. While WRB has been linked to cognitive impairment, it is unclear how it relates to self-harm behaviour. Finally, we found evidence for a suggestive association between STK10 and self-harm behaviour. STK10 has been linked with childhood cognitive ability83 and observed to be hyper-hydroxy-methylated upon acute stress84. Altogether these results provide promising candidate genes associated with self-harm ideation and behaviour. Future analyses should focus on replicating these findings and assessing their underlying mechanistic and possible translational roles on self-harm.
While the SNP-heritability of self-harm ideation and behaviour was significant—and some proportion of variance on the studied phenotypes was explained by PRS—, the percentage of variance explained was still far from the heritability estimates for NSSH (h2~37–59%), suicidal ideation (h2~47–66%) and suicide attempt (h2~55%) reported in twin and family studies7,15,85. Although the UKB recruitment process does not represent a random sample of the UK population86—and there is evidence of genetic factors associated with completion of the mental health section87— our PRS results provide evidence that the genetic associations discovered have some predictive power (albeit still a small one) over self-harm related phenotypes on an independent population. The above observations call for novel, well powered genetic studies of self-harm which will be required in order to obtain accurate SNP effect sizes88. A recent study suggest that even after well powered GWA studies have been conducted, most of the missing heritability for a phenotype is tagged by variants with a low MAF that cannot be easily imputed89. Therefore, whole genome sequencing studies of self-harm and suicidality could be paramount to achieve a complete understanding of the genetic architecture underlying self-harm.
The fact that a PRS for self-harm thoughts was able to explain up to 0.27% of the variance of NSSH is consistent with a previous twin study reporting a significant co-heritability between suicide ideation and NSSH7. Further, the follow up regressions correcting for suicide thoughts when predicting NSSH, and for NSSH when predicting suicide thoughts showed a reduction of the amount of variance explained. This observation is consistent with a partial shared genetic aetiology between them. To the best of our knowledge, this is the first study to date to report on a positive genetic prediction of NSSH. Notably, a previous PRS study assessing depression and self-harm in our sample34 identified an association between the genetic predisposition for depression and suicidal ideation, but no robust association with suicide attempt or non-suicidal self-harm. Thus, two possible explanations exist: (i) the previously stated genetic7 and phenotypic90 correlations between suicide thoughts and NSSH should be explained by depression-independent genetic factors, or (ii) the study by Maciejewski et al. (2017) was underpowered, possibly due to the low accuracy of the available summary statistics at that time and the number of cases in the target population, which limited its ability to detect an association between NSSH and the MDD-PRS.
Our results support the existence of a genetic overlap between NSSH and suicide thoughts. We also detected genetic overlap between suicide thoughts and suicide attempt, which was evidenced by the association between the PRS for self-harm behaviour and suicide thoughts disappearing after correcting for suicide attempt. Finally, evidence of a shared genetic aetiology between suicide attempt and NSSH did not reach statistical significance, as no PRS simultaneously (robustly) predicted both of them. A possible explanation is that genetic predisposition to NSSH is not associated with suicide attempt predisposition, in spite of the known overlap between NSSH and suicidality11,91. Another plausible explanation is a lack of power either on the discovery (due to either number of cases or a high noise to signal ratio) or target samples to detect an overlap between NSSH and suicide attempt due to a low prevalence of both behaviours.
Self-harm behaviours are likely to share genetic aetiology with several other traits and disorders. We have detected some of these candidate traits, such as neuroticism, nervousness and a risk-taking personality by assessing their genetic correlation with self-harm thoughts and behaviours. Interestingly, a negative genetic correlation of both self-harm phenotypes with age at first birth was detected, and this was also observed for the most recent GWAS on depression58. Moreover, another study identified a negative correlation between maternal age at child birth and suicidality. Although the design of this study corrected for genetic confounding92, the association would support the hypothesis that genetic factors predisposing to changes in maternal age could impact on depression and therefore on suicidality and self-harm. The usage of PRS for independent traits to predict self-harm results in a tool of great power to test for pleiotropy and genetic overlap between them. For example, a significant genetic correlation of suicide attempt with insomnia has been reported35, and we have detected insomnia to also correlate with broad sense self-harm. The existence of GWAS summary statistics for insomnia65 makes this hypothesis testable using the approach implemented herein.
