Abstract
Plant microbiota colonize all organs of a plant and play crucial roles including supplying nutrients to plants, stimulating seed germination, promoting plant growth, and defending plants against biotic and abiotic stress. Because of the economic importance, interactions between citrus and microbes have been studied relatively extensively, especially citrus-pathogen interactions. However, the spatial distribution of microbial taxa in citrus trees remains under-studied. In this study, Citrus reticulata cv. Chachiensis was examined for the spatial distribution of microbes by sequencing 16S rRNA genes. More than 2.5 million sequences were obtained from 60 samples collected from soil, roots, leaves, and phloem. The dominant microbial phyla from all samples were Proteobacteria, Actinobacteria and Acidobacteria. The composition and structure of microbial communities in different samples were analyzed by PCoA, CAP, Anosim and MRPP methods. Variation in microbial species between samples were analyzed and the indicator microbes of each sample group were identified. Our results suggested that the microbial communities from different tissues varied significantly and the microenvironments of tree tissues could affect the composition of its microbial community.
Similar content being viewed by others
Introduction
Plants host diverse microbes that colonize to, on and in their tissues1. Based on their habitats, plant-associated microbial communities are referred to as rhizosphere microbiome, rhizoplane microbiome, phyllospher microbiome, and endosphere microbiome, respectively2,3,4,5. Different microbiomes interact with host plants and impact plants in various ways. In general, plant-associated microbial organisms can potentially have both positive and negative impacts on plant growth, development, and health1. Direct impacts on plant growth and development by microorganisms include improved nutrient accessibility such as nitrogen fixation and phosphate solubilization; altered microenvironments such as changed acidity (pH); and hormonal stimulation (phytohormone production)3,6. Microorganisms are also involved in the promotion or suppression of plant diseases either directly (such as antibiotics production) or indirectly (via disease resistance)7. Accordingly, characterization of microbial compositions and dynamics in different plant tissues and elucidation of functions associated with specific microbes or microbial communities will provide the basis for developing commercially probiotics for plants or designing strategies to manipulate microbes or microbial communities for economic or environmental benefits8,9,10.
Recent advances on culture-independent, high-throughput sequencing of the variable regions of 16 S rRNA genes in microbes and total genome assembly from metagenomic sequence reads has revolutionized our research on the structures and diversity of microbiomes in different plant tissues. As a result, microbiome data on various types of plants have been accumulated rapidly in recent years. Two independent studies on the microbial community in the root of the model plant Arabidopsis thaliana revealed consistent results on the core microbiome, with Actinobacteria and a few families from Proteobacteria enriched consistently in the endosphere compared with rhizosphere11,12. Microbiomes from various tissues of many crops such as sugarcane, rice, tomato, maize, sorghum, soybean, and cotton have been characterized to various degrees13,14,15,16,17,18,19. Microbiomes from certain tissues of fruits and trees including pear, banana, and apple have initially analyzed as well20,21,22.
Orange (Citrus × sinensis) is one of the most important fruits for humans worldwide. In Brazil alone, 35.6 million tons of oranges were produced based on the most recently available data in 2013. Like other crops and fruits, citrus production faces many hurdles and challenges that need to be overcome to achieve high yield and high quality of oranges. Characterization of microbiomes in different tissues of citrus trees may yield useful information for the improvement of orange production. There is fragmented information available on microbiomes of citrus trees23. Most previous investigation on citrus-associated microbiomes are analyses on huanglongbing-infected, or specially-treated samples24,25,26,27,28,29.
Citrus reticulata cv. Chachiensis is an important citrus variety widely cultivated around the world and the orange peel of the fruit was regarded as the valuable material which could be processed as food, tea drinks, and seasoning30. Besides, the orange peel owned antioxidant properties and extremely high medicinal value was also used as herb-medicine31. In China, C. reticulata cv. Chachiensis has grown in Xinhui District, Jiangmen City, Guangdong Province was considered as one of the most important cash crops for locals and the orange peel named genuine Pericarpium Citri Reticulatae (Guang Chen Pi) was admired as the best Chen pi which possesses the excellent clinical efficacy32. Plant-related microorganisms could protect the host from pathogens and pests, help plants use nutrients, and improve plant stress resistance33, while the microbial community of C. reticulata cv. Chachiensis remains unknown. In this study, endosphere microbiomes in soils, roots, leaves, and phloem of C. reticulata cv. Chachiensis were systematically investigated by sequencing 16S rRNA genes. The composition and structure of the microbial community in different tissues were performed by PCoA, CAP, Anosim and MRPP methods. Variation in microbial compositions among citrus tissues were analyzed and indicator microbes for different samples were identified. Our results suggested that the microbial community from different tissues of C. reticulata cv. Chachiensis in south china varied significantly. Our results laid a foundation for future studies on microbial communities on citrus and their implications on orange production.
