Abstract
Meloidogyne incognita is a plant-parasitic root-knot nematode (RKN, PPN) responsible for causing damage to several crops worldwide. In Caenorhabditis elegans, the DAF-16 and SKN-1 transcription factors (TFs) orchestrate aging, longevity, and defense responses to several stresses. Here, we report that MiDaf16-like1 and MiSkn1-like1, which are orthologous to DAF-16 and SKN-1 in C. elegans, and some of their targets, are modulated in M. incognita J2 during oxidative stress or plant parasitism. We used RNAi technology for the stable production of siRNAs in planta to downregulate the MiDaf16-like1 and MiSkn1-like1 genes of M. incognita during host plant parasitism. Arabidopsis thaliana and Nicotiana tabacum overexpressing a hairpin-derived dsRNA targeting these genes individually (single-gene silencing) or simultaneously (double-gene silencing) were generated. T2 plants were challenged with M. incognita and the number of eggs, galls, and J2, and the nematode reproduction factor (NRF) were evaluated. Our data indicate that MiDaf16-like1, MiSkn1-like1 and some genes from their networks are modulated in M. incognita J2 during oxidative stress or plant parasitism. Transgenic A. thaliana and N. tabacum plants with single- or double-gene silencing showed significant reductions in the numbers of eggs, J2, and galls, and in NRF. Additionally, the double-gene silencing plants had the highest resistance level. Gene expression assays confirmed the downregulation of the MiDaf16-like1 and MiSkn1-like1 TFs and defense genes in their networks during nematode parasitism in the transgenic plants. All these findings demonstrate that these two TFs are potential targets for the development of biotechnological tools for nematode control and management in economically important crops.
Similar content being viewed by others
Introduction
Plant-parasitic nematodes (PPNs) are one of the major agricultural pathogens worldwide1. PPNs disturb plant roots by altering the cell cycle, increasing the size of parasitized cells, and causing cell hyperproliferation. This process results in the compatible interaction and development of nematode feeding sites (galls)2,3,4,5,6, which disrupt the uptake of water and nutrients and reduce plant growth and yield7,8,9. Root-knot nematodes (RKNs) are obligate sedentary endoparasites from the genus Meloidogyne spp.10. Meloidogyne incognita is one of the most commonly reported species, causing damage in several crops of economic importance worldwide11. Its life cycle comprises of six stages, namely, egg, J1 (first-stage juvenile), J2 (second-stage juvenile), J3 (third-stage juvenile), J4 (fourth-stage juvenile), and adults (female and male). The J3, J4, and females are typically sedentary endophytes, while the eggs and J2 are exophytes11. The limited range of available control agents or resistant cultivars has limited the efficiency of nematode control and management1,12. Thus, the development of new biotechnology tools is of great importance to overcome these challenges.
Plant-nematode interactions involve an extensive molecular immunity network involved in both defense and counter-defense13. After recognition of PPN elicitors, the host plants increase the production of reactive oxygen and nitrogen species (e.g., hydrogen peroxide-H2O2), and other toxic molecules derived from secondary metabolism14,15,16,17. In contrast, PPNs increase the production and release of antioxidant and detoxifying compounds (e.g., ROS scavengers, glutathione peroxidases, peroxidases, peroxiredoxins, and catalases)18,19,20, and effector proteins to overcome the host defense21,22,23. Thus, nematodes minimize plant cell damage, develop a feeding site, and promote cellular reproduction in susceptible plants24,25.
In Caenorhabditis elegans, the insulin/IGF-1 signaling (IIS) pathway acts as a major mediator between nematode development and stress responses26. The IIS pathway modulates oxidative stress responses by activating the phosphorylation cascade of PDK-1/AKT proteins, leading to the sequestration of DAF-16 and SKN-1 transcription factors (TFs) in the cytoplasm27,28. The C. elegans DAF-2 protein acts as a membrane receptor for insulin, which negatively regulates DAF-16 and SKN-129,30. In contrast, miR-71 expression inhibits this phosphorylation cascade, allowing the nuclear translocation of DAF-16 and SKN-131,32. Daf-16 and Skn-1TFs are large families of genes containing a highly conserved DNA binding region (FOXO and bZIP domains, respectively). These TFs are responsible for the modulation of up to 500 and 846 genes, respectively, which code for numerous protein families with some overlapping targets in the nematode secretome13. These target proteins are implicated in the regulation of the antioxidant and detoxification pathways and the unfolding protein response33,34,35,36,37. The genome and transcriptome sequence data from PPNs revealed that the orthologues of the Daf-16 and Skn-1 genes from C. elegans are also present in M. incognita11,38,39. Thus, it is postulated that DAF-16 and SKN-1 can also orchestrate some important immune adaptive responses to environmental stresses in PPNs during both oxidative stress and plant parasitism13,19,20. These features suggest that the Daf-16 and Skn-1 genes are interesting targets for the development of RNA interference (RNAi)-based new biotechnological tools (NBTs) for nematode control.
Herein, working with orthologous genes, we used RNAi technology for the in planta production of engineered siRNAs to downregulate the MiDaf16-like1 and MiSkn1-like1 genes of M. incognita. The A. thaliana and N. tabacum overexpressing a dsRNA that targets these genes were generated. Then, T2 plants were challenged with M. incognita, and the resistance level of these plants was evaluated. Our results showed that both the MiDaf16-like1 and MiSkn1-like1 genes and some other genes downstream of these TFs are upregulated in M. incognita J2 during oxidative stress or plant parasitism. Single- or double-gene silencing plants of A. thaliana or N. tabacum showed significant reductions in the number of eggs, J2, and galls, and a decrease in the nematode reproduction factor (NRF) compared with wild-type (WT) plants. The double-gene silencing plants had the highest resistance level, which was correlated with efficient downregulation of the two TFs, and lower expression of genes involved in oxidative stress response, detoxification, and the antioxidant pathway. As a result of our study, we have characterized for the first time the functions of DAF-16 and SKN-1 TFs in the PPN M. incognita.
Results
In silico analysis reveals potential Daf-16 and Skn-1 orthologous genes in M. incognita
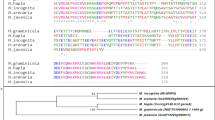
The DAF-16 and SKN-1 TFs were first identified in C. elegans and associated with the defense system against oxidative stress and increased nematode lifespan40,41,42. The family of these TFs is considered highly conserved not only in nematodes but also in mammals34,43,44. We used in silico analyses to search for putative orthologous genes in the M. incognita genome and identified 19 Daf-16-like and 4 Skn-1-like genes (Supplemental Table 1). The Daf-16 orthologous (designated MiDaf16-like1 to 19) genes showed low nucleotide (Fig. 1A) or amino acid (Fig. 1B) sequence identity when compared with those the C. elegans, other phytonematodes (Meloidogyne hapla, Globodera pallida and Bursaphelenchus xylophilus), the free-living nematode Pristionchus pacificus and the human parasite Strongyloides stercoralis. However, the FOXO domain, which corresponds to the core of the DNA binding region, was highly conserved in almost all 19 MiDaf16-like genes, with the exception of the MiDaf16-like4 and 5 genes (Fig. 1C). A phylogenetic analysis using nucleotide sequences of these MiDaf16-like genes suggested that MiDaf16-like2 could be considered an orthologue of the Daf-16 gene from C. elegans (Fig. 1D). However, phylogenetic analyses using the full amino acid sequence (Fig. 1E) or only the FOXO domain (Fig. 1F) sequences do not clearly show that the MiDaf16-like2 gene can unambiguously be considered the orthologue. In contrast, the MiSkn1-like1 to 4 genes showed greater identity in their nucleotide (Fig. 2A), amino acid (Fig. 2B), or only bZIP domain (Fig. 2C) sequences among themselves or when compared with sequences from other phytonematodes, but they had low sequence identity with the Skn-1 gene of C. elegans. Similar to the MiDaf16-like genes, the core of the DNA binding region of the bZIP domain from the MiSkn1-like genes was highly conserved compared with those of C. elegans and other phytonematodes (Fig. 2D). Phylogenetic analyses using nucleotide (Fig. 2E) or only the bZIP domain (Fig. 2F) sequences showed that the MiSkn1-like1 to 4 genes are clustered in a different clade from the Skn-1 gene of C. elegans. Thus, among multiple orthologous putative genes in M. incognita, we designated MiDaf16-like1, 2, 3, 11, and 12 as potential orthologues of the Daf-16 gene of C. elegans, while for the MiSkn1-like genes, it was not evident that may be considered the orthologue of the Skn-1 gene of C. elegans.