Some limitations relevant to this study must be acknowledged. First, this study focused only on a sample of European Ancestry, an approach that allows to avoid biases due to population stratification. However, this hinders our ability to extrapolate to other populations. Furthermore, the samples used in this study were mostly comprised of adults in their late 50 s for the UK-B or in their 40 s for the Australian sample. This limits our ability to understand adolescent and childhood related self-harm. This is important because children and adolescents present a higher prevalence of self-harm compared to adults. Moreover, self-harm is highly complex and heterogeneous93, comprising a variety of acts such as physical injury and poisoning. Additionally, the self-reported nature of the phenotypes assessed in this study, could be affected by participant specific recall bias, which would bias our results towards the null. In the present study, we modelled a broad self-harm liability regardless of suicidal intent and performed analyses to unveil its underlying aetiology. Several traits, such as depression and other psychiatric disorders are associated with an increased risk for self-harm, as evidenced by the high genetic correlations identified. Our findings might be potentially identifying factors related to the genetic liability to depression and psychopathology in general. Nonetheless, the fact that we identified a positive prediction after accounting for MDD-PRS, and that previous depression PRS were unable to predict NSSH in our target sample34 would suggest our findings to be related to self-harm.
In summary, we performed GWAS of self-harm ideation and self-harm behaviour and identified associations with two genetic loci and eleven genes. We characterized the SNP heritability and estimated genetic correlations between the two traits of interest and with a range of other psychiatric, behavioural and physiological traits. Our results suggested an association between the genetic predisposition to broad sense self-harm (both ideation and behaviour) with suicide thoughts. However, no genetic overlap between PRS for self-harm ideation and suicide attempt was detected. The PRS for self-harm behaviour was associated with suicide attempt but the association with NSSH did not reach statistical significance. In addition, our results support a partially common genetic aetiology for NSSH and suicide thoughts and for suicide thoughts and attempt, but no statistically significant evidence for a shared genetic aetiology between NSSH and suicidal attempt. Future studies should leverage novel statistical genetic approaches such as genomic structural equation modelling, to aid in the deconvolution of unique and shared genetic factors between suicidal and non-suicidal self-harm.
References
WHO. Suicide Data, http://www.who.int/mental_health/prevention/suicide/suicideprevent/en/ (2017).
American Psychiatric Association. Diagnostic and statistical manual of mental disorders (DSM-5®). (American Psychiatric Pub, 2013).
Nock, M. K. et al. Cross-national prevalence and risk factors for suicidal ideation, plans and attempts. The British Journal of Psychiatry 192, 98–105 (2008).
Klonsky, E. D., Oltmanns, T. F. & Turkheimer, E. Deliberate self-harm in a nonclinical population: Prevalence and psychological correlates. American journal of Psychiatry 160, 1501–1508 (2003).
Swannell, S. V., Martin, G. E., Page, A., Hasking, P. & St John, N. J. Prevalence of nonsuicidal self‐injury in nonclinical samples: Systematic review, meta‐analysis and meta‐regression. Suicide and Life‐Threatening Behavior 44, 273–303 (2014).
Lim, K.-S. et al. Global lifetime and 12-month prevalence of suicidal behavior, deliberate self-harm and non-suicidal self-injury in children and adolescents between 1989 and 2018: a meta-analysis. International journal of environmental research and public health 16, 4581 (2019).
Maciejewski, D. F. et al. Overlapping genetic and environmental influences on nonsuicidal self-injury and suicidal ideation: different outcomes, same etiology? JAMA psychiatry 71, 699–705 (2014).
