Abstract
Signaling elicited by the stem cell factors SOX2, OCT4, KLF4, and MYC not only mediates reprogramming of differentiated cells to pluripotency but has also been correlated with tumor malignancy. In this study, we found SOX2 expression signifies poor recurrence-free survival and correlates with advanced pathological grade in bladder cancer. SOX2 silencing attenuated bladder cancer cell growth, while its expression promoted cancer cell survival and proliferation. Under low-serum stress, SOX2 expression promoted AKT phosphorylation and bladder cancer cells’ spheroid-forming capability. Furthermore, pharmacological inhibition of AKT phosphorylation, using MK2206, inhibited the SOX2-mediated spheroid formation of bladder cancer cells. Gene expression profiling showed that SOX2 expression, in turn, induced IGF2 expression, while SOX2 silencing inhibited IGF2 expression. Moreover, knocking down IGF2 and IGF1R diminished bladder cancer cell growth. Lastly, pharmacological inhibition of IGF1R, using linsitinib, also inhibited the SOX2-mediated spheroid formation of bladder cancer cells under low-serum stress. Our findings indicate the SOX2–IGF2 signaling affects the aggressiveness of bladder cancer cell growth. This signaling could be a promising biomarker and therapeutic target for bladder cancer intervention.
Similar content being viewed by others
Introduction
Bladder cancer arises from the urinary bladder’s epithelial lining, called the urothelium, and is one of the most common urinary system malignancies1. Most patients are diagnosed with non-invasive bladder cancer, and surgery is the typical treatment option2. However, just over half of these patients will experience tumor recurrence3. Chemotherapy is the conventional treatment for patients with advanced stage bladder cancer, but few can be cured4. Therefore, novel biomarkers for monitoring, and therapeutic targets for targeting, bladder cancer progression are urgently needed.
Transcription factors involved in embryonic stem cell (ESC) and induced pluripotent stem cell (iPSC) signaling, such as SOX2, OCT4, MYC, and KLF4, have been linked to tumor progression in various cancers5,6. Among them, SOX2 is essential to self-renewal and differentiation of several adult tissue progenitor cells7,8,9. SOX2 expression has been implicated in human breast and lung cancers10,11,12. Genomic amplification of SOX2 has been observed in lung squamous cell carcinoma13, while genomic amplification of SOX4, another SOX family member, has been reported in bladder cancer14,15. SOX2 promotes abnormal proliferation of lung cancer cells and controls tumor initiation in skin squamous cell carcinoma16,17. Although there is high SOX2 expression in bladder cancer18, the oncogenic mechanism underlying SOX2-mediated tumor malignancy remains unclear.
Insulin-like growth factor 2 (IGF2), a mitogenic peptide hormone, is highly expressed during embryonic development19, and overexpressed in tumors associated with more aggressive status20. Previous studies suggest that IGF2 binds to its receptor, IGF1R, to initiate tumorigenesis of breast and lung cancers21,22 and promote progression of endometrial and gastric cancers23,24. Loss of imprinting contributes to overexpression of IGF2 in cancers of the prostate and colon and cancers with stem cell-like features25,26,27,28, while deregulation of IGF2 in cancers is also attributable to abnormal expression of transcription factors29,30,31. Insulin-like growth factor-binding protein 1 (IGFBP1) is a secreted protein serving as a negative regulator that competes with IGF ligands, thus preventing ligand–receptor activation32,33,34. Although IGF2/IGF1R signaling enhances tumor progression in several cancers, it is unclear whether IGF2/IGF1R signaling contributes to bladder cancer progression.
In this study, we found SOX2 is a prognostic marker in bladder cancer patients, signifying poor survival. We also found SOX2 promotes AKT phosphorylation in bladder cancer cells by inducing IGF2 and IGF1R expression and suppressing IGFBP1. We further characterized the potential of IGF1R signaling as a biomarker and therapeutic target in treating bladder cancer.
Results
SOX2 expression is correlated with tumor malignancy in bladder cancer
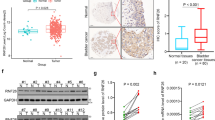
Because factors in ESC signaling and iPSC reprogramming have been linked to tumor malignancy, we used the Cox’s proportional hazards model to analyze the link between SOX2, KLF4, MYC and OCT4 expression and recurrence-free survival outcome for bladder cancer patients (Fig. 1a). Both univariate and multivariate regression analyses revealed that only SOX2 expression correlated with poor recurrence-free survival (Fig. 1a, and Supplementary Table 1). Box-and-whisker plots showed that SOX2 expression was also associated with advanced tumor grade of bladder cancer (Fig. 1b). Immunohistochemistry was used to verify SOX2 expression in primary bladder tumors, which showed SOX2 expression was high in tumors with “poorly differentiated” malignant grade (Fig. 1c). These data highlight SOX2’s potential involvement in bladder cancer tumor malignancy.
SOX2 is associated with poor histologic differentiation of bladder cancer. (a) Univariate and multivariate analyses for recurrence-free survival based on the expression of stem cell factors SOX2, KLF4, MYC, and OCT4 in bladder cancer patients from GSE32894 database. *P < 0.05. (b) Gene expression analysis to assess SOX2, KLF4, MYC, and OCT4 levels and their correlation with histologic grade of bladder tumors from GSE32894 database. One Way ANOVA and Tukey’s multiple comparison analysis were used to determine statistical significance: *P < 0.05; **P < 0.01; ***P < 0.001. (c) Representative images (upper) of immunohistochemical analysis for SOX2-negative and SOX2-positive staining in bladder transitional cell carcinoma. Scale bars: 500 μm (whole section); 50 μm (inset, 20× magnification). Fisher’s exact test (lower) for association between SOX2 expression and tumor grade in bladder transitional cell carcinoma (n = 22).