In silico analysis of Daf-16 (Dauer Formation-16) genes from nematodes. Pairwise sequence identity matrix from nucleotide (A) and amino acid (B) sequences generated using the Sequence Demarcation Tool Version 1.2 software68. (C) Positional conservation of the FOXO (Forkhead box) domain generated from multiple sequence alignment by the Color Align Conservation software69. Evolutionary analysis from nucleotide (D), amino acid (E) and FOXO domain (F) sequences generated from the Phylogeny.fr web service71. M. incognita (Mi), C. elegans (Ce), P. pacificus (Pp), M. hapla (Mh), G. pallida (Gp), S. stercoralis (Ss) and B. xylophilus (Bx). MiDaf16-like1 (Minc3s02528g30466), MiDaf16-like2 (Minc3s00293g09565), MiDaf16-like3 (Minc3s06738g40249), MiDaf16-like4 (Minc3s02143g28529), MiDaf16-like5 (Minc3s03756g34708), MiDaf16-like6 (Minc3s06700g40200), MiDaf16-like7 (Minc3s00670g15892), MiDaf16-like8 (Minc3s00896g18634), MiDaf16-like9 (Minc3s05371g38122), MiDaf16-like10 (Minc3s01745g26020), MiDaf16-like11 (Minc3s01171g21384), MiDaf16-like12 (Minc3s00913g18806), MiDaf16-like13 (Minc3s00459g12679), MiDaf16-like14 (Minc3s09607g43370), MiDaf16-like15 (Minc3s03624g34358), MiDaf16-like16 (Minc3s00600g14903), MiDaf16-like17 (Minc3s01319g22739), MiDaf16-like18 (Minc3s00100g04542), MiDaf16-like19 (Minc3s02176g28694), MhDaf16 (contig353.frz3.gene4), SsDaf16 (AAQ23177), CeDaf16 (R13H8.1c), PpDaf16 (AGA16632), BxDaf16 (BXY_0566400), and GpDaf16 (GPLIN_001276900).
In silico analysis of Skn-1 (Skinhead-1) genes from nematodes. Pairwise sequence identity matrix from nucleotide (A), amino acid (B) and Basic leucine zipper (bZIP) domain (C) sequences generated using Sequence Demarcation Tool version 1.2 software68. (D) Positional conservation of the bZIP domain generated from multiple sequence alignment by Color Align Conservation software69. Evolutionary analysis from nucleotide (E) and bZIP domain (F) sequences generated from the Phylogeny.fr web service71. M. incognita (Mi), C. elegans (Ce), P. pacificus (Pp), M. hapla (Mh), G. pallida (Gp), St. stercoralis (Ss) and B. xylophilus (Bx). MiSk1-like1 (Minc3s02028g27861), MiSkn1-like2 (Minc3s02028g27862), MiSkn1-like3 (Minc3s08604g42418), MiSkn1-like4 (Minc3s03116g32841), MhSkn1 (MhA1_Contig1686.frz3.gene3), SsSkn1 (SSTP_0000496400), CeSkn1 (CCD62212), PpSkn1 (PPA13755), BxSkn1 (BXY_0018200), and GpSkn1 (GPLIN_000599400).
MiDaf16-like1 and MiSkn1-like1 expression are modulated during oxidative stress and plant parasitism
To confirm the association of these two TFs in the defense response of M. incognita against oxidative stress and plant parasitism, we selected MiDaf16-like1 and MiSkn1-like1 for further study. Initially, we determined that 0.8 mM H2O2 is the lethal concentration (LC50) for newly hatched M. incognita J2 (Fig. 3A). Then, we exposed 5,000 newly hatched M. incognita J2 to 0.1 mM, 0.4 mM, and 0.8 mM H2O2 for 4 and 12 hours, collected them, isolated the RNA, and evaluated the expression profile of these two genes over time in response to the different concentrations of H2O2, compared to newly hatched J2 maintained in Milli-Q water. Real-time quantitative PCR assays showed that both genes were in fact modulated in response to oxidative stress (Fig. 3B). The MiDaf16-like1 gene was gradually upregulated in J2 exposed from 0.1 mM to 0.8 mM H2O2, maintaining a constant expression level over time. In contrast, the MiSkn1-like1 gene was highly upregulated in J2 exposed to 0.4 mM H2O2 for 4 h, while its expression level decreased after 12 h of exposure. Regarding the expression profile in the preparasitic phase or during the parasitism of host plants, we observed that these two genes are also modulated throughout the parasitic cycle of the nematode in both A. thaliana (Fig. 3C) and N. tabacum (Fig. 3D) WT plants. The highest level of expression of both genes was observed after 15 days post-inoculation (dpi), showing that they can act simultaneously in the defense process of M. incognita against multiple stresses during the parasitic phase. Then, we evaluated the expression profiles of some genes in the networks of these two TFs. Among them, the MiSod3-like1, MiGst1-like1, and MiTTL5-like1 genes, which potentially act in the M. incognita antioxidant pathway, in the detoxification pathway, and as an effector secreted by the nematode in response to oxidative stress, respectively. Real-time PCR assays showed that these genes are also modulated in M. incognita J2 when they are exposed to oxidative stress (Fig. 3E). Here, we again used H2O2 as the oxidative stress-inducing agent and exposed newly hatched M. incognita J2 to this stress at different concentrations and exposure times. The MiSod3-like1 and MiGst1-like1 genes had higher expression than MiTTL5-like1, which increased in parallel with the increasing H2O2 concentrations and the time of exposure of the nematode. These findings provide evidence that the MiDaf16-like1 and MiSkn1-like1 genes are indeed upregulated in M. incognita under oxidative stress, and that these genes modulate defense genes in the nematode. Next, we evaluated the expression profiles of these two TFs in the different life stages of M. incognita. Initially, we mined transcriptome datasets available in a public database (NCBI SRA) generated from the egg mass, J2, J3, J4, and M. incognita females39. These in silico results showed that the two genes are potentially expressed in all life stages of the nematode, with the MiDaf16-like1 gene having the highest number of reads (RPM) mapped in the J2 stage, while the MiSkn1-like1 gene had the largest number of reads mapped in stage J3 (Fig. 3F). In addition, we used these same transcriptome datasets to evaluate the expression profiles of all 19 MiDaf16-like and 4 MiSkn1-like genes in the different life stages of M. incognita. Our in silico results showed that all the MiDaf16-like genes are potentially expressed at all life stages of M. incognita, except MiDaf16-like7, for which no reads were mapped to its transcript (Supplemental Fig. 1A to E). The expression profiles of these genes were differentially modulated throughout the life cycle of the nematode, especially MiDaf16-like1, 2, 12, 16, and 19, which had more reads mapped to their transcripts, suggesting that they had a higher level of expression than the other genes. In addition, the highest level of expression of these genes was observed at the J2 stage, whereas the MiDaf16-like1 gene had a similar expression profile at all life stages, except for the J2 stage, which had a 5-fold greater number of reads mapped to its transcript than the other stages, suggesting a higher expression level in this stage. Regarding the MiSkn1-like genes, the four genes also showed potential expression throughout the life cycle of M. incognita, with greater expression in the J2 and J3 stages (Supplemental Fig. 1F). The MiSkn1-like2 gene had the highest number of reads mapped to its transcript compared to the others overall, whereas MiSkn1-like1 had the highest level of reads mapped to its transcript in the J3 stage. Real-time PCR assays using the total RNA isolated from the different plant-parasitic stages of M. incognita showed that the expression profiles of these two genes are indeed upregulated throughout the cycle of parasitism. The MiDaf16-like1 gene showed a greater expression level in the J2 stage (Fig. 3G), while the MiSkn1-like1 gene had a greater expression level in the J3/J4 stage (Fig. 3H). Finally, our results confirm that these genes are indeed associated with the defense response to oxidative stress and plant parasitism. In addition, their expression profile is orchestrated throughout their entire life cycle and in all phases of plant parasitism.