Pedersen, N. L. & Fiske, A. Genetic influences on suicide and nonfatal suicidal behavior: twin study findings. European Psychiatry 25, 264–267 (2010).
Chesney, E., Goodwin, G. M. & Fazel, S. Risks of all‐cause and suicide mortality in mental disorders: a meta‐review. World Psychiatry 13, 153–160 (2014).
Rihmer, Z. Suicide risk in mood disorders. Current opinion in psychiatry 20, 17–22 (2007).
Nock, M. K., Joiner, T. E. Jr, Gordon, K. H., Lloyd-Richardson, E. & Prinstein, M. J. Non-suicidal self-injury among adolescents: Diagnostic correlates and relation to suicide attempts. Psychiatry research 144, 65–72 (2006).
Roy, A., Segal, N. L., Centerwall, B. S. & Robinette, C. D. Suicide in twins. Archives of General Psychiatry 48, 29–32 (1991).
Roy, A. & Segal, N. L. Suicidal behavior in twins: a replication. Journal of Affective Disorders 66, 71–74 (2001).
Baldessarini, R. J. & Hennen, J. Genetics of suicide: an overview. Harvard Review of Psychiatry 12, 1–13 (2004).
Statham, D. J. et al. Suicidal behaviour: an epidemiological and genetic study. Psychological Medicine 28, 839–855 (1998).
Brent, D. A., Bridge, J., Johnson, B. A. & Connolly, J. Suicidal behavior runs in families: a controlled family study of adolescent suicide victims. Archives of general psychiatry 53, 1145–1152 (1996).
Mann, J. J., Waternaux, C., Haas, G. L. & Malone, K. M. Toward a clinical model of suicidal behavior in psychiatric patients. American journal of Psychiatry 156, 181–189 (1999).
Muehlenkamp, J. J. & Gutierrez, P. M. Risk for suicide attempts among adolescents who engage in non-suicidal self-injury. Archives of Suicide Research 11, 69–82 (2007).
Glenn, C. R. & Klonsky, E. D. Social context during non-suicidal self-injury indicates suicide risk. Personality and Individual Differences 46, 25–29 (2009).
Wichstrom, L. Predictors of non-suicidal self-injury versus attempted suicide: similar or different? Archives of Suicide Research 13, 105–122 (2009).
Zai, C. C. et al. A genome-wide association study of suicide severity scores in bipolar disorder. Journal of Psychiatric Research 65, 23–29 (2015).
Willour, V. L. et al. A genome-wide association study of attempted suicide. Molecular Psychiatry 17, 433 (2012).
Stein, M. B. et al. Genomewide association studies of suicide attempts in US soldiers. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 174, 786–797 (2017).
Schosser, A. et al. Genomewide association scan of suicidal thoughts and behaviour in major depression. PloS One 6, e20690 (2011).
Perroud, N. et al. Genome-wide association study of increasing suicidal ideation during antidepressant treatment in the GENDEP project. The Pharmacogenomics Journal 12, 68 (2012).
Perlis, R. H. et al. Genome-wide association study of suicide attempts in mood disorder patients. American Journal of Psychiatry 167, 1499–1507 (2010).
Menke, A. et al. Genome-wide association study of antidepressant treatment-emergent suicidal ideation. Neuropsychopharmacology 37, 797 (2012).
Laje, G. et al. Genome-wide association study of suicidal ideation emerging during citalopram treatment of depressed outpatients. Pharmacogenetics and Genomics 19, 666 (2009).
Galfalvy, H. et al. A pilot genome wide association and gene expression array study of suicide with and without major depression. The World Journal of Biological Psychiatry 14, 574–582 (2013).
Galfalvy, H. et al. A genome‐wide association study of suicidal behavior. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 168, 557–563 (2015).
Erlangsen, A. et al. Genetics of suicide attempts in individuals with and without mental disorders: a population-based genome-wide association study. Molecular Psychiatry, 1 (2018).
Mullins, N. et al. Genome-wide association study of suicide attempt in psychiatric disorders identifies association with major depression polygenic risk scores. bioRxiv, 416008 (2018).
