Abstract
Colorectal cancer (CRC) represents the third most frequently diagnosed malignancy worldwide and is the second most common cause of tumor-associated mortalities in Korea. Due to the disease’s aggressive behavior, the 5-year survival rate for CRC patients remains unpromising. Well-characterized cell lines have been used as a biological model for studying the biology of cancer and developing novel therapeutics. To assist in vitro studies, 18 CRC cell lines (SNU-1566, SNU-1983, SNU-2172, SNU-2297, SNU-2303, SNU-2353B, SNU-2359, SNU-2373B, SNU-2407, SNU-2423, SNU-2431, SNU-2465, SNU-2493, SNU-2536C, SNU-2621B, SNU-NCC-61, SNU-NCC-376, and SNU-NCC-377) derived from Korean patients were established and characterized in the present study. General characteristics of each cell line including doubling time, in vitro morphology, mutational profiles, and protein expressions of CRC-related genes were described. Whole exome sequencing was performed on each cell line to configure mutational profiles. Single nucleotide variation, frame shift, in-frame deletions and insertions, start codon deletion, and splice stop codon mutation of various genes were found and classified based on their pathogenicity reports. In addition, cell viability was assayed to measure their sensitivities to 24 anti-cancer drugs including anti-metabolites, kinase inhibitors, histone deacetylase inhibitors, alkylating inhibitors, and topoisomerase inhibitors, all widely used for various cancers. On testing, five CRC cell lines showed MSI, of which MLH1 or MSH6 gene was mutated. These newly established CRC cell lines can be used to investigate biological characteristics of CRC, particularly for investigating gene alterations associated with CRC.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) represents the third most frequently diagnosed tumor worldwide and is the second most common cause of tumor-associated mortalities in Korea1,2. It remains the second most perpetual type of tumor in both genders (men: 12.4%; women: 10.1%), and the number of CRC cases continues to increase. Approximately 25% of CRC cases are diagnosed in stage IV, and recurrence with distance metastasis follows after primary resection in nearly 50% of CRC patients3. Due to its aggressive behavior, the 5-year relative survival rate remains disapproving4. Neoadjuvent therapy is generally performed before surgical resection as single- or multi-agent chemotherapy to improve prognosis5. While roughly 50% of CRC patients respond to customary chemotherapy, the majority develop drug resistance through the course of treatment, and relapse or distance metastasis often follows. In recent years, novel anti-cancer agents that target surface growth factor receptors have been developed as adjuvant therapy to decrease the risk of the cancer recurrence6.
Well-characterized cell lines have been used as models for studying the biology of cancer and developing novel therapeutics7. However, most of the widely used CRC cell lines were derived from Caucasian and African American populations. Accordingly, inter-heterogeneity from ethnic diversity has been biased toward Western countries. To address this need, we established and characterized 18 novel CRC cell cultures (SNU-1566, SNU-1983, SNU-2172, SNU-2297, SNU-2303, SNU-2353B, SNU-2359, SNU-2373B, SNU-2407, SNU-2423, SNU-2431, SNU-2465, SNU-2493, SNU-2536C, SNU-2621B, SNU-NCC-61, SNU-NCC-376, and SNU-NCC-377) from 18 Korean CRC patients. Characterization includes cellular phenotypes, growth rates, mutations of CRC-driver genes, and sensitivities to 24 anti-colorectal cancer drugs that are approved by the National Cancer Institute. The 24 drugs are categorized as anti-metabolites (TAS-102, Capecitabine, 5-FU), kinase inhibitors (Regorafenib, Apitolisib, MK-5108, AZD2014, Afatinib, Buparlisib, Trametinib), histone deacetylase inhibitors (Belinostat, SAHA), alkylating inhibitors (Oxaliplatin), topoisomerase inhibitors (Irinotecan), growth factor receptor inhibitors (Cetuximab, Bevacizumab), natural compounds (Resveratrol, Curcumin, Baicalein, Genistein), and miscellaneous (Lecouvorin calcium, ICG-001, Olaparib). These newly established 18 cell lines can be used to study the molecular biology of CRC, specifically to investigate genomic alterations related to CRC.
Materials and Methods
Establishment of cell lines and cell culture
Eighteen human CRC cell lines (SNU-1566, SNU-1983, SNU-2172, SNU-2297, SNU-2303, SNU-2353B, SNU-2359, SNU-2373B, SNU-2407, SNU-2423, SNU-2431, SNU-2465, SNU-2493, SNU-2536C, SNU-2621B, SNU-NCC-61, SNU-NCC-376 and SNU-NCC-377) were established from pathologically proven colorectal tumor tissues acquired from Korean CRC patients. Each participant was given informed consent before cell line establishment and experiment. The detailed procedure was described previously8. These novel 18 CRC cell lines were deposited at Korean Cell Line Bank (Seoul, Korea).