Expression profiles of MiDaf16-like1 (Minc3s02528g30466) and MiSkn1-like1 (Minc3s02028g27861) in response to oxidative stress and plant parasitism. (A) Survival percentage (%) of M. incognita J2 race 3 after overnight exposure to different concentrations of H2O2, as described by Vicente et al.61. Error bars represent confidence intervals corresponding to three biological replicates. Different letters above the columns indicate significant differences (p-value <0.05) between different survival percentages in each H2O2 treatment, according to Tukey’s test. (B) Expression profile of MiDaf16-like1 and MiSkn1-like1 in M. incognita J2 race 3 after 4 and 12 h of exposure to different concentrations of H2O2. (C) Expression profile of MiDaf16-like1 and MiSkn1-like1 during M. incognita parasitism (5 to 30 days post-inoculation; dpi) in (C) A. thaliana and (D) N. tabacum. (E) Expression profile of MiSod3-like1, MiTTL5-like1, and MiGst1-like1 during exposure of M. incognita J2 race 3 to H2O2. Newly hatched J2 were used as controls for all assays of gene expression level. Error bars represent confidence intervals corresponding to three biological replicates. (F) Expression profiles of MiDaf16-like1 and MiSkn1-like1 in different life stages of M. incognita using transcriptome datasets (BioProject number: PRJNA39055939); retrieved from the BioSample database (NCBI). Error bars represent confidence intervals corresponding to three libraries per life stage of the nematode. Expression profile measured by real-time PCR of the MiDaf16-like1 (G) and MiSkn1-like1 genes in different life stages of M. incognita race 3. The relative expression levels were normalized with the β-tubulin (MiTUB) and glyceraldehyde 3-phosphate dehydrogenase (MiGAPDH) endogenous reference genes (Supplemental Table 3). Error bars represent confidence intervals corresponding to three biological replicates.
Downregulation of the MiDaf16-like1 and MiSkn1-like1 genes by RNAi
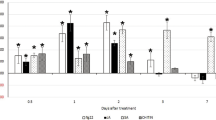
To evaluate the effect of downregulation of the MiDaf16-like1 and MiSkn1-like1 genes in the phytopathogenicity of M. incognita, we generated several transgenic plants of A. thaliana and N. tabacum by overexpressing an engineered dsRNA to accumulate siRNAs in these plants. These dsRNAs were engineered to downregulate MiDaf16-like1 or MiSkn1-like1 (single-gene silencing), or both genes simultaneously (double-gene silencing) (Fig. 4A). For the modulation of the MiDaf16-like1 gene, the dsRNA was constructed with a nucleotide sequence from the FOXO domain of this gene (Supplemental File 1), while for the modulation of the MiSkn1-like1 gene, we used a sequence from the bZIP domain (Supplemental File 2). Because we used a relatively conserved region for each of these two genes (Figs. 1C and 2D) to direct the in planta production of siRNAs, the MiDaf16-like12 and 15 (Supplemental Fig. 2A), and MiSkn1-like 2 to 4 (Supplemental Fig. 2B) genes could also be potentially downregulated during parasitism in these plants. In A. thaliana, we generated 14 independent transformants to drive the downregulation of the MiDaf16-like1 gene, nine transformants for downregulation of the MiSkn1-like1 gene, and ten double-gene silencing transformants for simultaneous downregulation of these genes (Fig. 4B). In contrast, six independent transformants were generated for each of these strategies (single- or double-gene silencing) in N. tabacum (Fig. 4C). Then, A. thaliana and N. tabacum plants of the T2 generation were challenged to evaluate their resistance to M. incognita. At 60 dpi of freshly hatched J2, the number of eggs, J2, and galls, and the NRF were determined. In almost all single- or double-gene silencing plants of A. thaliana, we observed a significant reduction in the number of eggs (Fig. 5A), J2 (Fig. 5B), and galls (Fig. 5C), and NRF (Fig. 5D) ranging from 20 to 80%, when compared to that of WT plants. In addition, the double-gene silencing plants presented an apparently better performance than the single-gene silencing plants (Supplemental Fig. 3A). In addition, we evaluated some of these same A. thaliana single- and double-gene silencing plants for resistance to the M. incognita J2 strain Morelos (Mexican isolate). Interestingly, we observed a high resistance level similar to that when inoculated with M. incognita race 3 with respect to the numbers of both galls (Supplemental Fig. 3D) and egg mass (Supplemental Fig. 3E). In the N. tabacum plants, we also observed reductions in the number of eggs (Fig. 5E), J2 (Fig. 5F), and galls (Fig. 5G), and in the NRF (Fig. 5H) in most transgenic plants compared to WT plants. Similar to the A. thaliana plants, the N. tabacum double-gene silencing plants showed an apparently better performance than the single-gene silencing plants (Supplemental Fig. 3B). Additionally, we evaluated the morphology of galls at 45 dpi in WT, single-gene silencing plants, and double-gene silencing plants of A. thaliana. In the WT line, we observed the end of the nematode life cycle, while the single-gene silencing lines showed giant cells lacking the typical dense cytoplasm that were apparently smaller and an apparent delay in nematode development. Similarly, the double-gene silencing lines showed giant cells lacking the characteristic dense cytoplasm but were apparently smaller (Supplemental Fig. 3C). Overall, our findings confirm that the potential in planta downregulation of the MiDaf16-like1 and MiSkn1-like1 genes disrupts the efficiency of M. incognita in parasitizing the host plant.
Binary vectors and genetic transformation of A. thaliana and N. tabacum. (A) Overview of the binary vectors for single knockdown of MiDaf16-like1 or MiSkn1-like1, and both simultaneously (double-gene silencing), which were used for in planta transformation mediated by Agrobacterium tumefaciens. (B) PCR detection of the transgenes in A. thaliana (B) and N. tabacum (C) T0 plants. Marker: 1.0-kb DNA ladder (Invitrogen® Cat. #10787018); NT: non-transgenic line used as a negative control for the PCR assay and bioassays in the greenhouse; C+: a transgenic plant used as a positive control for the PCR assay.
Evaluation of the resistance levels of A. thaliana (A to D) and N. tabacum (E to H) T2 plants to infection with M. incognita race 3. The number of eggs and J2 per gram of root, number of galls per plant, and NRF were evaluated 60 days post-inoculation. Error bars represent confidence intervals corresponding to three technical replicates. Asterisks indicate significant differences based on Tukey’s test at 5%.