Results
Distribution of 16s sequences among samples
More than 2.5 million sequence reads were obtained from 60 samples, with 15 samples from each type of the following sources: soils, roots, leaves, and phloem. All the raw data of samples were submitted to the SRA database with the submission number of SUB6069689. Specifically, 639,200 (24.90%) were from soils, 688,345 (26.81%) from roots, 639,373 (24.91%) from leaves, and 599,685 (23.4%) from phloem. Among the total sequence reads, 1,454,442 (56.7%) were assigned to chloroplast and mitochondrial reads, 1,085,802 (42.3%) assigned to other classifiable reads, and 26,359 (1.0%) were unclassifiable reads. Among the chloroplast and mitochondrial reads, 42.81%, 37.09%, 19.89%, and 0.21% were from leaf, phloem, root, and soil samples, respectively. Most other classifiable reads, 57.00%, and 36.17%, respectively, were from soil and root samples, and only 1.39 and 5.44% were from leaf and phloem samples (Tables 1 and S1).
Distribution of OTUs
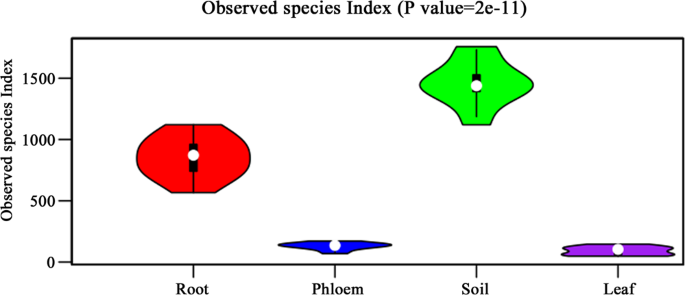
After excluding chloroflexi and mitochondrial reads and unclassifiable unique reads, the remaining reads were assembled into OTUs. The distribution of total OTUs were 4733, 3520, 421, and 583, respectively, for soil, root, leaf, and phloem samples. Based on our technical reproducibility, we set the criterion for ‘measurable OTUs’ as ≥20 reads in at least three samples of the same type. Based on this criterion, the measurable OTUs were 2245, 1034, 55, and 111 for soil, root, leaf, and phloem samples (Fig. 1).
β diversity analysis of samples
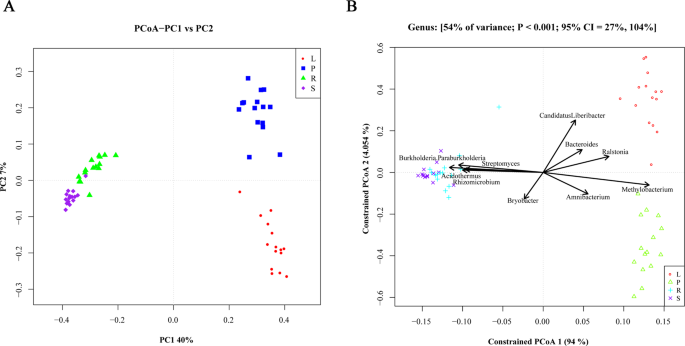
Principal co-ordinates analysis (PCoA) of samples from different parts of citrus trees were performed. The samples with the closer distance of Unweighted Unifrac indicated higher similarity between microbial communities. As shown in Fig. 2A, the microbial community in soil samples was more similar to that in root samples with all samples clustered together. The microbial communities in leaf and phloem samples differed significantly. The microbial communities differed significantly among the groups of samples from leaves and phloem. Canonical analysis of principal coordinates (CAP) yielded results similar to PCoA results. CAP analyses might reflect sample-to-sample variation between different sample groups, and the results indicated that all samples were divided into three clusters: 1) soil and root, 2) leaf, 3) phloem (Fig. 2B). The compositions of microbiota among different samples correlated among groups, with major species including Burkholderia Paraburkholderia, Streptomyces, Bryobacter, Acidothermus and Rhizomicrobium for the soil and root groups; Methylobacterium and Amnibacterium for phloem groups; and Ralstonia, Bacteroides, and Candidatus Liberibacter for leaf groups (Table 2).