Mycoplasma test
Mycoplasma contamination test was performed as described previously using e-MycoTM kit (iNtRON Biotechnology, INC., Gyeonggi, Korea)9. Samples were arranged in the kit, including positive and negative controls for mycoplasma contamination. Mycoplasma control DNA served as a positive control and sterilized distilled water was used as the negative control. PCR amplification was performed under the following conditions: denaturation, 94 °C; annealing, 58 °C; and extension, 75°C. To confirm the specificity of contamination, PCR products were analyzed with gel electrophoresis using 2% agarose gel. PCR amplification and contaminated products had sizes of 570 bp and 260 bp, respectively.
Growth properties and morphology in vitro
Cell growth rate was measured with same method described previously9. For growth properties, cells were seeded into 96-well plates at a density of 2.0 × 103 cells/well and were treated with EZ-cytox (DAEIL Lab, Seoul, Korea), a water-soluble tetrazolium salt solution that could be reduced by succinate-tetrazolium reductase to produce formazan dye. After incubating at 37 °C for 2 h, optical density (OD) was assessed at 450 nm using a Multiskan™ GO Microplate Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The number of cells was analyzed in triplicate at 24-hour intervals for at least 7 days. The doubling time of the cells was calculated from the growth phase. Growth curve and growth properties were drawn and calculated using GraphPad Prism software with normalized OD values. Cell morphology was assessed using an Axiovert 100 microscope at 100× magnification.
DNA fingerprinting
DNA fingerprinting analysis was performed as decreased before10. Briefly, total DNA was isolated from cell pellet by using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to manufacturer’s protocol. Quantified and diluted gDNA solution was added to reaction mixture consisted of Amp FISTR PCR reaction mix, Taq DNA polymerase, and Amp FISTR identifier primer set (Applied Biosystems, CA, USA). DNA was amplified using a GeneAmp PCR System 9700 (Applied Biosystem) with annealing temperature set to 59°C. Gene Scan-500 Rox standard (0.05 μl) and 9 μl oHi-Di Formamide (Applied Biosystem) were added to 1 μl of PCR product of each cell line and denatured at 95 °C for 2 min. The mixture was then analyzed with a 3500 xL Genetic Analyzer (Applied Biosystems).
Drug sensitivity test
At density of 2 × 105 cells/well, tumor cells were seeded into a 96-well plate. Optimal concentrations of anti-cancer drugs were then used to treat 18 CRCs. These concentrations were: 100 μg/ml of TAS-102, 100 μg/ml of Regorafenib, 1000 μg/ml of Leucovorin calcium, 1000 μg/ml of Capecitabine, 50 μg/ml of Apitolisib, 100 μg/ml of Belinostat, 50 μg/ml of Trametinib, 50 μg/ml of Cyclopamine, 100 μg/ml of ICG-001, 100 μg/ml of Buparlisib, 50 μg/ml of SAHA, 50 μg/ml of Afatinib, 5 μg/ml of AZD2014, 100 μg/ml of MK-5108, 50 μg/ml of Olaparib, 100 μg/ml of Irinotecan, 50000 μg/ml of 5-FU, 100 μg/ml of Oxaliplatin, 100 μg/ml of Baicalein, 100 μg/ml of Curcumin, 100 μg/ml of Genistein, 200 μg/ml of Resveratrol, 1000 μg/ml of Cetuximab, and 1000 μg/ml of Bevacizumab. The 96-well plate containing anti-cancer drugs was incubated for 72 h at 37 °C. After incubation, 10 ul EZ-Cytox solution was applied to each well. After the plate was incubated for 2 h at 37 °C, optical density value was assessed at 450 nm with a Multiskan™ GO Microplate Spectrophotometer (Thermo Fisher Scientific).