The M. incognita susceptibility is correlated with downregulation of its defense genes
To confirm whether transgenic plant parasitism results in siRNA uptake and the consequent downregulation of MiDaf16-like1 and MiSkn1-like1, we harvested galls at 60 dpi on WT and transgenic lines, isolated total RNA and evaluated the expression profiles of these genes. In addition, we evaluated the expression profiles of some genes from the DAF-16 and SKN-1 networks involved in nematode defense against stresses. Real-time PCR assays showed that MiDaf16-like1 (Supplemental Fig. 5A), MiSkn1-like1 (Supplemental Fig. 5B), or both simultaneously (Supplemental Fig. 5C) were downregulated in M. incognita during parasitism in single- or double-gene silencing plants, respectively. In addition, we observed that at least some defense genes from the Daf-16 and Skn-1 networks were consequently also downregulated during the parasitism of nematodes in single- or double-gene silencing plants (Supplemental Fig. 5D). In M. incognita infecting both single- or double-gene silencing plants, the MiPRDX2-like1, MiSod3-like1, MiGPX-like1, MiGst1-like1, and MiSod1-like1 genes were simultaneously downregulated at a level of approximately 80%, with the exception of the MiGst1-like1 gene in one of the double-gene silencing plants. The in silico analyses using the same transcriptome datasets available in the public database (NCBI SRA) generated from the egg mass, J2, J3, J4, and females of M. incognita39 showed that these defense genes were differentially modulated in all life stages of the nematode (Supplemental Fig. 4A to F). Given this, our findings confirm the efficient uptake of siRNAs during plant parasitism and downregulation of these TFs and genes in their networks, resulting in a decreased ability of M. incognita to infect host plants.
Discussion
The insulin/IGF-1 signaling (IIS) pathway was first shown to regulate dauer formation and resistance to multiple stresses in C. elegans26,45. Subsequently, the IIS pathway was also characterized in other organisms (e.g., PPN, Drosophila melanogaster, mice, and humans), confirming its role associated with the aging, longevity and defense response to stresses43,46. In C. elegans, Daf-2 and Age-1 genes coding to IGF-1 insulin receptor and phosphatidylinositol-3-OH kinase (PI3K) of the IIS pathway, respectively. The inhibition of these two genes results in lifespan extension in the nematode30,47,48. The signal transduction in the IIS pathway is orchestrated by sequential events and modulated by environmental conditions (e.g., high insulin content, no stresses, oxidative stress, stress from the environment). In favorable conditions (e.g., high insulin content and no stresses), the IIS pathway is activated, resulting in the normal development of nematodes. The binding of insulin or insulin-like peptides to the DAF-2 receptor results in the activation of the AGE-1 and PI3K genes, increasing the level of phosphatidylinositol(3,4,5)-trisphosphate (PIP3). PIP3 accumulation is balanced by DAF-18/PTEN phosphatase, promoting its conversion to phosphatidylinositol(4,5)-bisphosphate (PIP2). Then, PIP3 activates the kinase signaling cascade, composed of 3-phosphoinositide-dependent protein kinase 1 (PDK-1), protein kinase B (AKT-1 and -2), and serum- and glucocorticoid-inducible kinase-1 (SGK-1). In turn, these components phosphorylate and inactivate the DAF-16 and SKN-1 TFs by sequestering them in the cytoplasm and preventing nuclear import29. In unfavorable conditions to the nematode (e.g., low insulin content, oxidative stress, adverse environment), the IIS pathway is deactivated, resulting in DAF-16 and SKN-1 TF activation by efficient translocation from the cytoplasm to the nucleus. In addition, miR-71 inhibits the phosphorylation cascade via posttranscriptional regulation of the Age-1 and Akt genes, allowing the efficient translocation of DAF-16 and SKN-1 to the nucleus31. In the oxidative stress response, the PMK-1 kinase from the p38 MAPK pathway phosphorylates SKN-1, leading to its translocation and nuclear accumulation49,50. On the other hand, the 14-3-3 scaffolding proteins bind to the phosphorylation sites of DAF-16 and contribute to its sequestration within the cytoplasm51,52. In the nucleus, the DAF-16 and SKN-1 TFs are responsible for the transcriptional activation of up to 500 and 846 genes, respectively13,53. These activated genes belong to several functional groups and are implicated in the aging and longevity process and the antioxidant, detoxification, and protein unfolding response pathways54,55,56,57. Thus, downregulation of the IIS pathway and the consequent upregulation/activation of the DAF-16 and SKN-1 TFs confer high resistance to a variety of stresses (e.g., heat, hypoxia, osmotic, UV, metal toxicity, and oxidative stresses) in C. elegans.
Genome and transcriptome studies from PPNs have revealed genes orthologous to Daf-16 and Skn-1 from C. elegans in the Meloidogyne, Pratylenchus, and Bursaphelenchus genera39,58,59,60. The high level of sequence conservation suggests similar functions to those observed in C. elegans13,35,59. Studies of comparative genomes in PPNs have identified numerous defense genes from the DAF-16 and SKN-1 networks involved in ROS scavenging, coding proteins linked to the antioxidant pathway, superoxide dismutases (SOD), catalases (CAT), glutathione S-transferases (GST), glutathione peroxidase (GPX), and peroxiredoxins (PRDX)11,18,20,58,61. In addition, plant-nematode interaction studies suggest the involvement of these genes in the nematode defense response, both in the control of endogenous oxidative stress and in the modulation of the host cell18,20,62. In this study, we identified at least 19 MiDaf16-like and 4 MiSkn1-like genes in the M. incognita genome, with a high degree of sequence conservation in the FOXO and bZIP domains, and almost all were expressed in all life stages of the nematode. In addition, we confirmed that the MiDaf16-like1 and MiSkn1-like1 genes are expressed in all life stages of M. incognita and are modulated in response to oxidative stress and during plant parasitism, confirming the IIS pathway activation and its functionality in M. incognita; we also showed that some genes in their networks are consequently also modulated. Interestingly, without targeting all the members of these families because they are highly conserved in nematodes and other animals, these two gene families may be potential targets for the development of NBTs to impair the defense pathways of PPNs during plant parasitism. M. incognita is a RKN10, an important plant pathogen causing economic losses in several crops worldwide11, and an excellent model system for obligate sedentary endoparasitic PPNs13,21. Our findings confirm that in planta downregulation of MiDaf16-like1, MiSkn1-like1 (single-gene silencing), or both genes simultaneously (double-gene silencing), makes nematodes more susceptible to stress conditions during parasitism, significantly reducing the number of galls and eggs, NRF, and, consequently, decreasing the source of inoculum. In addition, we also found that the nematodes maintained in transgenic plants with either single- or double-gene silencing constructs present an apparent delay in nematode development, while their giant cells are smaller. These results can be explained by a depletion of nematode defense or counter-defense mechanisms during plant parasitism and potentially by a disruption of the fine-tuning between the nematode defense and development pathways. In agreement with this hypothesis, we have confirmed that downregulation of the MiDaf16-like and MiSkn1-like1 genes results in strong downregulation of genes from their networks during parasitism of M. incognita in single- or double-gene silencing plants. In this respect, the two superoxide dismutases (MiSod1-like1 and MiSod3-like1), peroxiredoxin (MiPRDX2-like1), glutathione S-transferase and peroxidase (MiGst1-like1 and MiGPX-like1), which act in the nematode antioxidant and detoxification pathways18,19,20,63,64, were indeed downregulated in M. incognita during parasitism in transgenic plants.
