Abstract
The traditional approach in neuroscience relies on encoding models where brain responses are related to different stimuli in order to establish dependencies. In decoding tasks, on the contrary, brain responses are used to predict the stimuli, and traditionally, the signals are assumed stationary within trials, which is rarely the case for natural stimuli. We hypothesize that a decoding model assuming each experimental trial as a realization of a random process more likely reflects the statistical properties of the undergoing process compared to the assumption of stationarity. Here, we propose a Coherence-based spectro-spatial filter that allows for reconstructing stimulus features from brain signal’s features. The proposed method extracts common patterns between features of the brain signals and the stimuli that produced them. These patterns, originating from different recording electrodes are combined, forming a spatial filter that produces a unified prediction of the presented stimulus. This approach takes into account frequency, phase, and spatial distribution of brain features, hence avoiding the need to predefine specific frequency bands of interest or phase relationships between stimulus and brain responses manually. Furthermore, the model does not require the tuning of hyper-parameters, reducing significantly the computational load attached to it. Using three different cognitive tasks (motor movements, speech perception, and speech production), we show that the proposed method consistently improves stimulus feature predictions in terms of correlation (group averages of 0.74 for motor movements, 0.84 for speech perception, and 0.74 for speech production) in comparison with other methods based on regularized multivariate regression, probabilistic graphical models and artificial neural networks. Furthermore, the model parameters revealed those anatomical regions and spectral components that were discriminant in the different cognitive tasks. This novel method does not only provide a useful tool to address fundamental neuroscience questions, but could also be applied to neuroprosthetics.
Similar content being viewed by others
Introduction
The traditional approach to investigating brain functions involves the presentation of different stimuli and the analysis of evoked brain response properties1. The latter are collected either through non-invasive (e.g., electroencephalography [EEG], magnetoencephalography [MEG] or functional magnetic resonance imaging [fMRI]) or invasive (e.g., electrocorticography [ECoG]) recording techniques2,3. Independently of the method used, the measured signals contain background noise arising from other biological processes and environmental interferences that need to be filtered out or attenuated4,5. Several preprocessing methods such as signal filtering or referencing can serve to limit neural noise or artifactual activity and are used to improve signals quality prior to the extraction of different brain features. One popular approach in the literature involves the decomposition of brain signals into distinct frequency bands (i.e., delta, theta, alpha, beta, gamma, and high-gamma) broadly divided into low-frequency components (LFC, below 40 Hz) and high-frequency activity. These different frequency bands have been extensively used as features to model brain phenomena6,7,8,9,10,11,12,13,14,15,16,17. For instance, LFCs, usually measured as low-pass filtered brain signal (below 40 Hz)18,19,20, have been used as informative features in several applications, including decoding of position, velocity and acceleration of executed motor tasks18,19,20,21,22,23,24,25,26. The use of this feature produces significant improvements in decoding and prediction compared to models restricted to power modulations in alpha and beta bands24,25,27. Given its enhanced signal to noise ratio compared with both EEG and MEG, ECoG can also exploit high-frequency components above 100 Hz. High-frequency band activity (HFB), usually measured as the average power changes in the band from 70 to 200 Hz (although some works include lower spectral components in the range of 60–90 Hz28), has been used for decoding in multiple tasks, including motor, auditory, and visual21,29,30,31,32,33,34. The extracted features are used to model brain responses for basic brain research, medical diagnostics, and rehabilitation applications35,36,37. In rehabilitation, brain signal’s features are used to control external devices that allow subjects to interact with the environment36,38. In this case, successful use of the devices requires modeling the relationship between the executed task/stimulus and the brain features3,39.
Models based on multilinear regression, support vector machines, probabilistic graphical models, and artificial neural networks have been proposed in combination with different types of features8,9,11,12,13,14,18,19,20,22,23,24,25,26,40,41,42. These features involve spatial patterns discovery28,38,43,44,45,46,47, which is argued to be critical for increasing signal-to-noise ratio and to improve interpretability of the observed brain activity. Approaches such as common average reference (CAR), spline Laplacian filters and common spatial patterns (CSP) have previously been proposed48,49,50,51,52, and more recent approaches based on Riemannian geometry have shown promising results in brain-computer interfaces by using covariance matrices for feature representation and learning45,53,54. Modeling of temporal dynamics through probabilistic approaches such as hidden Markov models42,55,56,57, conditional random fields11,12,13,14,21 and recurrent neural networks30,31,58 have also successfully been used. In addition, features based on frequency decomposition of brain signals performed through either Fourier or wavelet analysis are well described in the literature, highlighting the importance of including patient-specific frequency bands in the design of brain-computer interfaces50,54. More recently, approaches that go beyond the classic approach based on second order statistics (power spectrum and cross power spectrum) have been introduced, allowing modeling of cross-frequency interactions in brain signals using bi-spectrum59,60.
Each method presents advantages and disadvantages. In multilinear regression with distributed lag, a widely used method in the literature19,23,39,61,62,63,64,65,66,67,68, the original feature set is expanded by including lagged versions of the original set. Without appropriate regularization, this can introduce model over-fitting39. In probabilistic graphical models and deep neural networks architectures, temporal relationships can be incorporated through the modeling of long-range dependencies21,58 and include prior information about the execution of the tasks42. However, these approaches require the use of a considerable amount of data that is usually not available in experiments with humans, limiting the performance of these methods69.
We hypothesize that modeling brain responses as realizations of a random rather than a stationary process should improve the identification of those features that are critical to their generation. We propose a decoding method based on complex coherence that accounts for different parameters such as frequency, phase, and spatial distribution of neural signals. Such an approach does not require manually predefining frequency bands of interest or phase relationships (i.e. lags) between stimulus and brain responses. The method is built on the notion that each experimental trial is a realization of a random process whose characteristics reflect the presented stimulus or executed task. In contrast with methods assuming signal stationary within a trial19,23,39,61,62,63,64,65,66,67,68, a statistically consistent phase difference between the neural signals and stimulus is expected across repeated trials of the same stimulus presentation, leaving all other parameters (e.g., specific frequency bands of interest or the phase relationships (i.e., lags values) between stimulus and brain features) hypothesis free. Although other approaches attempted to solve the non-stationarity problem by modeling the temporal dynamics of brain signal’s features21,42,58, their performance is limited by the amount of data available for training models. Our results show that the proposed approach significantly improves decoding performance and reduces the computational load compared to other methods that require explicit tuning of hyper-parameters.
The remainder of this document is organized as follows. We first describe the proposed method, experimental tasks, and evaluation criteria, and then present a thorough analysis of the obtained results. We discuss the main aspects of the proposed method based on the results and compare this to other approaches in the literature, including caveats and cautions on the use of the proposed method.