Western blotting analysis
Detailed procedure was described previously9. Cells were harvested with a cell scraper after washing with cold PBS. Whole protein was extracted with EzRIPA buffer (ATTO Co., Tokyo, JAPAN) supplied with 1% protease inhibitor and 1% phosphatase inhibitor in accordance with the cell viability assay time frame. The volume of lysis buffer was adjusted to the number of cells collected in each vial. The protein concentration was determined by SMARTTM micro BCA protein assay kit (Intron biotechnology, Gyeonggi, Korea). Proteins in equal amounts were loaded on a 4-12% Bis-Tris gel (Invitrogen) and run at 50 volts for 2 h. Proteins on gel were then transferred to a PVDF membrane (Invitrogen) by electro-blotting with constant current of 80 mA at 4 °C overnight. Proteins on transferred membrane were blocked by incubating with 1.5% to 2.0% skim milk in 0.05% Tween 20-TBS buffer including 1 mM MgCl2 at room temperature for an hour. The membrane was then incubated with primary antibodies against EGFR (abcam, Cambridge, United Kingdom) (1:2000), HER2 (abcam, Cambridge, United Kingdom) (1:1000), MLH1 (Santa Cruz Biotechnology, TX, USA) (1:500), MSH2 (Santa Cruz Biotechnology, TX, USA) (1:500), EpCAM (Santa Cruz Biotechnology, TX, USA) (1:1000), E-cadherin (abcam, Cambridge, United Kingdom) (1:1000), vimentin (abcam, Cambridge, United Kingdom) (1:2000), and β-actin (Santa Cruz Biotechnology, TX, USA) (1:100) followed by incubation with mouse or rabbit IgG 2nd antibody (Jackson Immunoresearch, PA, USA) (1:5000) conjugated with peroxidase that matched with the primary antibody used. Chemiluminescent working solution WESTZOLTM (Intron biotechnology) was then used to treat the membrane which was then exposed to Fuji RX film (Fujifilm, Tokyo, Japan) for 1-5 minutes.
Whole exome sequencing
Detailed procedure was described previously9. SureSelect sequencing libraries were prepared using SureSelect Human All Exon 50 Mb Kit (Agilent) according to manufacturer’s instructions using a Bravo automated liquid handler. Three micrograms of genomic DNA were fragmented to a median size of 150 bp using a Covaris-S2 instrument (Covaris, MA, USA). Adapter ligated DNA was amplified by PCR. PCR product quality was then assessed by capillary electrophoresis. Hybridization buffer and DNA blocker mix were incubated at 95 °C for 5 minutes and 65 °C for 10 min in a thermal cycler. The hybridization mixture was then added to a bead suspension and incubated at RT for 30 min while mixing. These beads were washed and DNA was eluted from beads with 50 ml SureSelect elution buffer (Agilent). The flow cell was then loaded on a HiSeq. 2500 sequencing system (Illumina).
MSI test
Detailed procedure was described previously11. For microsatellite instability (MSI) analysis, BAT25 and BAT26 (two mononucleotide microsatellite markers) were evaluated using a capillary-based sequencing analysis8. PCR was performed as described above except that forward primers were labeled with a fluorescent dye. Labeled samples were run on an ABI 3730 genetic analyzer (Applied Biosystems). GeneMapper software v4.0 (Applied Biosystems) was used to calculate the size of each fluorescent PCR product. For gel-based MSI analysis, desired fragments were amplified in the presence of [a-P32] deoxycytidine triphosphate. PCR products were denatured and separated on 6 M urea/7%polyacrylamide gels run at 60 W.
Ethics approval and consent to participate
The study protocol was approved by the Institutional Review Board of Seoul National University Hospital (IRB No. H-1102-098-357). The study was performed in accordance with the Declaration of Helsinki.
Results
General characteristics of CRC cell lines
Human specimens were obtained from CRC patients who underwent surgeries at Seoul National University (SNU) Hospital from 1999 to 2008. Eighteen colorectal carcinoma cell lines (SNU-1566, SNU-1983, SNU-2172, SNU-2297, SNU-2303, SNU-2353B, SNU-2359, SNU-2373B, SNU-2407, SNU-2423, SNU-2431, SNU-2465, SNU-2493, SNU-2536C, SNU-2621B, SNU-NCC-61, SNU-NCC-376, and SNU-NCC-377) were established in RPMI 1640 medium supplemented with 10% FBS. In vitro and in vivo characteristics of newly established 18 CRC cell lines are summarized in Tables 1 and 2. All cell lines were free of contamination by mycoplasma (data not shown).
Morphology and growth properties of CRC cell lines
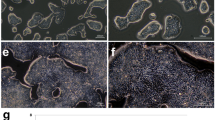
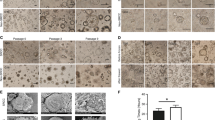
Cell images were acquired using Axiovert 100 microscope at 100× magnification (Fig. 1). On in vitro cultivation, sixteen CRC cell lines (SNU-1566, SNU-1983, SNU-2172, SNU-2297, SNU-2303, SNU-2353B, SNU-2373B, SNU-2407, SNU-2423, SNU-2431, SNU-2465, SNU-2536C, SNU-2621B, SNU-NCC-61, and SNU-NCC-377) grew as monolayers of substrate-adherent cells. SNU-NCC-61cell line showed spindle morphology while other cell lines showed polygonal morphology. SNU-2359 and SNU-2493 grew as floating clumps. SNU-NCC-376 cell line formed floating and adherent aggregates (Fig. 1). The majority of tumor cells displayed a polygonal shape and had exhibited round-to-oval nuclei with prominent single-to-double nucleoli. Each cell line was passaged at least three times prior to characteristic analysis. Population doubling times ranged from 32 to 138 hours.