The results of PCoA and CAP analyses in all the samples. (A) The PCoA analysis of microbial species among the total samples. The abscissa (x) represents the first principal component, the percentage represents the contribution of the first principal component to the sample difference; the ordinate (y) represents the second principal component, and the percentage represents the contribution of the second principal component to the sample difference. Each point in the figure represents a sample, and samples of the same group are represented by the same color. (B) The CAP analysis of microbial species among the total samples. The numbers at the top of the figure represent the variance contribution of these factors, P value, and the confidence interval (95% CI) of the variability, respectively. Species with a higher degree of association with the group are indicated by arrows. The length of the line represents the degree of correlation between an environmental factor and the distribution of the community and the distribution of the species. The longer the arrow, the greater the correlation.
Core microbial communities
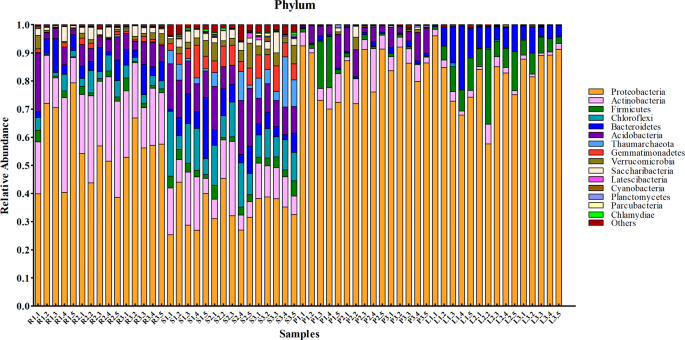
Twenty-three phyla of microbes were identified from all samples. The dominant phyla were Proteobacteria, Actinobacteria, and Acidobacteria. The core microbial community, defined as the shared microbial species among all independent samples within the same group, is shown in Fig. 3. Ten of the most abundant microbial species in the root and soil groups were from Proteobacteria, Actinobacteria, Acidobacteria, Bacteroidetes, Firmicutes, Chloroflexi, Gemmatimonadetes, Verrucomicrobia, Saccharibacteria, and Thaumarchaeota. However, the proportion of microbes from different phyla showed significant differences between these two groups. For the phloem groups, the major phyla were similar to those from root and soil groups except for Gemmatimonadetes. Core microbial species in the leaf group were Proteobacteria, Actinobacteria, Acidobacteria, Bacteroidetes, Firmicutes, Chloroflexi, Cyanobacteria, and Fusobacteria. Proteobacteria contained the majority of microbial species in both the phloem and leaf samples, representing more than 80% of identified species. The core families and genus of microbial communities were shown in Supplemental Fig. 1.
Similarity and difference among microbial community structures
The analytical methods Anosim and Multi-Response Permutation Procedure (MRPP) were used to compare potential similarities and differences among community structures of different sample groups. ANOSIM analyses (analysis of similarities) revealed that all R-values were greater than 0 and all P-values were 0.001, indicating significant differences between microbial community structures among different sample groups (Table 3). MRPP analyses also revealed significant differences with values of expecting delta ranged from 0.08305 to 0.3433 and values of significance 0.001 (Table 4).
Microbial species analyses
The metastat software based on Fisher exact test was used to analyze the differences in microbial species between different sample groups. The species with significant differences between different groups were filtered out according to the P and Q values (Table S2). Rhodopila was unique in the phloem group. Halomonas and Ralstonia were enriched in the leaf and phloem groups (P < 0.01). The abundance of these two microbes in the leaf group was significantly higher than that in the phloem group (P < 0.01). Methylobacterium and Sphingomonas were also very abundant in both the leaf and phloem groups (P < 0.01), with higher abundance in the phloem group. Streptomyces, Burkholderia-Paraburkholderia, and Acidibacter were mainly in the root and soil groups with higher abundance in the root group between the two (P < 0.05). Nitrosopumilus was enriched in the soil group compared to the other three groups (P < 0.01). Rhizobium was more abundant in the root group (P < 0.01).