Mullins, N. et al. Genetic relationships between suicide attempts, suicidal ideation and major psychiatric disorders: A genome‐wide association and polygenic scoring study. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics 165, 428–437 (2014).
Maciejewski, D. F. et al. The association of genetic predisposition to depressive symptoms with non-suicidal and suicidal self-injuries. Behavior genetics 47, 3–10 (2017).
Ruderfer, D. M. et al. Significant shared heritability underlies suicide attempt and clinically predicted probability of attempting suicide. Molecular psychiatry, 1 (2019).
Sudlow, C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS medicine 12, e1001779 (2015).
Davis, K. A. et al. Mental health in UK Biobank: development, implementation and results from an online questionnaire completed by 157 366 participants. BJPsych open 4, 83–90 (2018).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203 (2018).
Loh, P.-R. et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nature genetics 47, 284 (2015).
Liam Abbott, S. B. et al. The Hail team. GWAS Results, http://www.nealelab.is/uk-biobank/ (2018).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS computational biology 11, e1004219 (2015).
Watanabe, K., Taskesen, E., Van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nature communications 8, 1826 (2017).
Knopik, V. S. et al. Genetic effects on alcohol dependence risk: re-evaluating the importance of psychiatric and other heritable risk factors. Psychological Medicine 34, 1519–1530 (2004).
Heath, A. C. et al. Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychological medicine 29, 1069–1081 (1999).
Dudding, T. et al. Genome wide analysis for mouth ulcers identifies associations at immune regulatory loci. Nature Communications 10, 1052 (2019).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nature Genetics 47, 291 (2015).
Ni, G. et al. Estimation of genetic correlation via linkage disequilibrium score regression and genomic restricted maximum likelihood. The American Journal of Human Genetics 102, 1185–1194 (2018).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nature Genetics 47, 1236 (2015).
Zheng, J. et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33, 272–279 (2017).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics 81, 559–575 (2007).
Colodro-Conde, L. et al. A direct test of the diathesis–stress model for depression. Molecular psychiatry (2017).
Yang, J., Zaitlen, N. A., Goddard, M. E., Visscher, P. M. & Price, A. L. Advantages and pitfalls in the application of mixed-model association methods. Nature Genetics 46, 100 (2014).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. The American Journal of Human Genetics 88, 76–82 (2011).
Chang, L.-H. et al. Association between polygenic risk for tobacco or alcohol consumption and liability to licit and illicit substance use in young Australian adults. Drug and alcohol dependence 197, 271–279 (2019).
Nyholt, D. R. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. The American Journal of Human Genetics 74, 765–769 (2004).
Zheng, J. et al. PhenoSpD: an atlas of phenotypic correlations and a multiple testing correction for the human phenome. bioRxiv, 148627 (2017).
Lloyd-Jones, L. R. et al. Improved polygenic prediction by Bayesian multiple regression on summary statistics. Nature communications 10, 1–11 (2019).
Wray, N. R. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nature genetics 50, 668 (2018).
Nock, M. K. Self-injury. Annual Review of Clinical Psychology 6, 339–363 (2010).
Rentería, M. et al. Subcortical brain structure and suicidal behaviour in major depressive disorder: a meta-analysis from the ENIGMA-MDD working group. Translational psychiatry 7, e1116 (2017).
Domínguez-Baleón, C., Gutiérrez-Mondragón, L. F., Campos-González, A. I. & Rentería, M. E. Neuroimaging studies of suicidal behaviour and non-suicidal self-injury in psychiatric patients: A systematic review. Frontiers in Psychiatry 9, 500 (2018).
Strawbridge, R. J. et al. Identification of novel genome-wide associations for suicidality in UK Biobank, genetic correlation with psychiatric disorders and polygenic association with completed suicide. EBioMedicine (2019).
Manitt, C. et al. DCC orchestrates the development of the prefrontal cortex during adolescence and is altered in psychiatric patients. Translational psychiatry 3, e338 (2013).