Methods
Proposed method
The proposed method is described in Fig. 1 using a finger movement task as an example. To model brain responses to stimulus/task- execution, each trial of an experiment is assumed to be a realization of a random process. Individual trials measure the superposition of the response due to the stimulus/task plus random noise that is assumed to be uncorrelated to the actual brain response of interest. The proposed method makes use of complex coherence to determine the relationship in amplitude and (more importantly) phase between stimuli and brain features at each frequency components across the training trials. In this approach, the coherence is estimated across trials, instead of across time windows in each trial. That is, it is assumed that the properties of the process are the same across trials, but not necessarily across the time course of individual trials. This allows relaxing assumptions about the stationarity of the brain features. For each electrode, a spectral filter is calculated.
Description of the coherence-based spectro-spatial filter. The diagram represents the recordings of a trial while the patient performs a movement with his/her thumb. (A) Brain features are used as input to a set of linear filters trained for each electrode. The filters represent the transfer function of a linear system that maps the brain features to the movement of the finger. The outputs of all filters are combined using linear regression to produce the final prediction of the movement. The signals shown in the figure correspond to LFCs components. (B) Example of one patient’s electrodes location. (C) Example of the magnitude response of one of the learned filters showing a peak in the low-frequency domain around 1 Hz.
The spectral filter extracts commonalities between stimuli and brain features at each frequency band. The output of these filters is then combined using multivariate linear regression, producing a final prediction that incorporates frequency, phase, and spatial features of the brain response. Importantly, spatial filtering results can be analyzed to determine the contribution of different recording electrodes on the prediction, examining for anatomical discriminability among different cognitive tasks or different stimuli.
Signal preprocessing and feature extraction
For all three data-sets the preprocessing stage is identical. All electrodes were band-pass filtered between 0.1 and 200 Hz using a zero-phase Butterworth filter of 4th order. A notch filter at 60 Hz was used to reduced the interference of the power line (See Supplementary Table S1). A common average reference (CAR) was used to reduce the effect of the reference electrode in all the recording electrodes. For this, the average across all electrodes is subtracted from the individual electrodes in the following fashion:
where s(t) = {s1(t), …, sN(t)} represent the brain signals measured for a trial at different electrode locations preprocessed with the bandpass filter (0.1–200 Hz) and the notch filter at 60 Hz. N is the number of electrodes.
The brain signals were separated within two frequency ranges: low frequency components (LFC) from 0.5 to 40 Hz, and HFB(70 to 170 Hz). LFCs were estimated by filtering the brain signals with a band-pass Butterworth filter of 4th order between 0.5 and 40 Hz. The envelope of high frequency band components (HFBE) was calculated as the magnitude of the analytic signal in the following fashion: assuming that the signal zbandpass(t) is the result of band-pass filtering z(t) in the HFB range, the analytic signal zanalytic(t) is calculated as:
where \( {\mathcal H} \) represents the Hilbert transform operation. The envelope of high frequency band components was low-pass filtered with a Butterworth filter of 4th order and a cut-off frequency of 40 Hz to reduce rapid changes in the amplitude of the signal. After this, LFCs and HFBE where down-sampled to 200 Hz in order to reduce redundant information. The stimulus features (speech envelope or finger movement dynamics) were accordingly down-sampled to 200 Hz.
Coherence-based spectro-spatial filter
The complex coherence allows for determining how well two signals correlate at each frequency component70. Given the random variables x(t) = {x1(t), …, xN(t)} representing the brain features extracted at each recording electrode (either LFCs, HFBE or the concatenation of both) and y(t) representing the dynamics of the stimuli that elicits the brain responses, the complex coherence between each xj(t) and y(t) is given by:
where \({P}_{{x}_{j},{x}_{j}}(f)\) and Py,y(f) are the power spectral density of xj(t) (brain signal features at electrode j) and y(t) respectively, and \({P}_{{x}_{j},y}(f)\) is the cross-power spectral density calculated between xj(t) and y(t). Power spectral densities (and therefore the complex coherence) were calculated based on the welch method, but rather than dividing each trial in several segments, we assume that each trial of the same class is a different realization of the same process and therefore each trial is one window in the calculation. The windows size is then the length of the trial, the window function used was a Hamming window, and the overlap was set to zero. The magnitude squared of the complex coherence has values between 0 and 1 and can be understood as the squared correlation between the two signals at each frequency component. Using the coherence as a measure of correlation between two signals at each frequency f, a linear filter is defined as:
Using the expression in Equation (3) for coherence we obtain:
where Hj(f) is the linear filter. Note that \({P}_{{x}_{j},y}(f)\) is a complex quantity, while \({P}_{{x}_{j},{x}_{j}}\) is real. Therefore the phase spectrum of Hj(f) is given by the phase differences between y(t) and xj(t). That is, for prediction of y(t) from xj(t), the phase differences at each frequency f are taken into consideration by the filter. Estimation of y(t) from xj(t) is then obtained by:
where hj(t) represent the inverse Fourier transform of Hj(f). Finally, different electrodes may contain different type of information, and should be accordingly combined forming a spatial filter, as follows:
where bj is a weight that determines how important the jth feature xj(t) is for the prediction of the signal y(t). The term n(t) is used to model the error in the prediction. The set of coefficients bj, can be understood as a spatial filter that provides information about which brain areas are involved in the processing of the stimuli or task executed. Combining h(t) with the coefficients in Equation (7) forms a filter that takes into account the frequency and phase spectrum of the signals, and the spatial patterns representing the contribution of different brain areas.
It is worth noting that the frequency response of the filters h(t) is defined by the signals xj(t) and y(t), and therefore should be calculated independently for each patient. Once the filters hj(t) are build, parameters bj can be learned using the least square solution for linear regression. For validation of the performance of the method, the proposed filters are constructed using portion of the available data (training set) and tested in the remaining portion (testing set), using a 5-fold cross-validation approach.
subsectionData-set description.