DNA fingerprinting of 18 CRC cell lines
Fifteen tetranucleotide repeat loci and the gender-determining marker amelogen were heterogeneously distributed in each cell line, without cross-contamination (Table 3). They were also matched with the STR profiles of cell lines with passage 0 or 1 (including original tissue mass) in order to confirm that the established cell lines were not cross-contaminated with other patient material (Supplementary Table 1).
Expression levels of growth factor receptor and EMT proteins in 18 CRC cell lines
Protein expressions of MLH1 and MSH2 of newly established cell lines were analyzed in accordance with their mutational profiles. Three cell lines (SNU-1983, SNU-2434 and SNU-3030) had pathogenic mutations in MLH1 and the protein expression was exclusively low accordingly. Two cell lines (SNU-2359 and SNU-2493) harbored benign mutation in MLH1 (c.655 A > G/p.Ile219Va), and protein structure was not affected. Although no pathogenic MSH2 mutation was present in the newly established CRC cell lines, the protein expression of MSH2 was varying, which implicated the protein expression of MSH2 was determined by RNA splicing or epigenetical alternations (Fig. 2a). Four cell lines (SNU-2359, SNU-2431, SNU-2465 and SNU-NCC-61) exhibited augmented EGFR level. SNU-2431 and SNU-2465 had increased expression of both EGFR and HER2 (Fig. 2b). Expression levels of EMT-related proteins, E-cadherin, EPCAM and vimentin were analyzed according to the in vitro molphology (Fig. 2c). E-cadherin was significantly decreased in SNU-2423, while EPCAM was expressed in all cell lines. Vimentin was exclusively expressed in SNU-2536C and SNU-NCC-61. Both cell lines grew as monolayers of substrate-adherent cells with adherent aggregates.
The expression level of the mismatch repair protein. (a) MLH1 and MSH2, in 18 CRC cell lines. The protein expression level was detected by western blotting assay. The expression level of growth factor receptor and EMT proteins. (b) The expression level of growth factor receptor, HER2 and EGFR was assessed by western blot analysis. (c) The expression level of the EMT proteins, EPCAM, E-cadherin and Vimentin was assessed by western blot analysis.
Genomic analysis
Fifteen genes in developing CRC were screened in the 18 newly established CRC cell lines. Using Clinvar database (www.ncbi.nlm.nih.gov/clinvar), we determined pathogenic mutations. Results are summarized in Fig. 3, Table 4 and Supplementary Table 2. Mutations included in the Fig. 3 are only pathogenic mutations indicated by Clinvar database. Supplementary Table 2 includes the entire mutations in which their clinical meanings were in question. The most common actionable alterations across the sample sets were TP53 (83%) and APC (67%). KRAS and SMAD4 mutations were also prevalent in the sample sets at 44%. The most hyper-mutated cell line was SNU-2621B (10 mutations). Genes that are related to DNA repair such as POLD1, MSH6, and PMS2 were mutated in the SNU-2621B cell line. Similarly, SNU-1983 was also hyper-mutated (9 mutations) and DNA repair genes such as MLH1 and POLD1 were mutated. The truncation mutations of MLH1 and MSH6 genes in SNU-1566, SNU-1983 and SNU-2621B cell lines were confirmed with Sanger sequencing (Table 4, Supplementary Figs. 1–3).
Anticancer drug response of 18 CRC cell lines
Areas under curve (AUCs) of 18 CRC cell lines in response to NIH approved 24 anti-cancer drugs, including anti-metabolite (TAS-102, Capecitabine, 5-FU), kinase inhibitor (Regorafenib, Apitolisib, MK-5108, AZD2014, Afatinib, Buparlisib, Trametinib), histone deacetylase inhibitor (Belinostat, SAHA), alkylating inhibitor (Oxaliplatin), topoisomerase inhibitor (Irinotecan), growth factor receptor inhibitor (Cetuximab, Bevacizumab), natural compounds (Resveratrol, Curcumin, Baicalein, Genistein), and miscellaneous (Lecouvorin calcium, ICG-001, Olaparib), were estimated (Fig. 3). CRC cell lines were uniformly sensitive to Apitolisib, Trametinib, Belinostat, 5-FU, and Buparlisib with exceptions of SNU-2423 and SNU-2465. and resistant to Cetuximab, Bevacizumab, Leucovorin calcium, Olapraib, cyclopamine, and Resveratrol.