Recently, Koutsovoulos et al.65 sequenced the genomes of 11M. incognita isolates race 1 to 4 infecting different host plant across Brazil (including the race 3, used in this study) and showed that cumulative fixed divergence across these Brazilian isolates and the reference genome (M. incognita strain Morelos) reached approximately 0.02% of the nucleotides. However, these few point variations between the isolates showed no significant association with the host species, geographical origin of the samples and crop on which they were collected, and there were no disruptive variations identified in the coding regions of genes. Thus, these authors suggested that other factors are more important for the adaptation of this species than these few point mutations. Consistent with this hypothesis, Castagnone‐Sereno et al.66 showed that convergent gene copy number variations (CNV) were associated with breaking down of resistance by M. incognita. In addition, these authors showed that CNV and speculated that expression levels of these genes are two major features associated with M. incognita pathogenicity in different hosts. This data suggests that MiDAF16-like and MiSKN1-like gene families may vary in copy number of these genes between different M. incognita isolates and races, or host crops. In contrast, these genes may present considerable sequences similarity between different nematode races or isolates. Thus, the use of this biotechnological strategy is of great relevance in crops or cultivars of economic interest (such as soybean, cotton, coffee, cocoa, and tomato, among others) considered to be susceptible to this nematode or cultivated in areas with a high incidence of this pathogen. In addition, these two gene families have great potential to be modulated by NBTs in other species of PPNs (e.g., Heterodera schachtii, B. xylophilus, G. pallida, and M. hapla)13 to develop new strategies for their management and control.
In conclusion, we identified the MiDaf16-like and MiSkn1-like gene families in the genome of M. incognita and confirmed its modulation in response to oxidative stress, its expression level in the different life stages of the nematode, and its upregulation during plant parasitism. Next, we showed the efficient in planta production of siRNAs, the successful uptake of siRNAs by the nematode, and the downregulation of the MiDaf16-like1 and MiSkn1-like1 genes and, consequently, the genes in their networks. Additionally, we observed that single- or double-gene silencing plants of A. thaliana or N. tabacum showed high resistance to M. incognita. In addition, considering the high conservation of these gene families, our data suggest that NBTs can also be developed for modulation of these target genes in other species of PPNs. Finally, our findings showed that these two TF families are powerful targets for the development of NBTs to control and manage nematodes in economically important crops.
Material and Methods
In silico analysis of Daf-16 and Skn-1 TFs from nematodes
All gene sequences of M. incognita were retrieved from BioProject ID PRJEB8714 (sample: ERS1696677)38 from the online WormBase database version WBPS1367. Pairwise identity matrices from nucleotide and amino acid sequences were generated using the Sequence Demarcation Tool Version 1.2 software68. Positional conservation of the FOXO (Forkhead box) domain was generated from multiple sequence alignment using Color Align Conservation software69. In addition, the conserved domains in gene sequences were checked using the Conserved Domain Database (CDD)70. Phylogenetic analyses were performed using the Phylogeny.fr web service71. For this, sequences were aligned with MUSCLE software72, and the alignment was curated by the Gblocks model. Then, phylogenetic analyses were performed using the maximum likelihood method by PhyML software using the Approximate Likelihood-Ratio test (aLRT) SH-like branch support and GTR and WAG substitution models for nucleotide and amino acid sequences, respectively. Phylogeny trees were generated and visualized by TreeDyn software also implemented in this same web service. In addition, the gene and protein sequences from other nematode species used in these sequence analyses were also retrieved from the WormBase database version WBPS13 database. The expression levels of MiDaf16-like1, MiSkn1-like1, and some genes from their networks at different stages of the M. incognita life cycle were determined using transcriptome datasets (BioProject number: PRJNA390559) retrieved from the BioSample database (NCBI) (Supplemental Table 2). Fifteen transcriptome libraries from M. incognita egg, J2, J3, J4, and female stages were generated by Choi et al.39 using the Truseq RNA Sample Prep Kit (Illumina), and mRNAs were paired-end sequenced (2×101 bp) using Illumina HiSeq. 2000 technology (Supplemental Table 2). The libraries were downloaded and trimmed, and the transcripts were mapped using the genome reference retrieved from the WormBase Parasite database (BioProject PRJNA 340324)73. The number of reads mapped in interest target genes was normalized in reads per million (RPM). Additionally, the expression profile of MiDaf16-like1 to 19 and MiSkn1-like1 to 4 in different nematode life stages was estimated from these same transcriptome libraries (Supplemental Fig. 1). Features of the MiDaf16-like and MiSkn1-like genes from M. incognita, such as conserved domains in the gene sequences were identified using CDD Database from NCBI74, and PFAM Database from EMBL-EBI75. NES motifs were predicted using NetNES 1.1 Server76, while NLS motifs were predicted using the NLStradamus online tool77.
Meloidogyne incognita J2 inoculum
M. incognita J2 race 3 and M. incognita J2 strain Morelos were obtained from tomato plants (Solanum lycopersicum cv. Santa Clara) inoculated and maintained for eight weeks under greenhouse conditions. Infected roots were washed and macerated using a blender after treatment with 0.5% sodium hypochlorite. Eggs were harvested, rinsed with tap water and subsequently separated from root debris using 100 to 550-μm sieves78. Then, the eggs were hatched under aerobic conditions at 28 °C, and J2 were harvested every two days, decanted and quantified under a microscope using counting chambers.
MiDaf16-like1 and MiSkn1-like1 expression profiles in response to oxidative stress, host plant parasitism, and different nematode life stages
96-well plates containing 40 freshly hatched M. incognita J2 in a final volume of 100 µl were exposed in a gradient ranging from 0 to 5 mM H2O2 as described by Vicente et al.61. After overnight exposure to H2O2 conditions, the nematode survival rate was evaluated under a stereomicroscope. Six biological replicates were performed per treatment. Nematodes were considered dead if no movements were observed after mechanical and light stimulation. Then, tubes containing approximately 5,000 freshly hatched J2 in a final volume of 1 ml were exposed to 0.1, 0.4 and 0.8 mM H2O2 and incubated in the dark without agitation at 28 °C for 4 and 12 hours. The J2 were harvested, and the total RNA was isolated using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA), mild sonication and TissueLyser II (Qiagen, Hilden, Germany). The RNA concentration was estimated using a spectrophotometer (NanoDrop 2000, Thermo Scientific, Massachusetts, USA), and integrity was evaluated with 1% agarose gel electrophoresis. RNA samples were treated with RNase-free RQ1 DNase I (Promega, Madison, Wisconsin, USA) according to the manufacturer’s instructions. Then, 2 μg of DNase-treated RNA was used as a template for cDNA synthesis using Oligo-(dT)20 primer and SuperScript III RT (Life Technologies, Carlsbad, CA, USA), according to the manufacturer’s instructions. The cDNA was quantified by spectrophotometry and diluted with nuclease-free water to 200 ng/µl. RT-qPCR assays were performed in an Applied Biosystems 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using 400 ng of cDNA, 0.2 µM of each gene-specific primer (Supplemental Table 3) and GoTaq® qPCR Master Mix (Promega, Madison, Wisconsin, USA). The conditions for qPCR included an initial 95 °C for 10 min, then 40 cycles of 95 °C for 15 s and 59 °C for 50 s, followed by a final melting curve analysis. The expression of the MiDaf16-like1 (Minc3s02528g30466) and MiSkn1-like1 (Minc3s02028g27861) genes was normalized using the Mi18S (GenBank accession U81578)79 and MiACT (Minc3s00730g16611) endogenous reference genes in A. thaliana and N. tabacum, respectively. In addition, MiSod-3like1, MiGst1-like1, and MiTTL-5-like1 gene expression were also evaluated after J2 exposure to H2O2 and normalized using the Mi18S gene. Newly hatched J2 in Milli-Q water were used as a control for relative expression levels for both J2 exposed to H2O2 and J2 during plant parasitism. All samples were carried out in technical triplicate reactions. Primer efficiencies and target-specific amplification were confirmed by a single and distinct peak in the melting curve analysis. The relative expression level (fold change) was calculated using the 2−∆Ct or 2−∆∆Ct method80. The MiDaf16-like1 and MiSkn1-like1 gene expression in different life stages of M. incognita was determined from the egg mass harvested from tomato roots, J2 newly hatched in Milli-Q water at 28°C, J3 harvested from potential galls (stained with fuchsin) of tobacco at 12 to 15 dpi, J4 harvested from potential galls (stained with fuchsin) of tobacco at 20 to 24 dpi, and females harvested from galls of tobacco at 35 to 40 dpi. The total RNA and cDNA were prepared as described above, while the relative expression level (fold change) was normalized with the β-tubulin (MiTUB) and glyceraldehyde 3-phosphate dehydrogenase (MiGAPDH) endogenous reference genes.