Identification of microbial indicators
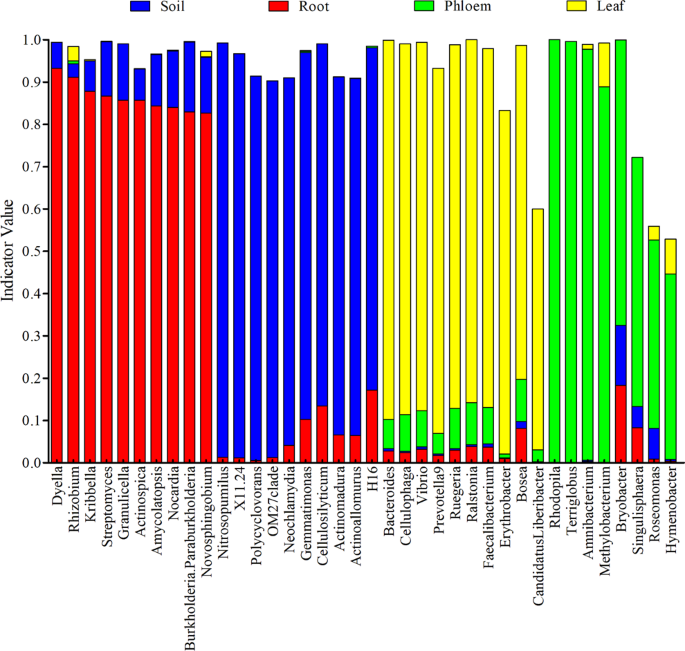
Indicator-values were used as criteria to access the uniqueness of microbial species in each group. As shown in Fig. 4, microbes specific to different groups were found. Among the unique microbes, Dyella, Rhizobium, Kribbella, Streptomyces, Granulicella, Actinospica, Amycolatopsis, Nocardia, Burkholderia. Paraburkholderia and Novosphingobium were identified as the indicators for the root group; Nitrosopumilus, X11.24, Polycyclovorans, OM27clade, Neochlamydia, Gemmatimonas, Cellulosilyticum, Actinomadura, Actinoallomurus and H16 were indicators for the soil group. Bacteroides, Cellulophaga, Vibrio, Prevotella9, Ruegeria, Ralstonia, Faecalibacterium, Erythrobacter, Bosea, and Candidatus Liberibacter were indicators for the leaf group; and Rhodopila, Terriglobus, Amnibacterium, Methylobacterium, Bryobacter, Singulisphaera, Roseomonas, and Hymenobacter were indicators for the phloem group.
The microbial indicators and indicator-values in different tissues of citrus trees. Different colors in the figure represent different groups. The indicator value of a species in a single group ranges from 0 to 1. When the indicator value is closer to 0, the species classification is almost non-existent in the group; while is closer to 1 indicates that the species is unique to the group and exists in almost all samples.
Discussion
Holobionts with high diversity residing in different plant tissues are regarded as plant microbiota1. The structure and function of microbial communities have attracted a great deal of attention because of increasing evidence suggesting critical roles of microbiomes in plant development and survival34. Plant-associated microorganisms could directly provide nutrition to plants via nitrogen fixation and phosphate solubilization35. Microbes can also induce resistance to other biotic and abiotic stresses through producing or degrading phytohormone35. Hence, it is necessary to investigate local microbial communities associated with plants to examine their impact on various aspects of a given plant species. Different plants host different microbial communities. Meanwhile, the microbial communities of plants could be affected by many factors. For example, the bacterium ‘Candidatus Liberibacter asiaticus’ (CLas) which induced the citrus devastating disease named Huanglongbing (HLB) could alter the root microbial community structure of Citrus limon and Citrus sinensis36. Administration of antibiotics including penicillin disrupted the interspecies microbiological connections and induced major changes in root bacterial community structure of grapefruit trees (Citrus paradise Macf.)37. Besides, the diversity of fungal endophyte communities in leaf, stem, trunk, and root tissues of C. reticulata cv. Siyahoo was observed38. However, a high similarity of dominant bacterial communities of Pericarpium Citri Reticulatae ‘Chachiensis’ obtained from different orchards in Xinhui District was observed39. In order to assess differences and dynamic changes in microbial communities among different tissues of C. reticulata cv. Chachiensis, 16S rRNA gene sequencing has been adapted to analyze microbes in samples derived from nearby soil, roots, leaves, and phloem. Our results demonstrated that the numbers and types of OTUs varied greatly among different sample groups. Among the microbes identified in this study, some genera have been well studied in the past few decades, whereas others were novel and unique. Identification of these microbes will facilitate comparative research on known microbes and functional studies on unique species. Furthermore, the microbial communities of many plant species were varied by geographical location. For example, the microbial diversity of C. reticulate blanco var. clementine in different regions was confirmed by 16S rDNA fingerprinting analysis40. For spruce (Picea spp.) trees, a significant difference of the microbial taxonomical composition in the phyllosphere was observed at three locations41. Moreover, the microbial community structure of malts from different cropping zones was variable. Among them, the effects of geographical location on the fungal community were more obvious than bacterial community42. Therefore, to study the microbial community of C. reticulata cv. Chachiensis more systematically, the effect of different geographical locations should be concerned and further studied.