Finger movements
The motor data-set consists of electrocorticographic recordings from five patients that underwent surgery for temporary placement of subdural electrodes due to intractable epilepsy. These data originally appeared in23. Clinical information is displayed in Table 1. Patients executed a repetitive finger movement task. During the task, patients were cued with a word displayed on a bedside monitor indicating which finger to move (Thumb, Index, Middle, Ring, and Pinky). Patients were asked to repetitively move the indicated finger during an interval of 2-second (trial). There were thirty trials for each finger. Recordings were done using a Neuroscan Synamps2 device with a sampling rate of 1000 Hz. In addition to ECoG, finger positions were recorded using a 5-degree-of-freedom data-glove sensor. The data-glove signals were originally sampled at 25 Hz and up-sampled at 1000 Hz to match the sampling rate of the ECoG signals. The data-glove signal for each finger is the signal to be predicted from the brain features. Only electrodes in the sensory-motor (S1) and Motor areas (M1) were included in the analysis (See Supplementary Fig. S1). The identification of these areas was based on brain-mapping using electrical stimulation on the patients during the surgery. All patients gave informed consent. The study was approved by the Institutional Review Board of the University of Washington School of Medicine.
Speech perception
The data-set consists of ECoG recordings from three patients that underwent placement of subdural electrodes due to intractable epilepsy. These data originally appeared in71. Clinical information is displayed in Table 2. During the experiment, patients were requested to listen to a taped female voice repeating six different words (battlefield, cowboys, python, spoon, swimming and telephone). Each trial started with a baseline period of 500 ms after which a word out of a total of six is randomly selected and played on speakers at the bedside of the patient. Based on the anatomical mapping, electrodes that responded to auditory stimulus were selected (see Supplementary Figure S2). Each word was repeated eighteen times. Recordings were done using a g.USBAMP (g.tec medical engineering GmbH, Austria) with a sampling rate of 9600 Hz -DC coupled-. The audio input was recorded in parallel with brain signals to achieve the minimum loss of synchronization, the selected sampling rate covers the essential portions of the voice spectrum. We use the speech envelope72,73 as the feature to be predicted from brain features using the proposed method. For each trial, we selected 1.5 s segments starting from the auditory onset. All patients volunteered and gave their informed consent. The experimental protocol was approved by the Albany Medical College Institutional Review Board and methods were carried out in accordance with the approved guidelines and regulations.
Speech production
The data-set consists of ECoG recordings from three patients (same patients as in the perception task with the same recording parameters). During the experiment, patients repeated a particular word presented to them (among six different words: battlefield, cowboys, python, spoon, swimming and telephone). Each trial started with a baseline period of 500 ms after which the patient repeated the word that he or she heard prior to the beginning of the trial. Based on clinical mapping, electrodes that responded speech tasks were selected and used for both speech perception and speech production analysis. Each word was repeated eighteen times. Technical details of the recordings are the same as described in the speech perception data-set. These data originally appeared in71. For each trial, we selected 1.5 s segments starting from speech production onset. This onset was determined using an audio signal which recorded along with the ECoG. The experimental protocol was approved by the Albany Medical College Institutional Review Board and methods were carried out in accordance with the approved guidelines and regulations.
Evaluation
For the finger movement data-set, we compare the results obtained with the proposed method with results available in the literature. 1. Pace-regression: used for finger movement detection in23, 2. A probabilistic graphical model presented in42 that uses prior information to model the smooth dynamics of the finger movement, 3. A mTRF (multivariate Temporal Response filter) presented in39 and, 3. A non-linear function fitting method based on neural networks (named ANNFit in the remaining of the document).
The methods based on Pace-regression and probabilistic graphical models used the same set of data used in this work. Details of the methods and parameter selection can be found in23 and42. mTRF39 learns a multivariate temporal response function that can be used to map brain signal features to stimulus properties or vice-versa, making use of regularized linear regression. In our case, the goal is to predict stimulus properties or task dynamics form the brain signal features. mTRF builds a highly dimensional feature set by adding lagged versions of the original features. The lags can be positive or negative (making the resulting filter non-causal). The brain signal features were lagged using values from −500 to 500 ms, with steeps of 1/fs where fs is the sampling rate of the features. In order to avoid over-fitting, given the large number of features used, mTRFs uses Ridge regression74,75, which uses L2 regularization; adding the square magnitude of the coefficients as a penalty to the loss function, favoring solutions with coefficients with small square magnitude. The amount of penalization is controlled by a hyper-parameter (λ). We perform a grid search using nested cross-validation within the training set. The range of values for λ was selected as presented in a recent study that compares regularization methods in forward and backward models for auditory attention decoding75. λ values range from 10−6 to 108 in 54 logarithmically-spaced steps, using the following formula (Equation 16 in75):
where λ0 = 10−6.
A non-linear method based on artificial neural networks was also implemented for comparison (ANNFit). The brain signal feature-set was expanded with lagged versions of the original feature set with lags from −500 ms to 500 ms as for the mTRF method. The first hidden layer is densely connected and uses a sigmoid activation function. In order to avoid over-fitting, a drop-out layer was incorporated with a dropping-out probability of 0.5. The output layer uses a linear combination of the output of the previous layer to predict the finger movement. The artificial neural network was implemented using the Keras library in Python with Tensorflow as backend. We used the mean-squared-error as loss function and 100 epochs. Adadelta was the selected learning algorithm given that it adapts learning rates based on a moving window of gradient updates, making it a robust optimizer compared to other alternatives as Adagrad76. The initial learning rate for Adadelta was set to 1.0.
For the listening and speech production data-set, the mTRF method and the ANNFit method were implemented with the same criteria described above for parameter learning and hyper-parameter selection.
The initial set of features input to the models was the same in all cases implemented in this work. While mTRF and the ANNFit models expand the feature set, the proposed method based on coherence does not need to do this, which reduces the probability of over-fitting and reduces the computational load.
We used a 5-fold cross-validation process to evaluate performances. Cross-validation was implemented by dividing the available data for each task in five blocks. In each fold, four blocks were used for training and the remaining block for testing. This process was repeated five times each time having a different block for testing; ensuring that the data in the testing set is not part of the training set within the same fold. For the finger movement dataset a model for each of the five fingers is learned, and a total of 30 trials per finger are available, leaving 24 trials for training and 6 trials for testing on each fold. For the speech datasets a model for each of the 6 words is learned and 18 trials per word are available, therefore in each fold 14 trials were used for training and 4 for testing. In all cases, training or testing, the brain features are accompanied by an external reference signal (data-glove sensor or audio recordings depending of the data-set). These reference signals are used to calculate the filters in the training stage and those in the testing set are employed as targets to validate the output of the proposed method.
Results concerning the prediction of the stimulus features (speech envelope) or task dynamics (finger movement tracking) were obtained using all electrodes previously selected based on anatomical mapping.