SOX2 regulates the growth of bladder cancer cells
Assessing SOX2 expression in bladder cancer cell lines showed its expression was considerably lower in T24 cells than in 5637 cells (Supplementary Figure S1). To investigate its role in bladder cancer oncogenesis, SOX2 was ectopically expressed in T24 cells using the lentiviral transduction system, and its expression was confirmed with immunoblotting and qPCR (Fig. 2a left). Trypan blue cell exclusion and alamarBlue proliferation analysis showed that SOX2 expression promoted cell proliferation (Fig. 2a right and Supplementary Figure S2a). Because 5637 represents a bladder cancer cell line with high SOX2 expression, we adopted the lentiviral shRNA system to knock down SOX2 in 5637 cells to further investigate the effect of eliminating SOX2 function. qPCR and immunoblotting assays indicated that endogenous SOX2 mRNA expression was suppressed by shSOX2 (Fig. 2b left). The trypan blue cell exclusion test, alamarBlue proliferation assay, and cell cycle analysis revealed that silencing SOX2 in 5637 cells inhibited cell proliferation due to S-phase arrest during cell cycle progression (Fig. 2b right and Supplementary Fig. S2b,c). In addition, clonogenic assays showed SOX2’s ectopic expression increased T24 cells’ colony-forming capability, whereas knockdown of SOX2 in 5637 cells weakened colony formation. (Fig. 2c). This suggests SOX2 expression promotes bladder cancer cell growth.
SOX2 mediates growth of bladder cancer cells. (a) qPCR (upper left) and immunoblotting (lower left) analysis to assess SOX2 mRNA and protein expression, respectively, in T24 cells transduced with the lentiviral vector encoding SOX2 cDNA (SOX2) or empty control vector (Ctrl). Trypan blue cell exclusion analysis of T24 cells transduced with the lentiviral vector encoding SOX2 cDNA (SOX2) or empty control vector (Ctrl) for the indicated days. Results are the average of three replicates and expressed as the mean ± S.D. **P < 0.01, ***P < 0.001. (b) qPCR (upper left) and immunoblotting (lower left) analysis of SOX2 expression in 5637 cells transduced with the lentiviral vector encoding shRNA against SOX2 (shSOX2) or scrambled control vector (SC). Trypan blue cell exclusion analysis of 5637 cells transduced with the lentiviral vector encoding shSOX2 or scrambled control vector (SC) for the indicated days. Results are the average of three replicates and expressed as the mean ± S.D. The #1 and #2 indicate the two distinct shRNAs that target different regions within SOX2. ***P < 0.001. (c) Clonogenic analysis (left) to assess the SOX2 expression effect on the colony-forming ability in T24 cells transduced with the lentiviral vector encoding SOX2 cDNA (SOX2) or empty control vector (Ctrl). Clonogenic analysis (right) to assess the SOX2 knockdown effect on the colony-forming ability in 5637 cells transduced with the lentiviral vector encoding shSOX2 or scrambled control vector (SC). Colonies were subjected to crystal violet staining and quantified by ImageJ analysis. Results are the average of three replicates and expressed as the mean ± S.D. *P < 0.05, **P < 0.01.
SOX2 promotes the survival of bladder cancer cells by activating AKT signaling
To test whether SOX2 plays a role in cell survival, we assessed SOX2 expression in T24 cells under a low-serum stress. Clonogenic analysis showed that SOX2 expression promoted T24 cell growth under a low-serum (1% FBS) condition (Fig. 3a). We further validated the effect of SOX2 expression on T24 cell-spheroid formation under low-serum stress. The T24 cells formed spheroids in a 3D culture system under the normal-serum (10% FBS) condition, wherein SOX2 expression did not affect spheroid formation (Fig. 3b). By contrast, long-term culturing of T24 spheroids under low-serum condition (1% FBS) attenuated the size of the spheroids; however, SOX2 expression sustained the T24 spheroid-forming capability under the low-serum condition, indicating SOX2 is involved in bladder cancer cell survival (Fig. 3b). In addition, the cell cycle analysis revealed that SOX2 expression sustained the S-phase in T24 cells under the low-serum condition (Fig. 3c and Supplementary Figure S2c bottom left). These findings suggest that SOX2 expression helps bladder cancer cells overcome low-serum stress.