Essex, M. J. et al. Epigenetic vestiges of early developmental adversity: childhood stress exposure and DNA methylation in adolescence. Child Development 84, 58–75 (2013).
Jansen, P. R. et al. Genome-wide Analysis of Insomnia (N = 1,331,010) Identifies Novel Loci and Functional Pathways. Nature Genetics 51, 394–403, https://doi.org/10.1038/s41588-018-0333-3 (2019).
Hysing, M., Sivertsen, B., Stormark, K. M. & O’connor, R. C. Sleep problems and self-harm in adolescence. The British Journal of Psychiatry 207, 306–312 (2015).
McCall, W. V. & Black, C. G. The link between suicide and insomnia: theoretical mechanisms. Current psychiatry reports 15, 389 (2013).
Fujii, T. et al. Possible association of the semaphorin 3D gene (SEMA3D) with schizophrenia. Journal of psychiatric research 45, 47–53 (2011).
Gilabert-Juan, J. et al. Semaphorin and plexin gene expression is altered in the prefrontal cortex of schizophrenia patients with and without auditory hallucinations. Psychiatry research 229, 850–857 (2015).
Nurnberger, J. I. et al. Identification of pathways for bipolar disorder: a meta-analysis. JAMA Psychiatry 71, 657–664 (2014).
Savage, J. E. et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nature Genetics 50, 912 (2018).
Coleman, J. et al. Functional consequences of genetic loci associated with intelligence in a meta-analysis of 87,740 individuals. bioRxiv, 170712 (2017).
Mars, B. et al. What distinguishes adolescents with suicidal thoughts from those who have attempted suicide? A population‐based birth cohort study. Journal of child psychology and psychiatry 60, 91–99 (2019).
Mick, E. et al. Genome-wide association study of proneness to anger. PloS one 9, e87257 (2014).
Dougherty, J. D. et al. The disruption of Celf6, a gene identified by translational profiling of serotonergic neurons, results in autism-related behaviors. Journal of Neuroscience 33, 2732–2753 (2013).
Roberson-Nay, R. et al. Twin Study of Early-Onset Major Depression Finds DNA Methylation Enrichment for Neurodevelopmental Genes. bioRxiv, 422345 (2018).
Pedrosa, E. et al. Analysis of protocadherin alpha gene enhancer polymorphism in bipolar disorder and schizophrenia. Schizophrenia research 102, 210–219 (2008).
Anitha, A. et al. Protocadherin α (PCDHA) as a novel susceptibility gene for autism. Journal of Psychiatry & Neuroscience: JPN 38, 192 (2013).
Vilariño-Güell, C. et al. LINGO1 and LINGO2 variants are associated with essential tremor and Parkinson disease. Neurogenetics 11, 401–408 (2010).
Andrews, J. L. & Fernandez-Enright, F. A decade from discovery to therapy: Lingo-1, the dark horse in neurological and psychiatric disorders. Neuroscience & Biobehavioral Reviews 56, 97–114 (2015).
Guillemin, C. et al. DNA methylation signature of childhood chronic physical aggression in T cells of both men and women. PloS one 9, e86822 (2014).
Gvion, Y. & Apter, A. Aggression, impulsivity, and suicide behavior: a review of the literature. Archives of Suicide Research 15, 93–112 (2011).
Davis, O. S. et al. A three-stage genome-wide association study of general cognitive ability: hunting the small effects. Behavior genetics 40, 759–767 (2010).
Li, S. et al. Genome-wide alterations in hippocampal 5-hydroxymethylcytosine links plasticity genes to acute stress. Neurobiology of Disease 86, 99–108 (2016).
Dutta, R. et al. Genetic and other risk factors for suicidal ideation and the relationship with depression. Psychological medicine 47, 2438–2449 (2017).
Keyes, K. M. & Westreich, D. UK Biobank, big data, and the consequences of non-representativeness. The Lancet 393, 1297 (2019).