Results
To address whether the coherence-based spectro-spatial filters method outperforms traditional approaches, we first compared its predictive power (in terms of the correlation between the predicted output and the actual stimulus/task dynamics) to the methods described in the Section 2.2. Evaluation of all methods implemented was performed in the same fashion, with the same features, using 5-folds cross-validation. Results show that for the tree data-sets the proposed method based on coherence and spatial filtering provides higher performance as displayed in Fig. 2. To assess the significance of performance gain, we performed a statistical test on the results. For the finger movement data-set, repeated-measures ANOVA on the performance results reveals significant differences between the five compared methods (DF = 4, F = 27.76, p < 0.0001). A post-hoc Tukey-Kramer multi-comparison test shows that the proposed method performs significantly better than all the other methods used for comparison (p < 0.001). For speech perception task (Listening) repeated measures ANOVA shows a significant difference between the three methods used (DF = 2, F = 95.03, p < 0.0001). A post-hoc Tukey-Kramer multi-comparison test shows that the proposed method performs significantly better than all the other methods (p < 0.0001). Similarly, for the speech production task repeated measures ANOVA shows significant differences in performance between the methods used (DF = 2, F = 18.53, p < 0.0001) and post-hoc Tukey-Kramer multi-comparison test show that the proposed method performs significantly better than the methods used for comparison (p < 0.0001). Detailed results for the correlation values between the actual and predicted feature dynamics in all patients across the three modalities are shown in Table 3, Tables 4 and 5 for finger movement task, listening and speech production respectively.
Furthermore, the proposed method enables to analyze the importance or weight of different recording electrodes in the prediction of the stimulus/task dynamics. As a result, the magnitude of coefficients bj (see Equation (7)) reflects the contribution of each electrode to the final prediction. In the proposed method, the combination of channels occurs after the filtering stage. Therefore, the output of each filter only contains components that are linearly related to the stimulus/task dynamics. Consequently, the set of coefficients bj can be understood as a set of spatial filters that contain discriminant information about the areas involved in task execution. These spatial filters can be plotted on the brain models for different tasks. Figure 3 displays the spatial distribution of the coefficients for LFC (panel B) and HFBE (panel A) components in one representative patient of the finger movement data-set. The results show that the proposed method leads to distinct spatial patterns, not only in response to different finger movements but also in the LFC and HFBE components. Similarly, the spatial filter for predicting the envelope of the perceived (Fig. 4) and produced (Fig. 5) speech revealed a level of discriminability across models learned for each word as well as between models learned using LFC and HFBE.
To assess the degree of discriminability within conditions (finger moved or word perceived/produced) revealed by the magnitude of the weights bj, we trained a set of models for each condition using a 5-fold cross-validation approach. We ranked the level of discriminability of the coefficients bj assigned to each electrode using K-means (k = 5 for finger movement dataset -five fingers- and 6 for speech data-sets -six words-) and fed an LDA (linear discriminant analysis) classifier with only the best two electrodes (selected using only the training set) to prevent over-fitting. The results revealed that the coefficients of the best two electrodes enabled discrimination of the models fitted to each finger movements with a high level of accuracy/Coen’s Kappa for both LFCs (Acc = 83%, kappa = 0.8) and HFB (Acc = 80%, kappa = 0.76). Similarly, we found that the coefficients discriminated among the models learned for different words in the speech perception (LFCs Acc = 90%, kappa = 0.88; HFBE Acc = 94%, kappa = 0.93) and production (LFCs Acc = 88%, kappa = 0.86; HFBE Acc = 93%, kappa = 0.92) tasks. These results show that the method learns for each condition a spatial filter that indicates the importance of a particular electrode in the decoding of the task dynamics. These results demonstrate that the calculated models for each condition are differentiable by the values of the coefficients, evidencing different spatial activations during the execution of different tasks. Using all the selected electrodes for the finger movement data-set we obtain higher classification performance (LFCs Acc = 95%,Kappa = 0.94; HFBE Acc = 92%, Kappa = 0.90), for speech perception task (LFCs Acc = 98%, Kappa = 0.97; HFBE Acc = 98%, Kappa = 0.97) and for speech production task (LFCs Acc = 94%, Kappa = 0.93; HFBE Acc = 99%, Kappa = 0.99). See Supplementary Figs S4–S6 for more details.
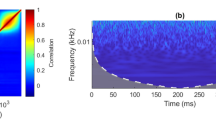
Figure 6 shows, for the finger movement task, the coherence between the brain features and the data-glove recordings in three patients for the movement of the Thumb. Colored areas indicate statistically non-significant values for coherence at each frequency component calculated using random phase test77. It is important to note that the magnitude squared coherence can be understood as the r2 at each frequency component between the signals into consideration. The peaks with higher value were found to be related to the average rate of finger flexion for individual patients, calculated using the data-glove signals. The High value in the magnitude squared coherence indicates that the brain features are highly informative about the executed task. Importantly, the frequency of peak coherence value is different for each patient. This is an expected observation as patients were cued to flex the finger freely during a particular amount of time. The proposed approach can then calculate the most informative frequency components within the LFC and the HFBE and use those to predict the dynamics of the task while reducing the impact of other components in the final prediction. Furthermore, Fig. 6 also shows a close relationship between the observed coherence when the LFCs and the HFBE are used.
Amplitude squared coherence between the data-glove signals and the brain features in the low frequency (LFC, top panel) and high frequency band envelope (HFBE, bottom). Colored areas show non-significant values for coherence. Note than only portions of the spectrum show significant coherence. The proposed approach appropriately attenuates frequency components that do not contain relevant information.
It must be noted that the proposed method does not require the brain features to be in phase across trials. It exploits the fact that the same stimulus will produce a response with the same underlying statistical properties. Therefore, what is exploited, is the consistency in the phase difference between the stimulus and the brain features at each frequency component. This means that even in cases when there is no precise information about the exact moment when the patient starts executing the task, the proposed method will perform well, as is the case for the finger movement data-set and the speech production data-set.
In cases when the stimulus presented is in itself highly non-stationary (as in the speech perception or speech production cases), the traditional approach of using short windows to calculate the coherence is inappropriate, as it assumes that across the time the statistical properties of the signals remain unchanged. In contrast, our approach uses each trial as a realization of the same process, model the relationships at the trial level in the Fourier domain, and then transform the signals back to the time domain. In such cases, physical interpretation of the frequency response of the coherence based filters may be obscured, but the mathematical modeling using Fourier analysis is still valid, and therefore useful for decoding purposes.
Although Person’s correlation is commonly used to report the similarity between the predicted output and the true targets, this index does not take reproducibility into account78. To allow future works to be compared with the results presented in this study we included results based on Concordance Correlation Coefficient78 (Tables S2–4).