Binary vectors and agrobacterium-mediated plant transformation
Three binary vectors were synthesized by the company Epoch Life Science (Sugar Land, TX, EUA) and subsequently transformed into the A. tumefaciens strain GV3101. The first binary vector was designed to negatively regulate the MiDaf16-like1 gene. Thus, two short fragments comprising the regions 578 to 676 and 2002 to 2108 from the MiDaf16-like1 gene were cloned in tandem, in both the sense and antisense orientations and separated by the PDK intron (Fig. 4A, and Supplemental File 1). The second binary vector was designed to negatively regulate the MiSkn1-like1 gene. Thus, one short fragment comprising the region 2283 to 2624 from the MiSkn1-like1 gene was cloned in sense and antisense and was also separated by the PDK intron. The third binary vector was designed to negatively regulate the MiDaf16-like1 and MiSkn1-like1 genes, simultaneously. For this, these same DNA fragments described above were also cloned into the double-gene silencing vector (Fig. 4A, and Supplemental File 2). It is expected that the siRNAs produced from the transgenes will target the highly conserved regions comprising the FOXO and bZIP domains, respectively (Supplemental Files 1 and 2).
Arabidopsis thaliana ecotype Col-0 was genetically transformed by the floral dip method81, while N. tabacum var. SR1 Petite Havana was transformed from young leaves of 4–5 weeks old, according to Park et al.82. N. tabacum plants were selected in vitro using 5 mg/L glufosinate-ammonium (FINALE, Liberty Link, Bayer). Both A. thaliana and N. tabacum were screened in vivo by glufosinate ammonium spraying, confirmed by conventional PCR using specific primers (Supplemental Table 3) and a quick test strip kit (QuickStix™ Kit for PAT/bar, EnviroLogix, Inc., USA) according to the manufacturer’s instructions. Several independent transformants were chosen for propagation, and several T2 lines of each construct were chosen for the bioassays.
Evaluation of resistance of the transgenic plants to M. incognita
The A. thaliana seedlings were transplanted to pots containing 45 g of autoclaved sand:substrate mixture (1:1; w-w) and maintained in a growth chamber with a 12 h photoperiod and 22 ± 2 °C temperature. Two weeks after transplanting, plants were inoculated with 500 M. incognita J2 race 3 or 200 J2 strain Morelos (suspended in distilled water). Fifteen plants per line were used, and the experiment was repeated at least two times, while WT plants were used as susceptible controls. At 60 days post-inoculation (dpi), the plants were evaluated for the number of eggs per gram of root, number of J2 hatched per gram of root, number of galls per plant, number of egg masses per plant, and NRF. The M. incognita NRF in transgenic plants was determined using Oostenbrink’s formula: NRF = final J2 number/initial J2 number or nematode final population/initial population83,84. In contrast, N. tabacum seedlings from T2 lines were transplanted to pots containing 125 g of sterile sand:soil mixture (1:1; w-w) and maintained in the greenhouse conditions. Eight days after transplanting, plants were inoculated with 1,000 M. incognita J2 (suspended in distilled water). Sixteen plants per transgenic line were used, while WT plants were used as susceptible controls. At 60 dpi, the plants were evaluated for the number of eggs per gram of root, number of J2 hatched per gram of root, number of galls per plant, and NRF. For morphological analysis, galls were harvested from nematode-infected roots of A. thaliana WT, single- or double-gene silencing plants at 45 dpi. Then, they were fixed in 2% glutaraldehyde in 50 mM PIPES buffer, pH 6.9, subsequently dehydrated and embedded in Technovit 7100 (Heraeus Kulzer, Wehrheim, Germany) as described by the manufacturer. Nematode-induced galls were sectioned (3 µm), stained in 0.05% toluidine blue and mounted in Depex (Sigma-Aldrich, St Louis, Missouri, USA). Microscopy analyses were carried out using bright-field optics, and images were acquired with a digital camera (Axiocam, Zeiss, Oberkochen, Germany).
Expression levels of MiDaf16-like1, MiSkn1-like1, and genes from their networks during M. incognita infection in transgenic plants
Galls from five A. thaliana plants (from WT and transgenic plants) of each line were harvested at 60 days post-inoculation and processed in pool form. Then, samples were ground in liquid nitrogen using a mortar and pestle and stored at −80 °C. RNA total was isolated using Concert Plant RNA Purification (Invitrogen, Carlsbad, CA, USA) plus PVP-40 according to the manufacturer’s specifications. Highly pure RNA was used to estimate the relative expression levels of the MiDaf16-like1 and MiSkn1-like1 genes during infection of transgenic plants, as described above. RNA isolated from galls harvested from WT A. thaliana was used as a positive control of the expression of the gene of interest. In addition, from these same RNA samples, we also validated the consequent downregulation of some network genes of the DAF-16 and SKN-1 TFs involved in nematode defense against stresses. The MiPRDX2-like1, MiSod3-like1, MiGPX-like1, MiGst1-like1, and MiSod1-like1 genes were evaluated in M. incognita harvested from four single- or double-gene silencing plants. The expression level was represented as the fold change calculated with the 2-∆CT formula using the glyceraldehyde 3-phosphate dehydrogenase (MiGAPDH) gene as the endogenous reference gene (Supplemental Table 3).
References
Bernard, G. C., Egnin, M. and Bonsi, C. The Impact of Plant-Parasitic Nematodes on Agriculture and Methods of Control, In Nematology - Concepts, Diagnosis and Control https://doi.org/10.5772/intechopen.68958 (2017).
de Almeida Engler, J. et al. Chapter Four - The Plant Cell Cycle Machinery: Usurped and Modulated by Plant-Parasitic Nematodes, in Advances in Botanical Research, Escobar, C. and Fenoll, C. Editors., Academic Press. p. 91–118 (2015).
Shukla, N. et al. Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-infected tomato (Solanum lycopersicum) roots reveals complex gene expression profiles and metabolic networks of both host and nematode during susceptible and resistance responses. Molecular Plant Pathology 19(3), 615–633 (2018).
Antonino de Souza Junior, J. D. et al. Application of Nuclear Volume Measurements to Comprehend the Cell Cycle in Root-Knot Nematode-Induced Giant Cells. Frontiers in Plant Science 8, 961 (2017).
de Almeida Engler, J. et al. Dynamic cytoskeleton rearrangements in giant cells and syncytia of nematode-infected roots. Plant Journal 38(1), 12–26 (2004).
de Almeida Engler, J. et al. CCS52 and DEL1 genes are key components of the endocycle in nematode-induced feeding sites. Plant Journal 72(2), 185–198 (2012).