SOX2 enhances bladder cancer cell survival via activation of AKT signaling. (a) Clonogenic analysis to assess the serum starvation effect on T24 cells transduced with the lentiviral vector encoding SOX2 cDNA (SOX2) or empty control vector (Ctrl) for 14 days. Colonies were cultured under low-serum (1% FBS) condition and subjected to crystal violet staining, followed by quantification of ImageJ analysis. Representative plates were photographed (bottom). Results are the average of three replicates and expressed as the mean ± S.D. *P < 0.05. (b) Spheroid formation analysis to assess low-serum spheroid forming ability of T24 cells transduced with the lentiviral vector encoding SOX2 cDNA (SOX2) or empty control vector (Ctrl). SOX2-expressing (SOX2) or control (Ctrl) T24 cells were cultured under the low-attached culture condition in normal-serum medium (10% FBS) for 7 days and subjected to low-serum medium (1% FBS) for another 17 days. Photos are representative images of the spheroids at day 1 and day 17 under low-serum condition (bottom). Results are the average of three independent experiments and expressed as the mean ± S.D. ***P < 0.001. (c) Flow cytometry analysis to assess cell-cycle distribution under low-serum condition in SOX2-expressing (SOX2) or control (Ctrl) T24 cells. The transduced cells were cultured under low-serum (1% FBS) condition for 48 hr and subjected to cell-cycle analysis. (d) Immunoblotting analysis to assess the expression of phosphorylated AKT at Ser473 and total AKT in SOX2-expressing (SOX2) or control (Ctrl) T24 cells under serum-free or low-serum condition (1% FBS) for 48 hr. (e) Immunoblotting analysis to assess the expression of phosphorylated AKT at Ser473 and total AKT expression in SOX2-expressing T24 cells under low-serum condition (1% FBS) in the presence or absence of MK2206 (1 μM) for 48 hr. (f) Clonogenic assay (left) and 3D colony-forming analysis (right) of SOX2-expressing T24 cells under low-serum condition (1% FBS) in the presence or absence of MK2206 (1 μM) for 14 days. Colonies were quantified by ImageJ analysis. Results are the average of three independent experiments and expressed as the mean ± S.D. *P < 0.05, ***P < 0.001.
Because AKT signaling contributes to cell survival, we examined SOX2’s effect on AKT (Ser473) phosphorylation. Immunoblotting analysis revealed that SOX2 expression in T24 cells sustained AKT phosphorylation under serum-free and low-serum (1% FBS) conditions (Fig. 3d). Moreover, knockdown of SOX2 decreased AKT phosphorylation in 5637 cells (Supplementary Figure S3a). To further verify whether AKT activation mediates SOX2-induced bladder cancer survival, we pharmacologically inhibited AKT activation with MK2206. MK2206 treatment reduced the AKT phosphorylation in 5637 cells as well as in SOX2-expressing T24 cells (Fig. 3e, Supplementary Figure S3b). Clonogenic and 3D colony-forming assays revealed MK2206 also reduced the colony numbers and sizes with respect to the SOX2-expressing T24 cells (Fig. 3f). These results highlight SOX2’s activation of AKT signaling to boost bladder cancer cell survival.
IGF2 expression is regulated by SOX2 under an epigenetic control
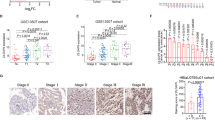
To identify the genes responsible for SOX2-mediated bladder cancer cell survival, gene expression profiling analysis was performed using the SOX2-expressing T24 cells and control T24 cells, and the result was uploaded as GSE145826. qPCR analysis confirmed IGF2 levels were considerably higher in SOX2-expressing T24 than in control T24 cells (Fig. 4a). Similarly, SOX2-high 5637 cells exhibited higher IGF2 level than SOX2-low T24 cells, while SOX2 silencing in 5637 cells inhibited IGF2 expression (Fig. 4b). Because SOX2 and IGF2 expression has been linked to embryonic development, we examined SOX2 and IGF2 expression in ESC and fibroblasts. RNA-seq and qPCR assays showed that both SOX2 and IGF2 levels were downregulated in the differentiated fibroblasts compared to ESC (Fig. 4c and Supplementary Figure S4). Since SOX2 mediates stem cell differentiation and iPSC reprogramming mainly via epigenetic regulation, whereby H3K4me3 marks sites with active gene expression35,36, we examined whether H3K4me3 signal on the IGF2 locus is affected by SOX2 expression in bladder cancer cells. ChIP-seq analysis showed a strong H3K4me3 signal on the IGF2 locus in ESC but not in fibroblasts (Supplementary Figure S5). ChIP-qPCR analysis revealed the H3K4me3 signal on the IGF2 locus increased because of SOX2 expression in T24 cells and decreased on account of SOX2 knockdown in 5637 cells (Fig. 4d). We also found IGFBP1 expression decreased because of SOX2 expression in T24 cells and increased on account of SOX2 knockdown in 5637 cells (Fig. 4e). These results indicate SOX2 expression regulates IGF2 signaling.
SOX2 induces IGF2/IGF1R signaling molecules in bladder cancer cells. (a) qPCR analysis to assess IGF2 expression in T24 cells transduced with the lentiviral vector encoding SOX2 cDNA (SOX2) or empty control vector (Ctrl). Results are the average of three replicates and expressed as the mean ± S.D. ***P < 0.001. (b) qPCR analysis to assess IGF2 expression in T24 versus 5637 cells (left) and in 5637 cells transduced with shSOX2 versus scrambled control (SC) vector (right). The #1 and #2 indicate the two distinct shRNAs that target different regions within SOX2. ***P < 0.001. (c) RNA-seq analysis of SOX2 and IGF2 expression in ESC (HUES3) and differentiated fibroblast cells from GSE73211 database. (d) ChIP-qPCR analysis to assess H3K4me3 levels of IGF2 region in SOX2-expressing T24 cell (left) and SOX2-silenced 5637 cells (right). **P < 0.01, ***P < 0.001. Primers targeting the H3K4me3 signal of IGF2 were designed at chr11: 2,158,962–2,159,038. (e) qPCR and immunoblotting analysis of IGFBP1 expression in SOX2-overexpressing T24 cells (left) and SOX2-silenced 5637 cells (right). Results are the average of three replicates and expressed as the mean ± S.D. *P < 0.05, ***P < 0.001.