In terms of computational load, the proposed method requires significantly less time to be trained. We calculated the average time necessary to train a model for the movement of the thumb finger with the proposed method and compared it to the mTRF and ANNFIT approaches. The results, after repeating this training 10 times, show a significant difference in the computational load. The proposed method require in average 0.65 +/− 0.07 seconds for training, ANNFIT requires an average of 24.11 +/− 1.4 seconds, and Ridge regression requires 26.1 +/− 1.02 seconds.
Discussion
Coherence-based spectro-spatial filter
We propose and assess a method to reconstruct stimuli or task dynamics from features extracted from brain responses, that does not require a priori manual specification of any signal parameters. Relevant parameters are calculated through the use of a reference signal in the training stage and used lately to make predictions based only on the brain features on a patient-specific basis. We found that the coherence-based method outperforms traditional predictive models and provides both a high-performance level (in terms of correlation) and consistency across motor and linguistic domains. The proposed method provides group averages of 0.74 for motor movements, 0.84 for speech perception, and 0.74 for speech production, while the best results obtained among all the other methods used for comparison are 0.7, 0.72 and 0.66 for motor movements, speech perception and speech production respectively. Notably, we demonstrate that the coherence-based method produces spatial filters that are discriminative of the task executed, revealing the importance of different brain areas for the execution of the tasks. The performance improvement relative to other methods is explained by several factors. The proposed method uses frequency decomposition and takes into account the phase relationships across trials. This allows for removing frequency components containing artifactual high power that does not show phase consistency between stimulus and brain features across trials. Such phase relationships are an essential factor because they reflect the latencies between the signals of interest at each frequency component. Multilinear regression-based approaches19,23,39,61,62,63,64,65,66,67,68 cannot account for phase relationships and requires expanding the set of regressors with lagged versions of the data which could lead to over-fitting, given the large number of features obtained. mTRF39 uses multilinear regression with lags but uses L2 regularization to avoid over-fitting issues, which require fine-tuning the regularization parameter λ (see 2.2). This makes the method computationally demanding as there is not closed-solution for the value of λ and it must be selected using grid search combined with cross-validation or grid search plus model selection approach such as Akaike information criterion (AIC) or Bayesian information criterion (BIC)79. This makes mTRF several folds slower than the proposed method for the same task. The proposed filter can be calculated in the Fourier domain using the FFTW algorithm80, which is highly efficient and widely implemented in commercial packages, (Python, Matlab, R). The ANNFit method used for comparison includes the possibility to incorporate non-linearities through sigmoid activation functions. However, it cannot model latencies among the signals of interest. Although expanding the regressors set with lagged versions of the data is possible as presented here, and in23,41,61,62, this may lead to over-fitting as the number of parameters increases while the amount of available data remains constant. To avoid these issues, we implemented a drop-out layer for the ANNFit method. However, the proposed method based on coherence not only outperforms the ANNFit method but is also computationally more efficient, despite the ANNFit being trained with a backend that uses parallel computing through graphical processing units (GPUs). Recently an approach based on canonical correlation analysis (CCA) has been proposed41 with good results. The CCA method produces a transformation of the input signals and also on the output signals (targets), which makes it impossible to compare in the scenario of the data-sets presented here. Nonetheless, the CCA also requires the inclusion of lagged versions of the input or to construct the input using multiple filtered versions of the brain signal features, which makes it prone to over-fitting. (see41)
We provided decoding results for LFCs and HFBE used both independently and in combination. For the finger movement data-set we found a high contribution from low frequency components (LFC), as previously reported in the literature25,81, which suggests that phase information is the most relevant information to be obtained from LFCs, which is consistent with other works showing selectively phase entrainment of the motor cortex on underlying rhythms in the low frequency range82. A recent work19 focused on LFCs showed that the phase of the brain features contains more information for decoding of kinematic parameters of the executed movements than the signal amplitude. It is also worth noting that the frequency range of the LFCs varies greatly across the literature, for instance, in81 the LFCs have a frequency content up to 20 Hz, while in24,25 the frequency content is up to 3 Hz. In83 LFCs were set to frequencies below 13 Hz for local field potentials, in84 frequencies below 2 Hz were selected for ECoG, and in85 a frequency content below 7 Hz was selected for EEG and MEG recordings. The approach in this work was to set a wide band (0–40 Hz) for the LFCs and determines based on coherence the frequencies that show phase consistency with the presented stimulus. The upper band was selected only to avoid contamination due to the power band at 60 Hz. In the case of the two speech data-sets, results show a greater contribution of HFBE compared to LFCs, in the case of speech perception where the brain activity expected is mainly sensory, we expect HFBE to provide better performance as it is highly correlated to the neuronal firing27. In the case of speech production there are motor components due to the vocal motor activity and auditory sensory components, therefore it is expected to obtain better performance combining those features. Furthermore, the difference in the performance between speech perception and speech production could be explained by the fact that auditory stimulus are highly consistent across trials as for a particular word, the same audio recording was presented to the patients, while in the speech production, variations in the speed at which the speech is produced were observed in the data. Such variations are difficult to model through linear systems which motivates the use of non-linear approaches as presented in71.
Given the limited amount of data available for each patient (30 trials per class for the finger movement data-set and 18 trials per class for the speech data-sets) electrode selection was guided by the clinical mapping, i.e. signal power increase during the relevant (motor or speech) tasks. This basic approach contributes to significantly reduce the dimensionality of the data and to limit over-fitting. While it likely produces interpretable results, it is not necessarily optimal in terms of decoding as areas that are not primarily included might also hold relevant information. When the amount of data is not limited, the spatial component of the proposed filter, can be combined with regularization techniques (i.e., L1 norm) that aim at finding sparse solutions (see86,87,88) allowing automatic selection of relevant electrodes.
The proposed coherence-based filter shows robust performances across different patients and operates well regardless of inter-individual differences and electrodes localization making it well suited for cases where the decoding goal is to decode what stimulus was presented to the patient. Using complex coherence, correlations between stimulus and brain features at each frequency component are calculated and are used to create a filter. Filter parameters are calculated under the assumption that each trial is a realization of a random process which allows employing the different trials to calculate a robust estimation of the cross-spectral density between brain features and the stimulus presented in training data. This permits extraction of components that are in phase with task dynamics across trials for each frequency and recording site. The resulting signal is then combined spatially to form a final prediction. The coherence-based spectro-spatial filter method has the advantage of including different dimensions of brain features such as phase, frequency, and space to handle the prediction of stimulus dynamics in an automated fashion. Importantly, the second stage of the proposed method combines different recordings electrodes. The difference between this and the methods used for comparison is that signals are combined after the coherence-based filtering, which ensures that the components combined have a linear relationship with the stimulus/task dynamics. This reduces the possibility that the coefficients learned (spatial filters) reflect a simple noise-canceling process. Evidence for this is provided by the clustering of the values of the coefficients for each task, enabling discrimination of the models learned by stimulus type (word presented) or task (finger moved), which provides information about the brain areas involved in the particular task.