Discussion
New CRC cases continue to increase. At the time of detection, many CRC cases have already progressed to stage IV, which makes surgical resection unfeasible, and nearly 50% of CRC cases have shown recurrence or distance metastasis after primary resection12. Although there has been much research on inventing novel therapeutics, the molecular basis of drug response and aggressive behavior remains obscure due to its genetic intricacy, and more comprehensive analysis is called for to refine regimes for treatment and prevention13.
The importance of human CRC cell lines lies in their similarity to original tissues and their renewability, which facilitate the study of human CRC. Several CRC cell lines such as HCT‐116, LoVo, SW‐480, and WiDr have accelerated the CRC research. Nevertheless, those accessible CRC cell lines are somewhat obsolete and possibly acquire genetic alternations as passaging14. Clinical correlation between original human materials and cancer cell lines can decrease due to the accumulation of genetic aberrations with increasing subculture numbers15,16,17,18,19. Therefore, novel CRC cell lines can deliver suitable biological models for investigating a broader spectrum of molecular characteristics of CRC. J. H. Oh et al. established 12 human CRC cell lines from 6 primary and 6 metastatic tumors of 11 Korean CRC patients10. In addition, J. L. Ku et al. established 13 CRC cell lines from 10 primary and 3 metastatic tumors of 13 Korean patients8. In this study, we established 18 novel CRC cell lines from 13 primary and 5 metastatic tumors of Korean patients who underwent surgical resection from 1999 to 2008 in SNU Hospital. Novel cell lines established through this study will be deposited to the Korean Cell Line Bank at various passages.
Nearly 15% of sporadic CRC cases show the MSI phenotype, which is prompted by inactivation of mismatch repair (MMR) genes such as MLH1, MSH2, and MSH620. Hereditary non-polyposis CRC, which accounts for 2–5% of all CRC cases is also concurrent with germline mutations in MMR genes. Nearly 90% of reported mutations in MMR genes were harbored in MLH1 and MSH221,22. In this study, five cell lines harbored pathogenic mutations in MMR genes. MLH1 was mutated in SNU-1983, SNU-2359, SNU-2434, SNU-2493, and SNU-3030. Among these cell lines, three (SNU-1983, SNU-3030, and SNU-2434) had pathogenic mutations in MLH1, and the protein expression was exclusively low accordingly. Interestingly, we found no pathogenic MSH2 mutation in the newly established CRC cell lines. Although Wei et al. reported that there were different patterns of MSH2 and MLH1 mutations between Asian and Caucasian population23, the prevalence of MLH1 mutation in comparison with MSH2 mutation in an Asian population has not been reported. Although we found no pathogenic MSH2 mutation, the protein expression of MSH2 varied, which implied that the protein expression of MSH2 was determined by RNA splicing or epigenetic alternations. Two (SNU-1566, SNU-2423) of these five cell lines were derived from patients with hereditary non-polyposis CRC.
APC, KRAS, and tp53 are frequently abberant genes in CRCs15, and these three genes were mostly mutated in the CRC cell lines characterized in this study as well. Most of the identified APC germline alternations are nonsense mutations or frameshift mutations near the 5’ end of the gene, which truncated the protein structure24. We considered APC mutations pathogenic when they were reported in Clinvar (https://www.ncbi.nlm.nih.gov/clinvar), and the types of pathogenic APC mutations we identified in this study were also nonsense or frameshift.
KRAS serves as a fundamental mediator in the transduction of several growth or differentiation factor stimuli25. Most aberrations in KRAS harbor codons 12, 13, 59, and 6126. In this study, KRAS mutations gene were harbored in 8 of 18 cell lines (44%). Two cell lines (SNU-1566 and SNU-2423) had a mutation at codon 13, and six lines had a mutation at codon 12. Mutation types were G to A or G to T transitions.