IGF2/IGF1R signaling is essential for SOX2-mediated bladder cancer cell survival
To understand whether IGF2/IGF1R signaling is responsible for high-SOX2 expressing bladder cancer cell growth, we knocked down IGF2 and IGF1R (Fig. 5a–d). Clonogenic assays showed that IGF2 and IGF1R knockdown weakened the colony-forming capability of the high-SOX2 expressing 5637 cells (Fig. 5b,d). Immunoblotting assays revealed that AKT phosphorylation decreased on account of knockdown of IGF2 or IGF1R in 5637 cells (Fig. 5a,c). Linsitinib, an IGF1R inhibitor, was used to block IGF2/IGF1R signaling-mediated AKT phosphorylation in SOX2-expressing bladder cancer cells. Immunoblotting assays revealed that linsitinib inhibited AKT phosphorylation in 5637 cells as well as in SOX2-expressing T24 cells (Fig. 5e and Supplementary Figure S3b). 3D colony-forming assays revealed that linsitinib reduced colony formation of SOX2-expressing T24 cells (Fig. 5f). This suggests that IGF2 and IGF1R are crucial for SOX2-mediated growth and survival of bladder cancer cells.
SOX2-induced IGF2/IGF1R signaling is essential for bladder cancer cells. (a) qPCR (upper) and immunoblotting (lower) analysis of 5637 cells transduced with the lentiviral vector encoding shRNA against IGF2 (shIGF2) or scrambled control vector (SC). The #1 and #2 indicate the two distinct shRNAs that target different regions within IGF2. ***P < 0.001. (b) Clonogenic assay of 5637 cells transduced with shIGF2 or scrambled control vector (SC). Colonies were subjected to crystal violet staining (top) and quantified by ImageJ analysis. Results are the average of three replicates and expressed as the mean ± S.D. **P < 0.01. (c) qPCR (upper) and immunoblotting (lower) analysis of 5637 cells transduced with the lentiviral vector encoding shRNA against IGF1R (shIGF1R) or scrambled control vector (SC). The #1 and #2 indicate the two distinct shRNAs that target different regions within IGF1R. ***P < 0.001. (d) Clonogenic assay of 5637 cells transduced with shIGF1R or scrambled control vector (SC). Colonies were subjected to crystal violet staining (top) and quantified by ImageJ analysis. Results are the average of three replicates and expressed as the mean ± S.D. **P < 0.01. (e) Immunoblotting analysis to assess the expression of phosphorylated AKT at Ser473 and total AKT expression in SOX2-expressing T24 cells under low-serum (1% FBS) condition in the presence or absence of linsitinib (5 μM) for 48 hr. (f) 3D colony-forming analysis to assess linsitinib effect on colony formation of SOX2-expressing T24. The cells were grown under the low-attached condition in low-serum medium (1% FBS) in the presence or absence of linsitinib (5 μM) for 7 days. Photos (left) are representative images of the colony and colony sizes were quantified (right). Results are the average of three independent experiments and expressed as the mean ± SD. ***P < 0.001.
IGF1R signaling serves as a prognostic biomarker for bladder cancer
Our present results confirmed that SOX2 regulates IGF2/IGF1R signaling in bladder cancer cells. SOX4, another SOX family member, is reported to be amplified in bladder cancer14,15. We confirmed that SOX4, but not SOX2, was mainly amplified in primary bladder tumors (Supplementary Figure S6). To further examine the potential of SOX2, SOX4, and IGF1R signaling in bladder cancer prognosis, we correlated the expression of these molecules with recurrence-free survival in primary bladder tumors. Kaplan-Meier analysis showed that SOX2, but not SOX4, expression correlated with a poor recurrence-free survival in bladder cancer patients, and the patients harboring a SOX2-high/SOX4-low signature had a worse recurrence-free survival outcome than those with SOX2-low/SOX4-high signature (Fig. 6a–c). Moreover, SOX4, but not SOX2, correlates with a good overall survival, and patients harboring the SOX4-low/SOX2-high signature show a worse overall survival than those with the SOX4-high/SOX2-low signature (Supplementary Figure S7). Furthermore, IGF1R expression was also associated with a poor recurrence-free survival outcome (Fig. 6d). We observed that the patients harboring a SOX2-high/IGF1R-high signature had a worse recurrence-free survival outcome than those with SOX2-low/IGF1R-low signature (Fig. 6e). Both univariate and multivariate regression assays of IGF signaling molecules showed that IGF1R is the only independent predictor of poor recurrence-free survival (Table 1). These findings suggest that IGF1R can be a potential biomarker for predicting poor survival outcomes.
SOX2 and IGF1R as prognostic factors in bladder cancer. (a,b) Kaplan–Meier analysis of SOX2 (a) and SOX4 (b) expression with recurrence-free survival in patients from TCGA_BLCA cohort. The significance was examined by log-rank test. *P < 0.05. (c) Kaplan–Meier analysis to assess the correlation of SOX2-high/SOX4-low and SOX2-low/SOX4-high signatures with recurrence-free survival in patients from TCGA_BLCA cohort. The significance was examined by log-rank test. *P < 0.05. (d) Kaplan–Meier analysis of IGF1R expression with recurrence-free survival in patients from TCGA_BLCA cohort. The significance was examined by log-rank test. **P < 0.01. (e) Kaplan–Meier analysis to assess the correlation of SOX2-high/IGF1R-high and SOX2-low/IGF1R-low signatures with recurrence-free survival in patients from TCGA_BLCA cohort. The significance was examined by log-rank test. ***P < 0.001. (f) The schematic model of SOX2-mediated oncogenesis of bladder cancer. SOX2 induces the expression of IGF2 and IGF1R, but suppresses that of IGFBP1, thus promoting AKT phosphorylation, with increased proliferation and survival of bladder cancer cells.