Caveats and caution
The proposed method is better suited for discrete tasks. For instance, speech perception experiments as those presented here, in which the patient listens to a word and a prediction of the acoustic envelope of the audio attended is made, is an excellent example of such a discrete task. The proposed method could be used for continuous prediction as long as the causality of the filters hj(t) is ensured. Although engineering methods for ensuring causality exists, particular implementations of these techniques are beyond the scope of this study. However, in cases where patients are exposed to long continuous stimuli, methods based on linear switching dynamics or modeling of sequential states like hidden Markov models (HMM), conditional random fields (CRF) and recurrent neural networks, are better options given that the non-stationarity can be modeled with the different states that make part of such models. Although we selected a linear model for spatial filtering, non-linear methods are possible. We selected a linear approach aiming to obtain interpretable results as shown in Figs. 3–5. Finally, as explained before, when the signals that need to be modeled are highly non-stationary, physical or physiological interpretation of the spectral shape of the coherence-based filters should be made with caution as in such cases the Fourier representations of the signals, although numerically correct, may have no meaningful interpretation. Nonetheless, the results are still useful for decoding, as shown in the results of this work, in particular for speech-related tasks.
Conclusion
We present a method capable of predicting from features extracted from brain signals, characteristic features of a stimulus. The proposed method employs complex coherence to extract common patterns among the brain features related to the dynamics of the presented stimulus. This includes spatial information forming a spectro-spatial filter that is capable of reconstructing the dynamics of the stimulus with high performance (in terms of the correlation coefficient). Analysis of the coefficients that form the learned spatial patterns showed discriminability among different conditions, indicating the involvements of different areas and frequency components during the execution of various cognitive tasks such as finger movement as well as speech perception and production. The anatomical discriminability revealed by the method can be exploited in the design of neuro-prosthesis as well as for investigating the normal brain function.
In order to allow reproducibility of the results, an implementation of the proposed coherence-based method has been made available in89.
Data availability
A Finger movement data-set was made available by Gerwin Schalk. A data-set with the same finger movement task is publicly available at “A library of human electrocorticographic data and analyses.” (https://exhibits.stanford.edu/data/catalog/zk881ps0522). Speech perception and speech production data-sets were provided by Robert T. Knight and Gerwin Schalk and may be provided by them to interested researchers upon request.
References
Handy, T. C. Event-related potentials: A methods handbook (MIT press, 2005).
Sitaram, R. et al. Fmri brain-computer interface: a tool for neuroscientific research and treatment. Computational intelligence and neuroscience 2007 (2007).
Dornhege, G. et al. Toward Brain-Computer Interfacing, chap. 4 (MIT Press, Cambridge, Massachusetts, 2007).
Urigüen, J. A. & Garcia-Zapirain, B. Eeg artifact removal–state-of-the-art and guidelines. Journal of neural engineering 12, 031001 (2015).
Murphy, K., Birn, R. M. & Bandettini, P. A. Resting-state fmri confounds and cleanup. Neuroimage 80, 349–359 (2013).
Hyafil, A., Giraud, A.-L., Fontolan, L. & Gutkin, B. Neural cross-frequency coupling: connecting architectures, mechanisms, and functions. Trends in neurosciences 38, 725–740 (2015).
Pefkou, M., Arnal, L. H., Fontolan, L. & Giraud, A.-L. Theta-and beta-band neural activity reflect independent syllable tracking and comprehension of time-compressed speech. Journal of Neuroscience 2882–16 (2017).
Belkacem, A. N., Nishio, S., Suzuki, T., Ishiguro, H. & Hirata, M. Neuromagnetic decoding of simultaneous bilateral hand movements for multidimensional brain-machine interfaces. IEEE Transactions on Neural Systems and Rehabilitation Engineering (2018).
Schirrmeister, R. T. et al. Deep learning with convolutional neural networks for eeg decoding and visualization. Human brain mapping 38, 5391–5420 (2017).
Ma, T. et al. The hybrid bci system for movement control by combining motor imagery and moving onset visual evoked potential. Journal of Neural Engineering 14, 026015 (2017).
Delgado Saa, J. & Cetin, M. Bayesian nonparametric models for synchronous brain-computer interfaces. arXiv preprint arXiv:1612.08642 (2016).
Delgado Saa, J. Probabilistic Graphical Models for Brain Computer Interfaces. Ph.D. thesis, Sabanci University (2014).
Delgado Saa, J. & Cetin, M. Discriminative methods for classification of asynchronous imaginary motor tasks from eeg data. IEEE Transactions on Neural Systems and Rehabilitation Engineering 21, 716–724 (2013).
Delgado Saa, J. & Çetin, M. A latent discriminative model-based approach for classification of imaginary motor tasks from eeg data. Journal of neural engineering 9, 026020 (2012).
Hasan, A. S. B. & Gan, J. Q. Conditional random fields as classifiers for three-class motor-imagery brain-computer interfaces. Journal of neural engineering 8, 025013 (2011).
Birbaumer, N. & Cohen, L. G. Brain computer interfaces: communication and restoration of movement in paralysis. The Journal of Physiology 579, 621–636 (2007).
Mina, R. T., Atiya, A., Owis, M. I. & Kadah, Y. M. Brain-Computer Interface Based on Classification of Statistical and Power Spectral Density Features. Biomedical Engineering 2–5 (2006).
Waldert, S. et al. A review on directional information in neural signals for brain-machine interfaces. Journal of Physiology-Paris 103, 244–254 (2009).
Hammer, J. et al. The role of ecog magnitude and phase in decoding position, velocity, and acceleration during continuous motor behavior. Frontiers in neuroscience 7, 200 (2013).
Jerbi, K. et al. Coherent neural representation of hand speed in humans revealed by meg imaging. Proceedings of the National Academy of Sciences 104, 7676–7681 (2007).
Delgado Saa, J., De Pesters, A. & Cetin, M. Asynchronous decoding of finger movements from ecog signals using long-range dependencies conditional random fields. Journal of neural engineering 13, 036017 (2016).
Flamary, R. & Rakotomamonjy, A. Decoding finger movements from ecog signals using switching linear models. Frontiers in neuroscience 6 (2012).
Kubánek, J., Miller, J. W., Ojemann, J. G., Wolpaw, J. R. & Schalk, G. Decoding flexion of individual fingers using electrocorticographic signals in humans. J Neural Eng 6, 066001 (2009).