Nearly 50% of CRC cases have several genetic alternations in tp5327. In this study, mutations of tp53 were present in 16 (88%) of the 18 cell lines. All tp53 mutations were at codons 72 (n = 12), 74 (n = 1), 176 (n = 1), 196 (n = 1), 213 (n = 1), 245 (n = 1), 337 (n = 1), 342 (n = 1), 800 (n = 1), and c.376-1 G > A in our study. Interestingly, pPro72Arg mutation is commonly found in gastric cancer28. This mutated codon is associated with colorectal cancer29. SMAD4 serves as the fundamental component of TGF-β signaling, and it is reported to be inactivated in many types of tumor including pancreas, stomach, and colon30. SMAD4 mutation has been found in 10~35% of CRC31,32,33,34. Similarly, we found mutation of SMAD4 in 2 (10.5%) of 18 cell lines. SMAD4 mutations were at codons 386 (n = 1) and 442 (n = 1). PTEN mutations are known to occur in 5–14% of CRC35,36,37. PTEN serves as an anti-oncogene. Over-activation of PI3K/AKT pathway is mainly associated with loss of PTEN38. In this study, we found mutation of PTEN in only 1 of the 18 lines (5.2%), SNU-1983. SNU-1983 had KRAS mutation without BRAF or PIK3CA mutation. Mutations in BRAF, specifically valine-to-glutamate change at residue 600 (V600E), account for approximately 10% of CRC cases39. The present study showed BRAF mutation with V600E in 4 (21%) of 18 lines. STK11 regulates cell polarity and is a tumor suppressor. This gene is mainly related to Peutz-Jeghers syndrome40. Cadherin-1 (CDH1), in the classical cadherin superfamily, is associated with cancer proliferation and invasiveness41. SNU-2621B had mutation in STK11 and CDH1, but no other cell lines had these mutations. SNU-2621B had one STK11 mutation (pGly163) and two CDH1 mutations (pArg74* and Arg800fs). Two CDH1 mutations (pArg74* and Arg800fs) usually occur in gastric cancer42.
All 18 CRC cell lines were sensitive to Apitolisib, Trametinib, Belinostat, 5-FU, and Buparlisib. Interestingly, SNU-2465 was resistant to Apitolisib, whereas all other lines were susceptible. All 18 CRC cell lines were resistant to Cetuximab, Bevacizumab, Leucovorin calcium, Olapraib, Cyclopamine, and Resveratrol. These novel cell lines will be deposited at the Korean Cell Line Bank and distributed worldwide for those who study colorectal cancer. These lines can be used as valuable materials to investigate biological properties of heterogeneous CRC.
References
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2016. CA: a cancer J. clinicians 66, 7–30, https://doi.org/10.3322/caac.21332 (2016).
Jung, K. W., Won, Y. J., Kong, H. J. & Lee, E. S. Cancer Statistics in Korea: Incidence, Mortality, Survival, and Prevalence in 2015. Cancer Res. treatment: Off. J. Korean Cancer Assoc. 50, 303–316, https://doi.org/10.4143/crt.2018.143 (2018).
Manhas, J. et al. Characterization of cancer stem cells from different grades of human colorectal cancer. Tumour biology: J. Int. Soc. Oncodev. Biol. Med. 37, 14069–14081, https://doi.org/10.1007/s13277-016-5232-6 (2016).
Amado, N. G. et al. Flavonoids and Wnt/beta-catenin signaling: potential role in colorectal cancer therapies. Int. J. Mol. Sci. 15, 12094–12106, https://doi.org/10.3390/ijms150712094 (2014).
Van Cutsem, E., Cervantes, A., Nordlinger, B. & Arnold, D. Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. oncology: Off. J. Eur. Soc. Med. Oncol. 25(Suppl 3), iii1–9, https://doi.org/10.1093/annonc/mdu260 (2014).
Khan, K., Wale, A., Brown, G. & Chau, I. Colorectal cancer with liver metastases: neoadjuvant chemotherapy, surgical resection first or palliation alone? World J. gastroenterology 20, 12391–12406, https://doi.org/10.3748/wjg.v20.i35.12391 (2014).
Butler, S. J., Richardson, L., Farias, N., Morrison, J. & Coomber, B. L. Characterization of cancer stem cell drug resistance in the human colorectal cancer cell lines HCT116 and SW480. Biochemical biophysical Res. Commun. 490, 29–35, https://doi.org/10.1016/j.bbrc.2017.05.176 (2017).
Ku, J. L. et al. Establishment and characterization of 13 human colorectal carcinoma cell lines: mutations of genes and expressions of drug-sensitivity genes and cancer stem cell markers. Carcinogenesis 31, 1003–1009, https://doi.org/10.1093/carcin/bgq043 (2010).
Kim, S. C. et al. Establishment and Characterization of Paired Primary and Peritoneal Seeding Human Colorectal Cancer Cell Lines: Identification of Genes That Mediate Metastatic Potential. Transl. Oncol. 11, 1232–1243, https://doi.org/10.1016/j.tranon.2018.07.014 (2018).