Discussion
Although signaling elicited by the stem cell factors SOX2, OCT4, KLF4, and MYC has been associated with cancer progression in several tumors, it has remained unclear how these signaling molecules mediate bladder cancer progression. We found that SOX2, but not OCT4, KLF4, or MYC expression, correlates with poor prognosis and histologic differentiation in bladder cancer. Moreover, we found that SOX2 promotes bladder cancer cell survival by inducing the IGF2/IGF1R pathway, thereby activating AKT survival signaling. Pharmacological inhibition of IGF1R or AKT inhibits bladder cancer cell survival. Our findings provide insights on SOX2-mediated oncogenesis in bladder cancer and highlight putative therapeutic targets.
Because the introduction of SOX2, OCT4, MYC, and KLF4 is sufficient to reprogram differentiated cells into iPSC with ESC properties, the combined expression of these factors has been hypothesized to initiate tumors and promote cancer progression37,38,39,40. On the one hand, tumors harboring ESC-like gene expression signature with active targets of SOX2, OCT4, and MYC were associated with poor pathological differentiation and poor prognosis in brain, breast, and bladder cancer patients6. On the other hand, tumor malignancy has been attributed only to MYC and not to other ESC factors41. Here, we observed that only SOX2 expression correlates with poor recurrence-free survival of bladder cancer patients. Moreover, we found that SOX2 expression is associated with poor pathological differentiation, emphasizing its involvement in bladder cancer malignancy. This is in accordance with the results of Ruan et al., who reported that SOX2 and Ki67 expression in T1 early stage, non-muscle, invasive bladder tumors correlate with poor recurrence-free survival18. Currently, SOX2 is thought to be a cancer stem cell marker in relation to bladder tumors and Sox2 knockout within primary invasive bladder cancer caused enhanced tumor regression42. SOX4, another SOX family member, is overexpressed in bladder tumors harboring the genomic 6p22 amplification15,43,44. Here, we observed that SOX4 is mainly amplified in primary bladder tumors, whereas only SOX2 expression is associated with poor recurrence-free survival in patients with bladder cancer. SOX2 expression did not affect SOX4 levels in T24 cells (Supplementary Figure S8). Together, these data suggest that SOX4 and SOX2 play distinct roles in tumor initiation and progression.
The molecular mechanism linking SOX2 expression to poor prognosis in bladder cancer has not been well understood. We found that SOX2 expression not only promotes cell proliferation but also enhances cell survival under low-serum stress. We discovered that, under low-serum stress, SOX2 expression induces AKT phosphorylation and sustains bladder cancer cells’ spheroid-forming capability. In many cancers, AKT phosphorylation promotes cell survival by inducing drug resistance and desensitizing radiation therapy. We observed that pharmacological inhibition of AKT phosphorylation attenuates bladder cancer cells’ SOX2-mediated survival and spheroid-forming capability. Furthermore, we identified IGF2 and IGF1R as AKT upstream molecules which induce AKT phosphorylation in SOX2-positive bladder cancer cells. IGF2 expression boosts cancer cell survival and tumor progression in colon cancer45,46. To search for SOX2-binding targets in colon cancer cells, Fang et al. performed ChIP-seq analysis and identified IGF1R as one of direct binding targets of SOX247. We observed that SOX2 expression in T24 cells increased IGF1R expression, whereas SOX2 knockdown in 5637 cells did not affect IGF1R expression (Supplementary Figure S9). In contrast, SOX2 expression increased IGF2 expression, and SOX2 knockdown decreased IGF2 level, supporting IGF2 as a potential downstream target of SOX2. Our ChIP-qPCR analysis did not show SOX2 binding to the IGF2 locus (data not shown). In addition to directly binding to target genes, SOX2 regulates gene expression via histone modifications during stem cell differentiation and lung cancer cell plasticity16,48. ChIP-qPCR analysis showed that the signal of H3K4me3, a predominant mark of active promoters, increased upon SOX2 expression in bladder cancer cells. Knockdown of IGF2 and IGF1R attenuated the growth of bladder cancer cells. All these data verify SOX2 promotes bladder cancer cell growth and survival by inducing IGF2/IGF1R signaling.
Currently, limited targeted therapies are available for treating this aggressive disease. Since our evidence suggests that SOX2 activates AKT survival signaling and promotes spheroid-forming ability by inducing IGF2/IGF1R in bladder cancer cells, IGF2 and IGF1R may be potential therapeutic targets for treating bladder cancer. IGF2 expression promotes aggressiveness in lethal prostate cancer and has been proposed to be a druggable target31. Indeed, we found that pharmacological inhibition of IGF2/IGF1R signaling with linsitinib decreased AKT phosphorylation and attenuated bladder cancer cells’ SOX2-mediated colony-forming ability. These findings highlight IGF2/IGF1R’s potential as therapeutic targets to treat bladder cancer. Moreover, we observed that IGF1R expression correlates with poor recurrence-free survival in bladder cancer patients. The tumors harboring high-SOX2/high-IGF1R signature are associated with the worst survival outcome in bladder cancer patients. These data suggest IGF1R may serve as a novel biomarker for monitoring bladder cancer progression.