Schalk, G. et al. Two-dimensional movement control using electrocorticographic signals in humans. Journal of Neural Engineering 5, 75 (2008).
Schalk, G. et al. Decoding two-dimensional movement trajectories using electrocorticographic signals in humans. Journal of Neural Engineering 4, 264 (2007).
Leuthardt, E. C., Miller, K. J., Schalk, G., Rao, R. P. & Ojemann, J. G. Electrocorticography-based brain computer interface-the seattle experience. IEEE Transactions on Neural Systems and Rehabilitation Engineering 14, 194–198 (2006).
Manning, J. R., Jacobs, J., Fried, I. & Kahana, M. J. Broadband shifts in local field potential power spectra are correlated with single-neuron spiking in humans. Journal of Neuroscience 29, 13613–13620 (2009).
Pfurtscheller, G., Graimann, B., Huggins, J. E., Levine, S. P. & Schuh, L. A. Spatiotemporal patterns of beta desynchronization and gamma synchronization in corticographic data during self-paced movement. Clinical neurophysiology 114, 1226–1236 (2003).
Akbari, H., Khalighinejad, B., Herrero, J. L., Mehta, A. D. & Mesgarani, N. Towards reconstructing intelligible speech from the human auditory cortex. Scientific reports 9, 874 (2019).
Angrick, M. et al. Speech synthesis from ecog using densely connected 3d convolutional neural networks. Journal of neural engineering 16, 036019 (2019).
Anumanchipalli, G. K., Chartier, J. & Chang, E. F. Speech synthesis from neural decoding of spoken sentences. Nature 568, 493 (2019).
Kuo, C.-H. et al. Context-dependent relationship in high-resolution micro-ecog studies during finger movements. Journal of neurosurgery 1, 1–9 (2019).
Martin, A. B. et al. Temporal dynamics and response modulation across the human visual system in a spatial attention task: an ecog study. Journal of Neuroscience 39, 333–352 (2019).
Isik, L., Singer, J., Madsen, J. R., Kanwisher, N. & Kreiman, G. What is changing when: Decoding visual information in movies from human intracranial recordings. NeuroImage (2017).
Smith, S. Eeg in the diagnosis, classification, and management of patients with epilepsy. Journal of Neurology, Neurosurgery & Psychiatry 76, ii2–ii7 (2005).
Birbaumer, N. Breaking the silence: brain–computer interfaces (bci) for communication and motor control. Psychophysiology 43, 517–532 (2006).
Xie, K. et al. Portable wireless electrocorticography system with a flexible microelectrodes array for epilepsy treatment. Scientific reports 7, 1–8 (2017).
Wolpaw, J. R., Birbaumer, N., McFarland, D. J., Pfurtscheller, G. & Vaughan, T. M. Brain–computer interfaces for communication and control. Clinical neurophysiology 113, 767–791 (2002).
Crosse, M. J., Di Liberto, G. M., Bednar, A. & Lalor, E. C. The multivariate temporal response function (mtrf) toolbox: a matlab toolbox for relating neural signals to continuous stimuli. Frontiers in human neuroscience 10, 604 (2016).
Vaidya, M. et al. Emergent coordination underlying learning to reach to grasp with a brain-machine interface. Journal of Neurophysiology 119, 1291–1304 (2018).
de Cheveigné, A. et al. Decoding the auditory brain with canonical component analysis. NeuroImage 172, 206–216 (2018).
Wang, Z., Ji, Q., Miller, K. J. & Schalk, G. Prior knowledge improves decoding of finger flexion from electrocorticographic (ecog) signals. Frontiers in Neuroscience 5 (2011).
Delisle-Rodriguez, D. et al. Adaptive spatial filter based on similarity indices to preserve the neural information on eeg signals during on-line processing. Sensors 17, 2725 (2017).
Cohen, M. X. Comparison of linear spatial filters for identifying oscillatory activity in multichannel data. Journal of neuroscience methods 278, 1–12 (2017).
Dähne, S. et al. Spoc: a novel framework for relating the amplitude of neuronal oscillations to behaviorally relevant parameters. NeuroImage 86, 111–122 (2014).
Wu, D., King, J.-T., Chuang, C.-H., Lin, C.-T. & Jung, T.-P. Spatial filtering for eeg-based regression problems in brain-computer interface (bci). IEEE Transactions on Fuzzy Systems (2017).
Wang, J., Feng, Z., Lu, N., Sun, L. & Luo, J. An information fusion scheme based common spatial pattern method for classification of motor imagery tasks. Biomedical Signal Processing and Control 46, 10–17 (2018).
Pfurtscheller, G. & Da Silva, F. L. Event-related eeg/meg synchronization and desynchronization: basic principles. Clinical neurophysiology 110, 1842–1857 (1999).
Ramoser, H., Muller-Gerking, J. & Pfurtscheller, G. Optimal spatial filtering of single trial eeg during imagined hand movement. IEEE transactions on rehabilitation engineering 8, 441–446 (2000).
Hashimoto, Y. & Ushiba, J. Eeg-based classification of imaginary left and right foot movements using beta rebound. Clinical neurophysiology 124, 2153–2160 (2013).
Blankertz, B., Tomioka, R., Lemm, S., Kawanabe, M. & Muller, K.-R. Optimizing spatial filters for robust eeg single-trial analysis. IEEE Signal processing magazine 25, 41–56 (2007).
Ang, K. K., Chin, Z. Y., Wang, C., Guan, C. & Zhang, H. Filter bank common spatial pattern algorithm on bci competition iv datasets 2a and 2b. Frontiers in neuroscience 6, 39 (2012).
Yger, F., Berar, M. & Lotte, F. Riemannian approaches in brain-computer interfaces: a review. IEEE Transactions on Neural Systems and Rehabilitation Engineering 25, 1753–1762 (2016).
Delisle-Rodriguez, D. et al. System based on subject-specific bands to recognize pedaling motor imagery: Towards a bci for lower-limb rehabilitation. Journal of neural engineering (2019).
D A. Moses, J. G. M., Matthew K. Leonard & Chang, E. F. Real-time decoding of question-and-answer speech dialogue using human cortical activity. Nature Communications 10 (2019).
Moses, D. A., Leonard, M. K. & Chang, E. F. Real-time classification of auditory sentences using evoked cortical activity in humans. Journal of neural engineering 15, 036005 (2018).
Milstein, D. et al. Multiscale semi-markov dynamics for intracortical brain-computer interfaces. In Advances in Neural Information Processing Systems, 868–878 (2017).