Oh, J. H. et al. Establishment and characterization of 12 human colorectal-carcinoma cell lines. International journal of cancer 81, 902–910, 10.1002/(sici)1097-0215(19990611)81:6<902::aid-ijc11>3.0.co;2-t (1999).
Kim, S. C. et al. Establishment and Characterization of 10 Human Pancreatic Cancer Cell Lines Including a HER2 Overexpressed Cell Line. Pancreas 48, 1285–1293, https://doi.org/10.1097/mpa.0000000000001420 (2019).
Van der Jeught, K., Xu, H. C., Li, Y. J., Lu, X. B. & Ji, G. Drug resistance and new therapies in colorectal cancer. World J. gastroenterology 24, 3834–3848, https://doi.org/10.3748/wjg.v24.i34.3834 (2018).
Hammond, W. A., Swaika, A. & Mody, K. Pharmacologic resistance in colorectal cancer: a review. Therapeutic Adv. Med. Oncol. 8, 57–84, https://doi.org/10.1177/1758834015614530 (2016).
Li, J. et al. Characterization of Human Cancer Cell Lines by Reverse-phase Protein Arrays. Cancer Cell 31, 225–239, https://doi.org/10.1016/j.ccell.2017.01.005 (2017).
Lengauer, C., Kinzler, K. W. & Vogelstein, B. Genetic instability in colorectal cancers. Nature 386, 623–627, https://doi.org/10.1038/386623a0 (1997).
Ma, L. & Li, W. Emodin inhibits LOVO colorectal cancer cell proliferation via the regulation of the Bcl-2/Bax ratio and cytochrome c. Exp. Ther. Med. 8, 1225–1228, https://doi.org/10.3892/etm.2014.1900 (2014).
Shan, B. E., Wang, M. X. & Li, R. Q. Quercetin inhibit human SW480 colon cancer growth in association with inhibition of cyclin D1 and survivin expression through Wnt/beta-catenin signaling pathway. Cancer investigation 27, 604–612, https://doi.org/10.1080/07357900802337191 (2009).
Huang, W. W. et al. Cantharidin induces G2/M phase arrest and apoptosis in human colorectal cancer colo 205 cells through inhibition of CDK1 activity and caspase-dependent signaling pathways. Int. J. Oncol. 38, 1067–1073, https://doi.org/10.3892/ijo.2011.922 (2011).
Chen, T. R., Drabkowski, D., Hay, R. J., Macy, M. & Peterson, W. Jr. WiDr is a derivative of another colon adenocarcinoma cell line, HT-29. Cancer Genet. cytogenetics 27, 125–134 (1987).
Thibodeau, S. N. et al. Microsatellite instability in colorectal cancer: different mutator phenotypes and the principal involvement of hMLH1. Cancer Res. 58, 1713–1718 (1998).
Thibodeau, S. N., Bren, G. & Schaid, D. Microsatellite instability in cancer of the proximal colon. Science 260, 816–819, https://doi.org/10.1126/science.8484122 (1993).
Wheeler, J. M., Bodmer, W. F. & Mortensen, N. J. DNA mismatch repair genes and colorectal cancer. Gut 47, 148–153, https://doi.org/10.1136/gut.47.1.148 (2000).
Wei, W. et al. Distinct mutations in MLH1 and MSH2 genes in hereditary non-polyposis colorectal cancer (HNPCC) families from China. BMB Rep. 44, 317–322, https://doi.org/10.5483/BMBRep.2011.44.5.317 (2011).
Nagase, H. et al. Correlation between the location of germ-line mutations in the APC gene and the number of colorectal polyps in familial adenomatous polyposis patients. Cancer Res. 52, 4055–4057 (1992).
Kawasaki, H., Saotome, T., Usui, T., Ohama, T. & Sato, K. Regulation of intestinal myofibroblasts by KRas-mutated colorectal cancer cells through heparin-binding epidermal growth factor-like growth factor. Oncol. Rep. 37, 3128–3136, https://doi.org/10.3892/or.2017.5520 (2017).
Bos, J. L. ras oncogenes in human cancer: a review. Cancer Res. 49, 4682–4689 (1989).
Greenblatt, M. S., Bennett, W. P., Hollstein, M. & Harris, C. C. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 54, 4855–4878 (1994).
Huang, Z. H., Hua, D., Li, L. H. & Zhu, J. D. Prognostic role of p53 codon 72 polymorphism in gastric cancer patients treated with fluorouracil-based adjuvant chemotherapy. J. cancer Res. Clin. Oncol. 134, 1129–1134, https://doi.org/10.1007/s00432-008-0380-8 (2008).