The limitation of this study is that we mainly used T24 and 5637 cells, which represent muscle invasive bladder cancer cell lines maintained in different media, to dissect the potential oncogenic role of SOX2 in bladder cancer. Thus, these data may not be sufficient to represent all bladder cancer. Through gene expression analysis, 5637 cells have been classified into basal subtype bladder cancer, which displays cancer stem cell markers and is associated with poor prognosis49,50,51. SOX2 has been characterized as s a marker for stem-like tumor cells in bladder cancer42. Hence, our findings that SOX2 activates IGF2/IGF1R signaling and predicts poor prognosis further link IGF2/IGF1R signaling to SOX2 mediated aggressiveness in bladder cancer.
In conclusion, we demonstrated that SOX2 stimulated IGF2 expression to activate AKT signaling, enhancing the survival and spheroid-forming capability of bladder cancer cells. Moreover, we observed SOX2 and IGF1R levels are significantly correlated with poor prognosis in bladder cancer patients. The fact that SOX2–IGF2/IGF1R signaling axis confers aggressiveness in bladder cancer suggests that IGF2/IGF1R may serve as therapeutic targets in treating bladder cancer.
Materials and Methods
Chemicals and reagents
Linsitinib (OSI-906) was purchased from Cayman Chem (Ann Arbor, MI, USA). MK2206 was ordered from LC Laboratories (Woburn, MA, USA).
Cell culture
T24 and 5637 cell lines were obtained in 2017 from BCRC (Bioresource Collection and Research Center, Taiwan). T24 and 5637 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) and RPMI-1640, respectively, supplemented with 10% fetal bovine serum (FBS).
RNA extraction and qPCR
Total RNA was extracted from cells using REzol reagent (Protech Technology Enterprise CO., Ltd., Taiwan) according to the manufacturer’s protocol. Quantitative real-time PCR (qPCR) was conducted with the StepOneTM Real-Time PCR system (Applied Biosystems; ABI, USA)52. Amplification of specific genes was detected using the UPL universal probe system (Roche Life Science, USA) or specific synthesized probes (Integrated Device Technology; IDT, USA). The data were normalized to the housekeeping gene 18 S rRNA. For the detailed RT-primer and probe sequences, see Supplementary Table S2.
Plasmids
SOX2 expressing vector was prepared as described previously11. pLKO.1-shSOX2 #1 (TRCN0000003252), pLKO.1-shSOX2 #2 (TRCN0000010772), pLKO.1-shIGF2 #1 (TRCN0000062430), pLKO.1-shIGF2 #2 (TRCN0000062432), pLKO.1-shIGF1R #1 (TRCN0000121193) and pLKO.1-shIGF1R #2 (TRCN0000121301) were obtained from the National RNAi Core Facility, Academia Sinica (Taipei, Taiwan). pLKO.1-Scrambled shRNA was obtained from Addgene. Lentiviral production and infection were performed using the previously described method53.
Trypan blue cell exclusion assay
Cells (105 cells/well) were seeded in 24-well plates and incubated in triplicate tests. In indicated times, cells were trypsinized and then counted using Trypan Blue staining in low magnification under the microscope.
Cell-cycle analysis by flow cytometry
Cells were plated at a density such that they would be 50% confluent on the day of analysis. Cells were trypsinized and fixed in cold 70% ethanol for 10 min and then stained with propidium iodide solution (1 mM) at room temperature for 30 min. The stained cells were analyzed using BD Accuri C6 flow cytometer with excitation at 488 nm and emission at 617 nm.
AlamarBlue cell proliferation assay
To perform the cell proliferation assay, cells were seeded at 5 × 102 cells per well in 96-well plate. The cell growth at indicated time interval was monitored using alamarBlue (Thermo Scientific) according to manufacturer’s instructions. Cell titers were calculated by measuring the absorbance at 585 nm by VICTOR2 D fluorometer (PerkinElmer).
Clonogenic assay
Cells were plated in 24-well plates with a density of 100 cells/well. Cells were incubated for 2 weeks, and the surviving colonies were fixed and stained with crystal violet. Colonies were quantified by ImageJ software.
Spheroid assay
24-well cell culture plates were coated with 0.7% agarose, and 1 × 105 cells/well were seeded. The seeded cells were then placed on an orbital shaker with shaking at 60 rpm for 7 days to form one spheroid/well. Spheroids were then subjected to low serum (1% FBS) treatment for the indicated time periods. Spheroid volumes were calculated by using the formula V = (4/3)*pi*r3, where the radius r was measured by the microscope.
3D Colony-forming assay
60-mm plastic tissue-culture petri plates were coated with 0.7% agarose, and 200 cells/plate were seeded. The seeded cells were then placed into the incubator and incubated for 7 days to form unattached floating spheroid colonies. The floating spheroid colonies were then subjected to the indicated treatment for another 7 days. The volumes of spheroid colonies were calculated by using the formula V = (4/3)*pi*r3, where the radius r was measured by the microscope.
Immunoblotting
Cells were harvested in RIPA lysis buffer supplemented with a protease inhibitor cocktail, followed by immunoblotting with SOX2 antibody (1:1000 dilution, GTX101506, GeneTex), p-AKT (Ser473, 1:1000 dilution, GTX28932, GeneTex), AKT (1:1000 dilution, #1085-1, EPITOMICS), IGF2 (1:1000 dilution, GTX129110, GeneTex), IGF1R (1:1000 dilution, GTX50433, GeneTex), IGFBP1 (1:1000 dilution, GTX129006, GeneTex), and GAPDH (1:10,000 dilution, GTX100118, GeneTex).