Xie, Z., Schwartz, O. & Prasad, A. Decoding of finger trajectory from ecog using deep learning. Journal of neural engineering 15, 036009 (2018).
Zhou, S.-M., Gan, J. Q. & Sepulveda, F. Classifying mental tasks based on features of higher-order statistics from eeg signals in brain-computer interface. Information Sciences 178, 1629–1640 (2008).
Shahjahan, S. & Girijesh, P. Bispectrum-based feature extraction technique for devising a practical braincomputer interface. Journal of Neural Engineering 8, 025014 (2011).
Crosse, M. J., Butler, J. S. & Lalor, E. C. Congruent visual speech enhances cortical entrainment to continuous auditory speech in noise-free conditions. Journal of Neuroscience 35, 14195–14204 (2015).
Golumbic, E. Z., Cogan, G. B., Schroeder, C. E. & Poeppel, D. Visual input enhances selective speech envelope tracking in auditory cortex at a cocktail party. Journal of Neuroscience 33, 1417–1426 (2013).
Di Liberto, G. M. & Lalor, E. C. Indexing cortical entrainment to natural speech at the phonemic level: methodological considerations for applied research. Hearing research 348, 70–77 (2017).
Broderick, M. P., Anderson, A. J., Di Liberto, G. M., Crosse, M. J. & Lalor, E. C. Electrophysiological correlates of semantic dissimilarity reflect the comprehension of natural, narrative speech. Current Biology 28, 803–809 (2018).
Di Liberto, G. M., Lalor, E. C. & Millman, R. E. Causal cortical dynamics of a predictive enhancement of speech intelligibility. Neuroimage 166, 247–258 (2018).
Sullivan, O., Crosse, A. E., Di Liberto, M. J., Lalor, G. M. & Visual, E. C. cortical entrainment to motion and categorical speech features during silent lipreading. Frontiers in human neuroscience 10, 679 (2017).
Fiedler, L. et al. Single-channel in-ear-eeg detects the focus of auditory attention to concurrent tone streams and mixed speech. Journal of neural engineering 14, 036020 (2017).
Holdgraf, C. R. et al. Encoding and decoding models in cognitive electrophysiology. Frontiers in systems neuroscience 11, 61 (2017).
Lotte, F. et al. A review of classification algorithms for eeg-based brain–computer interfaces: a 10 year update. Journal of neural engineering 15, 031005 (2018).
Petre Stoica, R. M. Spectral Analysis of Signals (Prentice Hall, Upper Saddle River, New Jersey, 2005).
Martin, S. et al. Word pair classification during imagined speech using direct brain recordings. Scientific reports 6, 25803 (2016).
Ghitza, O., Giraud, A.-L. & Poeppel, D. Neuronal oscillations and speech perception: critical-band temporal envelopes are the essence. Frontiers in Human Neuroscience 6, 340 (2013).
Peelle, J. E. & Davis, M. H. Neural oscillations carry speech rhythm through to comprehension. Frontiers in psychology 3, 320 (2012).
Hoerl, A. E. & Kennard, R. W. Ridge regression: Biased estimation for nonorthogonal problems. Technometrics 12, 55–67 (1970).
Wong, D. D. et al. A comparison of regularization methods in forward and backward models for auditory attention decoding. Frontiers in neuroscience 12, 531 (2018).
Zeiler, M. D. Adadelta: an adaptive learning rate method. arXiv preprint arXiv:1212.5701 (2012).
Ebisuzaki, W. A method to estimate the statistical significance of a correlation when the data are serially correlated. Journal of Climate 10, 2147–2153 (1997).
Lawrence, I. & Lin, K. A concordance correlation coefficient to evaluate reproducibility. Biometrics 255–268 (1989).
Aho, K., Derryberry, D. & Peterson, T. Model selection for ecologists: the worldviews of aic and bic. Ecology 95, 631–636 (2014).
Frigo, M. & Johnson, S. G. The design and implementation of FFTW3. Proceedings of the IEEE 93, 216–231 (2005).
Stavisky, S. D., Kao, J. C., Nuyujukian, P., Ryu, S. I. & Shenoy, K. V. A high performing brain–machine interface driven by low-frequency local field potentials alone and together with spikes. Journal of neural engineering 12, 036009 (2015).
Miller, K. J. et al. Human motor cortical activity is selectively phase-entrained on underlying rhythms. PLoS computational biology 8, e1002655 (2012).
Rickert, J. et al. Encoding of movement direction in different frequency ranges of motor cortical local field potentials. Journal of Neuroscience 25, 8815–8824 (2005).
Ball, T., Schulze-Bonhage, A., Aertsen, A. & Mehring, C. Differential representation of arm movement direction in relation to cortical anatomy and function. Journal of neural engineering 6, 016006 (2009).
Waldert, S. et al. Hand movement direction decoded from meg and eeg. Journal of Neuroscience 28, 1000–1008 (2008).
Gurve, D. et al. Subject-specific eeg channel selection using non-negative matrix factorization for lower-limb motor imagery recognition. Journal of Neural Engineering (2019).
Ghaemi, A., Rashedi, E., Pourrahimi, A. M., Kamandar, M. & Rahdari, F. Automatic channel selection in eeg signals for classification of left or right hand movement in brain computer interfaces using improved binary gravitation search algorithm. Biomedical Signal Processing and Control 33, 109–118 (2017).
Yu, T., Yu, Z., Gu, Z. & Li, Y. Grouped automatic relevance determination and its application in channel selection for p300 bcis. IEEE Transactions on Neural Systems and Rehabilitation Engineering 23, 1068–1077 (2015).
Delgado Saa, J. F. et al. Implementation of coherence-based spectro-spatial filters for stimulus features prediction from electrocorticographic recordings (2018).
Acknowledgements
This work was performed thanks for the Swiss National Funds project grant to ALG 320030_163040, and to the EU FET-BrainCom project and NINDS R3723115. The authors thanks Dr. Gerwin Schalk and Dr. Kai Miller for providing access to the data-sets used in this work.
Author information
Authors and Affiliations
Contributions
J.D.S. processed the experimental data, performed the analysis, and designed the figures. J.D.S., A.C., S.M., B.P., R.K., and A.L.G. discussed the results and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
All authors declare that they have no significant competing financial, professional, or personal interests that might have influenced the performance or presentation of the work described in this manuscript.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Delgado Saa, J., Christen, A., Martin, S. et al. Using Coherence-based spectro-spatial filters for stimulus features prediction from electro-corticographic recordings. Sci Rep 10, 7637 (2020). https://doi.org/10.1038/s41598-020-63303-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-63303-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.