Carneiro, R. G. et al. Uptake and translocation of nitrogen, phosphorus and calcium in soybean infected with Meloidogyne incognita and M. javanica. Fitopatologia Brasileira 27, 141–150 (2002).
Melakeberhan, H. et al. Effect of Meloidogyne incognita on Plant Nutrient Concentration and Its Influence on the Physiology of Beans. Journal of Nematology 19(3), 324–330 (1987).
Lu, P. et al. Physiological Effects of Meloidogyne incognita Infection on Cotton Genotypes with Differing Levels of Resistance in the Greenhouse. Journal of Nematology 46(4), 352–359 (2014).
Trudgill, D. L. & Blok, V. C. Apomictic, polyphagous root-knot nematodes: exceptionally successful and damaging biotrophic root pathogens. Annual Review of Phytopathology 39, 53–77 (2001).
Abad, P. et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nature Biotechnology 26, 909 (2008).
Seo, Y. & Kim, Y. H. Control of Meloidogyne incognita Using Mixtures of Organic Acids. The Plant Pathology Journal 30(4), 450–455 (2014).
Gillet, F. X. et al. Plant-parasitic nematodes: towards understanding molecular players in stress responses. Annals of Botany 119(5), 775–789 (2017).
Melillo, M. T. et al. ROS and NO production in compatible and incompatible tomato-Meloidogyne incognita interactions. European Journal of Plant Pathology 130(4), 489–502 (2011).
Manosalva, P. et al. Conserved nematode signalling molecules elicit plant defenses and pathogen resistance. Nature Communications 6, 7795 (2015).
Holscher, D. et al. Phenalenone-type phytoalexins mediate resistance of banana plants (Musa spp.) to the burrowing nematode Radopholus similis. PNAS 111(1), 105–110 (2014).
Kong, L.-A. et al. Large-scale identification of wheat genes resistant to cereal cyst nematode Heterodera avenae using comparative transcriptomic analysis. BMC Genomics 16(1), 801 (2015).
Shinya, R. et al. Secretome Analysis of the Pine Wood Nematode Bursaphelenchus xylophilus Reveals the Tangled Roots of Parasitism and Its Potential for Molecular Mimicry. PLoS One 8(6), e67377 (2013).
Dubreuil, G. et al. Peroxiredoxins from the plant parasitic root-knot nematode, Meloidogyne incognita, are required for successful development within the host. International. Journal of Parasitology 41(3-4), 385–96 (2011).
Bellafiore, S. et al. Direct identification of the Meloidogyne incognita secretome reveals proteins with host cell reprogramming potential. PLoS Pathogens 4(10), e1000192 (2008).
Bournaud, C. et al. Meloidogyne incognita PASSE-MURAILLE (MiPM) Gene Encodes a Cell-Penetrating Protein That Interacts With the CSN5 Subunit of the COP9 Signalosome. Frontiers in Plant Science 9(904) (2018).
Xie, J. et al. A Novel Meloidogyne incognita Effector Misp12 Suppresses Plant Defense Response at Latter Stages of Nematode Parasitism. Frontiers in Plant Science 7, 964–964 (2016).
Lin, B. et al. A novel nematode effector suppresses plant immunity by activating host reactive oxygen species-scavenging system. The New phytologist 209(3), 1159–1173 (2016).
Gheysen, G. & Mitchum, M. G. How nematodes manipulate plant development pathways for infection. Current Opinion in Plant Biology 14(4), 415–421 (2011).
Goverse, A. & Smant, G. The Activation and Suppression of Plant Innate Immunity by Parasitic Nematodes. Annual Review of Phytopathology 52(1), 243–265 (2014).
Schindler, A. J. & Sherwood, D. R. Should I stay or should I go? Identification of novel nutritionally regulated developmental checkpoints in C. elegans. Worm 3(4), e979658 (2014).
Lin, K. et al. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nature Genetics 28(2), 139–145 (2001).
Henderson, S. T. & Johnson, T. E. Daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Current Biology 11(24), 1975–1980 (2001).
Tullet, J. M. et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132(6), 1025–1038 (2008).
Larsen, P. L. Aging and resistance to oxidative damage in Caenorhabditis elegans. PNAS 90(19), 8905–8909 (1993).
Boulias, K. & Horvitz, H. R. The C. elegans microRNA mir-71 acts in neurons to promote germline-mediated longevity through regulation of DAF-16/FOXO. Cell metabolism 15(4), 439–450 (2012).
de Lencastre, A. et al. MicroRNAs both promote and antagonize longevity in C. elegans. Current Biology 20(24), 2159–2168 (2010).
Webb, A. E., Kundaje, A. & Brunet, A. Characterization of the direct targets of FOXO transcription factors throughout evolution. Aging cell 15(4), 673–685 (2016).
Nakagawa, S. et al. DNA-binding specificity changes in the evolution of forkhead transcription factors. PNAS 110(30), 12349–12354 (2013).
Choe, K. P., Leung, C. K. & Miyamoto, M. M. Unique structure and regulation of the nematode detoxification gene regulator, SKN-1: implications to understanding and controlling drug resistance. Drug Metabolism Reviews 44(3), 209–223 (2012).
Blackwell, T. K. et al. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radical Biology & Medicine 88(Pt B), 290–301 (2015).
Zou, C.-G. et al. The DAF-16/FOXO Transcription Factor Functions as a Regulator of Epidermal Innate Immunity. PLoS Pathogens 9(10), e1003660 (2013).
Blanc-Mathieu, R. et al. Hybridization and polyploidy enable genomic plasticity without sex in the most devastating plant-parasitic nematodes. PLoS genetics 13, e1006777, https://doi.org/10.1371/journal.pgen.1006777 (2017).
Choi, I. et al. RNA-Seq of Plant-Parasitic Nematode Meloidogyne incognita at Various Stages of Its Development. Frontiers in Genetics 8, 190–190 (2017).
Tullet, J. M. A. et al. The SKN-1/Nrf2 transcription factor can protect against oxidative stress and increase lifespan in C. elegans by distinct mechanisms. Aging Cell 16(5), 1191–1194 (2017).
Ogawa, T. et al. Natural thioallyl compounds increase oxidative stress resistance and lifespan in Caenorhabditis elegans by modulating SKN-1/Nrf. Scientific Reports 6, 21611 (2016).
Sun, X., Chen, W.-D. & Wang, Y.-D. DAF-16/FOXO Transcription Factor in Aging and Longevity. Frontiers in Pharmacology, 8(548) (2017).
Obsil, T. & Obsilova, V. Structural basis for DNA recognition by FOXO proteins. Biochimica et Biophysica Acta 1813(11), 1946–1953 (2011).
Hu, M. et al. Structural and functional characterisation of the fork head transcription factor-encoding gene, Hc-daf-16, from the parasitic nematode Haemonchus contortus (Strongylida). International Journal for Parasitology 40(4), 405–415 (2010).
Fielenbach, N. & Antebi, A. C. elegans dauer formation and the molecular basis of plasticity. Genes & Development 22(16), 2149–2165 (2008).
Altintas, O., Park, S. & Lee, S. J. The role of insulin/IGF-1 signaling in the longevity of model invertebrates, C. elegans and D. melanogaster. BMB Reports 49(2), 81–92 (2016).
Honda, Y. & Honda, S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. The FASEB Journal 13(11), 1385–1393 (1999).
McElwee, J. J. et al. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. The Journal of Biological Chemistry 279(43), 44533–44543 (2004).
Inoue, H. et al. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes &. Development 19(19), 2278–2283 (2005).