Immunohistochemistry (IHC) staining
IHC was conducted as previously described16. The primary SOX2 antibody used in the IHC was SOX2 antibody (1:400 dilution, PM056, MBL). The immunoreactivity pattern and histologic appearance of the tissue array (BLC661, US BIOMAX) were examined and scored by the pathologist (Supplementary Table S3). The Allred scoring system was used to give the staining scores for the expression of SOX2 based on the sum of the percentage of cells that were stained by IHC (on a scale of 0–5) and the intensity of the staining (on a scale of 0–3). The final score was determined by the sum of the intensity scores and the positivity scores of stained cells.
ChIP-qPCR
ChIP-qPCR was performed using the protocol as previously described16, with the antibodies against H3K4me3 (GTX128954, GeneTex) or control IgG (GTX35035, GeneTex). For the detailed RT-primer and probe sequences used in ChIP-qPCR, see Supplementary Table S2.
Public domain data analysis
Public gene expression profiling datasets of bladder cancer patients were accessed from TCGA_BLCA database via UCSC cancer browser, and GSE32894 and GSE73211 via GEO browser. The means of SOX2, SOX4, KLF4, MYC, OCT4, and IGF1R expression were used as cut-off points for statistics analysis. The ChIP-seq results of H3K4me3 epigenetic regulation in embryonic stem cells and fibroblasts were collected from ENCODE (https://www.encodeproject.org/). The genetic alteration rates of SOX2 and SOX4 in bladder cancer were analyzed from cBioPortal (http://www.cbioportal.org/). For the used public domain databases, see Supplementary Table S4.
Statistical analysis
The attribution of different gene expression and clinical associated variables in death risks of bladder cancer were calculated using the Cox’s proportional hazards regression analysis and demonstrated in forest plot or tables. Recurrence-free survival and overall survival curves of bladder cancer patients were analyzed by the Kaplan-Meier plots and compared the difference between groups by the log-rank test. All statistics analysis was presented using SAS software, version 9.4. Significance difference was set in which P value was less than 0.05.
References
Yaxley, J. P. Urinary tract cancers: An overview for general practice. J Family Med Prim Care 5, 533–538 (2016).
Burger, M. et al. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol 63, 234–241 (2013).
Witjes, J. A. et al. EAU guidelines on muscle-invasive and metastatic bladder cancer: summary of the 2013 guidelines. Eur Urol 65, 778–792 (2014).
Stenzl, A. et al. Treatment of muscle-invasive and metastatic bladder cancer: update of the EAU guidelines. Eur Urol 59, 1009–1018 (2011).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Ben-Porath, I. et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40, 499–507 (2008).
Fong, H., Hohenstein, K. A. & Donovan, P. J. Regulation of self-renewal and pluripotency by Sox2 in human embryonic stem cells. Stem Cells 26, 1931–1938 (2008).
Tompkins, D. H. et al. Sox2 is required for maintenance and differentiation of bronchiolar Clara, ciliated, and goblet cells. PLoS One 4, e8248 (2009).
Hu, Q. et al. The EGF receptor-sox2-EGF receptor feedback loop positively regulates the self-renewal of neural precursor cells. Stem Cells 28, 279–286 (2010).
Chen, Y. et al. The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. J Biol Chem 283, 17969–17978 (2008).
Chou, Y. T. et al. The emerging role of SOX2 in cell proliferation and survival and its crosstalk with oncogenic signaling in lung cancer. Stem Cells 31, 2607–2619 (2013).
Lu, Y. et al. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS One 5, e11022 (2010).
Bass, A. J. et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nature genetics 41, 1238–1242 (2009).
Shen, H. et al. 6p22.3 amplification as a biomarker and potential therapeutic target of advanced stage bladder cancer. Oncotarget 4, 2124–2134 (2013).
Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315-322 (2014).
Lin, S. C. et al. Epigenetic Switch between SOX2 and SOX9 Regulates Cancer Cell Plasticity. Cancer Res 76, 7036–7048 (2016).
Boumahdi, S. et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 511, 246–250 (2014).
Ruan, J. et al. Predictive value of Sox2 expression in transurethral resection specimens in patients with T1 bladder cancer. Med Oncol 30, 445 (2013).
Stylianopoulou, F., Efstratiadis, A., Herbert, J. & Pintar, J. Pattern of the insulin-like growth factor II gene expression during rat embryogenesis. Development 103, 497–506 (1988).
Livingstone, C. IGF2 and cancer. Endocr Relat Cancer 20, R321–339 (2013).
Bates, P. et al. Mammary cancer in transgenic mice expressing insulin-like growth factor II (IGF-II). Br J Cancer 72, 1189–1193 (1995).
Moorehead, R. A., Sanchez, O. H., Baldwin, R. M. & Khokha, R. Transgenic overexpression of IGF-II induces spontaneous lung tumors: a model for human lung adenocarcinoma. Oncogene 22, 853–857 (2003).
Pavelic, J., Radakovic, B. & Pavelic, K. Insulin-like growth factor 2 and its receptors (IGF 1R and IGF 2R/mannose 6-phosphate) in endometrial adenocarcinoma. Gynecol Oncol 105, 727–735 (2007).
Pavelic, K. et al. Gastric cancer: the role of insulin-like growth factor 2 (IGF 2) and its receptors (IGF 1R and M6-P/IGF 2R). J Pathol 201, 430–438 (2003).
Jarrard, D. F., Bussemakers, M. J., Bova, G. S. & Isaacs, W. B. Regional loss of imprinting of the insulin-like growth factor II gene occurs in human prostate tissues. Clin Cancer Res 1, 1471–1478 (1995).
Leick, M. B., Shoff, C. J., Wang, E. C., Congress, J. L. & Gallicano, G. I. Loss of imprinting of IGF2 and the epigenetic progenitor model of cancer. Am J Stem Cells 1, 59–74 (2012).