Adams, M. et al. Factors associated with sharing email information and mental health survey participation in two large population cohorts. BioRxiv, 471433 (2018).
Wray, N. R. et al. Pitfalls of predicting complex traits from SNPs. Nature Reviews Genetics 14, 507 (2013).
Wainschtein, P. et al. Recovery of trait heritability from whole genome sequence data. bioRxiv, 588020 (2019).
Knorr, A. C., Ammerman, B. A., Hamilton, A. J. & McCloskey, M. S. Predicting status along the continuum of suicidal thoughts and behavior among those with a history of nonsuicidal self-injury. Psychiatry Research (2019).
Andover, M. S. & Gibb, B. E. Non-suicidal self-injury, attempted suicide, and suicidal intent among psychiatric inpatients. Psychiatry research 178, 101–105 (2010).
Bjørngaard, J. H. et al. Maternal age at child birth, birth order, and suicide at a young age: a sibling comparison. American journal of epidemiology 177, 638–644 (2013).
Taylor, P. J. et al. A meta-analysis of the prevalence of different functions of non-suicidal self-injury. Journal of Affective Disorders 227, 759–769 (2018).
Acknowledgements
This research was conducted using data from the UK Biobank resource under application number 25331. AC-G is supported by a UQ Research Training Scholarship from The University of Queensland (UQ). MER thanks the support of the NHMRC and Australian Research Council (ARC), and National Health and Medical Research Council (NHMRC) through a Research Fellowship (GNT1102821) and NHMRC Centre for Research Excellence in Suicide Prevention (GNT1042580). This research was made possible thanks to support from the US National Institutes of Health (grants AA013326, AA07535, AA0758O, AA07728, AA10249, AA13320, AA13321, AA14041, AA11998, AA17688, DA00272, DA012854, DA07261, DA018267, DA018660, DA23668 and DA019951); the Australian National Health and Medical Research Council (241944, 339462, 389927, 389875, 389891, 389892, 389938, 442915, 442981, 496739, 552485, 552498, 628911 1047956); the Australian Research Council (A7960034, A79906588, A79801419, DP0770096, DP0212016 and DP0343921). MH and KD are funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. MH is an NIHR Senior Investigator. KJHV is supported by the Foundation Volksbond Rotterdam. We would like to acknowledge Scott Gordon and Richard Parker for their support, and the twins and their families for their participation. We gratefully acknowledge all the studies and databases that made GWAS summary data available (a full list of which is available at http://ldsc.broadinstitute.org/about/).
Author information
Authors and Affiliations
Contributions
A.I.C. and M.E.R. conceived, designed the study and wrote the first draft of the manuscript. A.I.C. conducted the analyses with supervision of M.E.R. and N.G.M. and input from K.J.H.V. and D.F.M., N.G.M., D.J.S., P.A.F.M. and A.C.H. secured funding and collected data for the Australian dataset. K.D., A.J. and M.H. designed the mental health questionnaire and self-harm assessment on the UK-B. All co-authors contributed to the interpretation of the results and provided feedback on the preliminary versions of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Campos, A.I., Verweij, K.J.H., Statham, D.J. et al. Genetic aetiology of self-harm ideation and behaviour. Sci Rep 10, 9713 (2020). https://doi.org/10.1038/s41598-020-66737-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-66737-9
This article is cited by
-
Unveiling Adolescent Suicidality: Holistic Analysis of Protective and Risk Factors Using Multiple Machine Learning Algorithms
Journal of Youth and Adolescence (2024)
-
Functional and molecular characterization of suicidality factors using phenotypic and genome-wide data
Molecular Psychiatry (2023)
-
Investigating the shared genetic architecture and causal relationship between pain and neuropsychiatric disorders
Human Genetics (2023)
-
Peripheral and neural correlates of self-harm in children and adolescents: a scoping review
BMC Psychiatry (2022)
-
Family patterns in suicidal behavior
Nature Reviews Psychology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.