Hoeven, R. et al. Ce-Duox1/BLI-3 generated reactive oxygen species trigger protective SKN-1 activity via p38 MAPK signaling during infection in C. elegans. PLoS Pathogens 7(12), e1002453 (2011).
Li, J. et al. The 14-3-3 protein FTT-2 regulates DAF-16 in Caenorhabditis elegans. Developmental Biology 301(1), 82–91 (2007).
Wang, Y. et al. C. elegans 14-3-3 proteins regulate life span and interact with SIR-2.1 and DAF-16/FOXO. Mechanisms of Ageing and Development 127(9), 741–747 (2006).
Oliveira, R. P. et al. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 8(5), 524–541 (2009).
Ewald, C. Y. et al. Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature 519, 97 (2015).
Klotz, L. O. et al. Redox regulation of FoxO transcription factors. Redox Biology 6, 51–72 (2015).
Pajares, M. et al. Redox control of protein degradation. Redox Biology 6, 409–420 (2015).
Chavez, V. et al. Oxidative stress enzymes are required for DAF-16-mediated immunity due to generation of reactive oxygen species by Caenorhabditis elegans. Genetics 176(3), 1567–1577 (2007).
Opperman, C. H. et al. Sequence and genetic map of Meloidogyne hapla: A compact nematode genome for plant parasitism. PNAS 105(39), 14802–14807 (2008).
Kikuchi, T. et al. Genomic Insights into the Origin of Parasitism in the Emerging Plant Pathogen Bursaphelenchus xylophilus. PLoS Pathogens 7(9), e1002219 (2011).
Burke, M. et al. The plant parasite Pratylenchus coffeae carries a minimal nematode genome. 17(6), 621 (2015).
Vicente, C. S. L. et al. Catalases Induction in High Virulence Pinewood Nematode Bursaphelenchus xylophilus under Hydrogen Peroxide-Induced Stress. PLoS One 10(4), e0123839–e0123839 (2015).
Zhang, W. et al. Enhancement of oxidative stress contributes to increased pathogenicity of the invasive pine wood nematode. Philosophical Transactions of the Royal Society B: Biological Sciences 374(1767), 20180323 (2019).
Olahova, M. & Veal, E. A. A peroxiredoxin, PRDX-2, is required for insulin secretion and insulin/IIS-dependent regulation of stress resistance and longevity. Aging Cell 14(4), 558–568 (2015).
Dubreuil, G. et al. Transcriptome analysis of root-knot nematode functions induced in the early stages of parasitism. New Phytologist 176(2), 426–436 (2007).
Koutsovoulos, G. D. et al. Population genomics supports clonal reproduction and multiple independent gains and losses of parasitic abilities in the most devastating nematode pest. Evolutionary Applications n/a, 1–16 (2020).
Castagnone-Sereno, P. et al. Gene copy number variations as signatures of adaptive evolution in the parthenogenetic, plant-parasitic nematode Meloidogyne incognita. Molecular Ecology 28, 2559–2572 (2019).
Lee, R. Y. N. et al. WormBase 2017: molting into a new stage. Nucleic Acids Research 46, D869–D874 (2017).
Muhire, B. M., Varsani, A. & Martin, D. P. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One 9, e108277–e108277 (2014).
Stothard, P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques 28(1102), 1104 (2000).
Marchler-Bauer, A. et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Research 45, D200–D203 (2017).
Dereeper, A. et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Research 36, W465–469 (2008).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797 (2004).
Szitenberg, A. et al. Comparative genomics of apomictic root-knot nematodes: hybridization, ploidy, and dynamic genome change. Genome Biology and Evolution 9, 2844–2861 (2017).
Marchler-Bauer, A. et al. CDD: NCBI’s conserved domain database. Nucleic Acids Research 43, D222–226 (2015).
El-Gebali, S. et al. The Pfam protein families database in 2019. Nucleic Acids Research 47, D427–D432 (2018).
la Cour, T. et al. Analysis and prediction of leucine-rich nuclear export signals. Protein Engineering, Design and Selection 17, 527–536 (2004).
Nguyen, B. A. N., Pogoutse, A., Provart, N. & Moses, A. M. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics 10, 202 (2009).
Hussey, R. S. & Barker, K. R. A comparison of methods of collecting inocula of Meloidogyne spp., including a new technique. Plant Disease Reports 57, 1025–1028 (1973).
Dubreuil, G. et al. Tobacco rattle virus mediates gene silencing in a plant parasitic root-knot nematode. Journal Experimental Botany 60, 4041–4050 (2009).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative CT method. Nature Protocols 3, 1101 (2008).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal 16, 735–743 (1998).
Park, S. H., Rose, S. C., Zapata, C., Srivatanakul, M. & Smith, R. H. Cross-protection and selectable marker genes in plant transformation. In Vitro Cellular & Developmental Biology - Plant 34, 117–121 (1998).
Windham, G. L. & Williams, W. P. Host Suitability of Commercial Corn Hybrids to Meloidogyne arenaria and M. incognita. Journal of Nematology 19, 13–16 (1987).
Oostenbrink, M. Major characteristics of the relation between nematode and plants. Meded. Landbouwhogeschool 66, 3–46 (1966).
Acknowledgements
We are grateful to EMBRAPA, UCB, CAPES, CNPq, INCT PlantStress Biotech, and FAP-DF for the financial support to scientific research. We also thank Dr. Bum-Soo Hahn for providing access to the transcriptome datasets of different M. incognita life stages. In addition, we thank Bruna Medeiros Pereira for kindly providing the initial inoculum of M. incognita race 3. M.F.B. is grateful to Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for a postdoctoral research fellowship (PDJ 150936/2018-4). I.T.L.T. and C.M.P. (CAPES-COFECUB SV.922/18) are grateful to the CAPES/Cofecub project for financial support in the researcher and students exchange programs between institutions.
Author information
Authors and Affiliations
Contributions
M.F.G.S. and J.A.E. were the leading researchers for all the work and provided intellectual inputs and financial support. M.F.B. and C.B. performed the in vitro oxidative stress assay. M.F.B. performed in silico analysis, plant transformation, and molecular assays. R.C.T. performed the data mining of 15 transcriptome libraries and then the differential expression profiles of the MiDaf16-like and MiSkn1-like genes. M.F.B., I.T.L.T., R.A.G.M. and C.E.M.P. performed the production of M. incognita inoculum and performed and evaluated all bioassays. M.F.G.S., L.L.P.M., J.A.E., C.B. and F.-X.G. provided intellectual input. M.F.B. wrote the manuscript. All authors read, provided input and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Basso, M.F., Lourenço-Tessutti, I.T., Mendes, R.A.G. et al. MiDaf16-like and MiSkn1-like gene families are reliable targets to develop biotechnological tools for the control and management of Meloidogyne incognita. Sci Rep 10, 6991 (2020). https://doi.org/10.1038/s41598-020-63968-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-63968-8
This article is cited by
-
Green synthesis and anthelmintic activity of silver nanoparticles using Morus Alba Fruit extract against different stages of equine strongyles
Veterinary Research Communications (2024)
-
In planta RNAi targeting Meloidogyne incognita Minc16803 gene perturbs nematode parasitism and reduces plant susceptibility
Journal of Pest Science (2024)
-
An ex vitro hairy root system from petioles of detached soybean leaves for in planta screening of target genes and CRISPR strategies associated with nematode bioassays
Planta (2024)
-
Selective control of parasitic nematodes using bioactivated nematicides
Nature (2023)
-
Overexpression of the GmEXPA1 gene reduces plant susceptibility to Meloidogyne incognita
Plant Cell Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.