Cui, H. et al. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science 299, 1753–1755 (2003).
Sakatani, T. et al. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science 307, 1976–1978 (2005).
Lui, J. C. & Baron, J. Evidence that Igf2 down-regulation in postnatal tissues and up-regulation in malignancies is driven by transcription factor E2f3. Proc Natl Acad Sci USA 110, 6181–6186 (2013).
Tada, Y. et al. The stem cell transcription factor ZFP57 induces IGF2 expression to promote anchorage-independent growth in cancer cells. Oncogene 34, 752–760 (2015).
Vidal, S. J. et al. A targetable GATA2-IGF2 axis confers aggressiveness in lethal prostate cancer. Cancer Cell 27, 223–239 (2015).
Kajimura, S., Aida, K. & Duan, C. Insulin-like growth factor-binding protein-1 (IGFBP-1) mediates hypoxia-induced embryonic growth and developmental retardation. Proc Natl Acad Sci USA 102, 1240–1245 (2005).
Clemmons, D. R. Use of mutagenesis to probe IGF-binding protein structure/function relationships. Endocr Rev 22, 800–817 (2001).
Firth, S. M. & Baxter, R. C. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev 23, 824–854 (2002).
Benayoun, B. A. et al. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 158, 673–688 (2014).
Zhou, C. et al. Comprehensive profiling reveals mechanisms of SOX2-mediated cell fate specification in human ESCs and NPCs. Cell Res 26, 171–189 (2016).
Chiou, S. H. et al. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res 70, 10433–10444 (2010).
Guo, Y. et al. Expression profile of embryonic stem cell-associated genes Oct4, Sox2 and Nanog in human gliomas. Histopathology 59, 763–775 (2011).
Luo, W. et al. Embryonic stem cells markers SOX2, OCT4 and Nanog expression and their correlations with epithelial-mesenchymal transition in nasopharyngeal carcinoma. PLoS One 8, e56324 (2013).
Yu, F. et al. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene 30, 2161–2172 (2011).
Wong, D. J. et al. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell 2, 333–344 (2008).
Zhu, F. et al. SOX2 Is a Marker for Stem-like Tumor Cells in Bladder Cancer. Stem Cell Reports 9, 429–437 (2017).
Aaboe, M. et al. SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res 66, 3434–3442 (2006).
Heidenblad, M. et al. Tiling resolution array CGH and high density expression profiling of urothelial carcinomas delineate genomic amplicons and candidate target genes specific for advanced tumors. BMC Med Genomics 1, 3 (2008).
Rogers, M. A., Kalter, V., Strowitzki, M., Schneider, M. & Lichter, P. IGF2 knockdown in two colorectal cancer cell lines decreases survival, adhesion and modulates survival-associated genes. Tumour Biol 37, 12485–12495 (2016).
Unger, C. et al. Stromal-derived IGF2 promotes colon cancer progression via paracrine and autocrine mechanisms. Oncogene 36, 5341–5355 (2017).
Fang, X. et al. ChIP-seq and functional analysis of the SOX2 gene in colorectal cancers. Omics 14, 369–384 (2010).
Boland, M. J., Nazor, K. L. & Loring, J. F. Epigenetic regulation of pluripotency and differentiation. Circ Res 115, 311–324 (2014).
Warrick, J. I. et al. FOXA1, GATA3 and PPAR Cooperate to Drive Luminal Subtype in Bladder Cancer: A Molecular Analysis of Established Human Cell Lines. Sci Rep 6, 38531 (2016).
Pan, S., Zhan, Y., Chen, X., Wu, B. & Liu, B. Identification of Biomarkers for Controlling Cancer Stem Cell Characteristics in Bladder Cancer by Network Analysis of Transcriptome Data Stemness Indices. Front Oncol 9, 613 (2019).
Choi, W. et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 25, 152–165 (2014).
Hwang, W. et al. Expression of Neuroendocrine Factor VGF in Lung Cancer Cells Confers Resistance to EGFR Kinase Inhibitors and Triggers Epithelial-to-Mesenchymal Transition. Cancer Res 77, 3013–3026 (2017).
Chou, Y. T. et al. CITED2 functions as a molecular switch of cytokine-induced proliferation and quiescence. Cell Death Differ 19, 2015–2028 (2012).
Acknowledgements
This work was supported by National Tsing Hua University, Shuang Ho Hospital, Taipei Medical University (106TMU-SHH-25), and Ministry of Science and Technology (104-2314-B-038-024- to Y.H. Wang, 106-2320-B-007-005-MY3 to Y.T. Chou), Executive Yuan, Taiwan.
Author information
Authors and Affiliations
Contributions
Y.-F. Chiu: collection and assembly of data, data analysis and interpretation, manuscript writing; C.-C. Wu: conception and design, data analysis and interpretation, manuscript editing; M.-H. Kuo, C.-C. Miao, M.-Y. Zheng, P.-Y. Chen, and S.-C. Lin: collection and assembly of data; J.-L. Chang: pathological assessment, data analysis and interpretation; Y.-H. Wang: conception and design, data analysis and interpretation, manuscript editing and final approval of manuscript; Y.-T. Chou: conception and design, data analysis and interpretation, manuscript writing and final approval of manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chiu, YF., Wu, CC., Kuo, MH. et al. Critical role of SOX2–IGF2 signaling in aggressiveness of bladder cancer. Sci Rep 10, 8261 (2020). https://doi.org/10.1038/s41598-020-65006-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-65006-z
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.