Oh, J. et al. Polymorphisms of the pri-miR-34b/c promoter and TP53 codon 72 are associated with risk of colorectal cancer. Oncol. Rep. 31, 995–1002, https://doi.org/10.3892/or.2013.2926 (2014).
Andrabi, S. et al. SMAD4 mutation segregating in a family with juvenile polyposis, aortopathy, and mitral valve dysfunction. American journal of medical genetics. Part. A 155a, 1165–1169, https://doi.org/10.1002/ajmg.a.33968 (2011).
De Bosscher, K., Hill, C. S. & Nicolas, F. J. Molecular and functional consequences of Smad4 C-terminal missense mutations in colorectal tumour cells. Biochemical J. 379, 209–216, https://doi.org/10.1042/bj20031886 (2004).
Koyama, M., Ito, M., Nagai, H., Emi, M. & Moriyama, Y. Inactivation of both alleles of the DPC4/SMAD4 gene in advanced colorectal cancers: identification of seven novel somatic mutations in tumors from Japanese patients. Mutat. Res. 406, 71–77 (1999).
Miyaki, M. & Kuroki, T. Role of Smad4 (DPC4) inactivation in human cancer. Biochemical biophysical Res. Commun. 306, 799–804, https://doi.org/10.1016/s0006-291x(03)01066-0 (2003).
Takagi, Y. et al. Somatic alterations of the DPC4 gene in human colorectal cancers in vivo. Gastroenterology 111, 1369–1372 (1996).
Berg, M. et al. DNA sequence profiles of the colorectal cancer critical gene set KRAS-BRAF-PIK3CA-PTEN-TP53 related to age at disease onset. PLoS One 5, e13978, https://doi.org/10.1371/journal.pone.0013978 (2010).
De Roock, W., De Vriendt, V., Normanno, N., Ciardiello, F. & Tejpar, S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet. Oncol. 12, 594–603, https://doi.org/10.1016/s1470-2045(10)70209-6 (2011).
Dicuonzo, G. et al. Colorectal carcinomas and PTEN/MMAC1 gene mutations. Clin. cancer research: an. Off. J. Am. Assoc. Cancer Res. 7, 4049–4053 (2001).
Salmena, L., Carracedo, A. & Pandolfi, P. P. Tenets of PTEN tumor suppression. Cell 133, 403–414, https://doi.org/10.1016/j.cell.2008.04.013 (2008).
Barras, D. BRAF Mutation in Colorectal Cancer: An Update. Biomarkers cancer 7, 9–12, https://doi.org/10.4137/bic.S25248 (2015).
Kopacova, M., Tacheci, I., Rejchrt, S. & Bures, J. Peutz-Jeghers syndrome: diagnostic and therapeutic approach. World J. gastroenterology 15, 5397–5408, https://doi.org/10.3748/wjg.15.5397 (2009).
Bustos-Carpinteyro, A. R. et al. CDH1 somatic alterations in Mexican patients with diffuse and mixed sporadic gastric cancer. BMC Cancer 19, 69, https://doi.org/10.1186/s12885-019-5294-0 (2019).
Kaurah, P. et al. Founder and recurrent CDH1 mutations in families with hereditary diffuse gastric cancer. Jama 297, 2360–2372, https://doi.org/10.1001/jama.297.21.2360 (2007).
Acknowledgements
This research was supported by the Korean Cell Line Research Foundation and Soon-Chan, Kim received a scholarship from the BK21-plus education program provided by the NRF.
Author information
Authors and Affiliations
Contributions
Soon-Chan Kim (S.C.K.) performed whole exome sequencing data analysis and AUC value analysis. S.C.K. took a pictures of cell lines and carried sanger sequencing. S.C.K. also wrote introduction, materials and method, result and discussion section. Hyun-Soo Kim (H.S.K.) performed western blot analysis and drug treatment. HSK also analyzed cell growth rate and took a pictures of cell lines. H.S.K. wrote introduction, materials and method and discussion section. Jae Hyeon Kim and Nahyun Jeong extracted genomic DNA from original patient material and performed sanger sequencing. Young-Kyoung Shin performed STR verification of newly established cell lines and cell line maintenance. Min Jung Kim and Ji Won Park provided human CRC specimens. Seung-Yong Jeong provided human CRC specimens and designed the study. Ja-Lok Ku established 18 CRC cell lines and designed the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, SC., Kim, HS., Kim, J.H. et al. Establishment and characterization of 18 human colorectal cancer cell lines. Sci Rep 10, 6801 (2020). https://doi.org/10.1038/s41598-020-63812-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-63812-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.