Large differences in community structures in different microbiota have been observed in other plants from previous studies14,43. Microenvironments of tree tissues are likely one of the major factors driving alteration in the composition of microbial communities44. Microenvironments in tissues of citrus trees might also play a major role in the variation of microbial communities observed here. As an open system, plants constantly obtain minerals and water from the surrounding soil. In plant-microbe interactions, a plant may play an active role in shaping its associated microbial communities based on its needs. In turn, microbes may also affect plant physiology. The dynamic interactions among plants, microbes, and other environmental factors determine the functioning of an ecological system45,46,47. Soil by far harbors the most biodiversity and is the largest reservoir of microbes. Soil microbes impact plant microbiota profoundly48. Roots of a plant are the initial and main sites for plant-microbe intimate interactions. Rhizoplane, one of the root-associated layers, serves as a critical gate that regulates microbial entry into roots49. A comprehensive analysis indicated that no difference existed in the microbial community structure of rhizosphere and associated bulk soil samples collected from twelve citrus varieties which distributed on six continents50. In this study, the microbial community structure in roots was also similar to that in soil, suggesting that root-associated microbes were mainly derived from the soil biome35. Alternatively, the exudates containing sugars, organic acids and amino acids secreted by plant roots have strongly affected the composition of microbes in surrounding soil. Proteobacteria, Actinobacteria, Acidobacteria and Bacteroidetes have been reported as the primary consumers of plant exudates and the predominant taxa of citrus50,51. Other reports supported the above results and the microbial community of other plants including maize, arabidopsis and tamarix consists of a few dominant phyla, such as Proteobacteria, Actinobacteria, and Bacteroidetes16,52,53. In this study, Proteobacteria, Actinobacteria, Acidobacteria, Bacteroidetes, and Firmicutes were ranked as the top five core microbes in soil and root samples, which was similar to the global citrus rhizosphere microbiome50. However, the core genera of global citrus rhizosphere microbiome including Pseudomonas, Agrobacterium, Cupriavidus, Bradyrhizobium, Rhizobium, Mesorhizobium, Burkholderia, Cellvibrio, Sphingomonas, Variovorax and Paraburkholderia differed with our results, and the different growth area, seasonal variation and environmental factors could be the reasons for the difference. Furthermore, the genus from these phyla including Dyella, Rhizobium, Kribbella, Streptomyces, Granulicella, Actinospica, Amycolatopsis, Nocardia, Burkholderia. Paraburkholderia and Novosphingobium were identified as the indicator microbes in the root group. These indicator microbes could play crucial functions to citrus trees, for example, supplying nutrients, conferring resistance against pathogens and parasites47.
The microbial communities in citrus leaves and phloem are far smaller in comparison with those in roots and soil in terms of the number of OUTs in each group based on our results. The significant differences could be related to microenvironments in different tissues. Microbes can be originated from different sources, including aerosols or dust45. Insects are another important source for plants to gain microbes, including viruses, phytoplasmas, fungi, and bacteria, which are commonly transferred by insects. For example, Diaphorina citri Kuwayama (Hemiptera) transmit the bacterium Candidatus Liberibacter asiaticus (CLas), which causes a destructive disease called Huanglongbing54. Therefore, different sources for plant tissues to gain microbes could be another reason for large differences in their microbial communities. As ground material, the central bacterial community in Pericarpium Citri Reticulatae ‘Chachiensis’ separated from the fresh fruit of C. reticulata cv. Chachiensis were investigated and Chloroplast and mitochondria were identified as the most abundant bacterial phylum39. The significant differences that occurred between our results and the results obtained from Pericarpium Citri Reticulatae ‘Chachiensis’ might due to the massive mitochondria and chloroplast of C. reticulata existed during the processes of microbes enrichment and DNA extraction39. In our knowledge, this is the first time to investigate the microbial community of citrus phloem, which could helpful for understanding the distribution of microbial communities in entire citrus plants. Besides, Methylobacterium and Amnibacterium were identified as the major genus in citrus phloem. Methylobacterium was demonstrated to reduce the proliferation of citrus pathogens including CLas55, while the function of Amnibacterium should be explained in further study. For the leaf samples, Proteobacteria, Firmicutes and Bacteroidetes were ranked as the top three phyla and counted for more than 96.9% of all the microorganisms. Moreover, Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria were the most abundant bacterial classes in leaf samples, which was similar to the results of microbial communities of leaves collected from citrus trees across Florida29. Furthermore, some microbes with uniqueness were specific to tissue and were taken as the indicators of that tissue. For example, Methylobacterium, Amnibacterium, Rhodopila, and Terriglobus are indicators for the phloem group. Ralstonia, Bacteroides and Prevotella are indicators for the leaf group. The roles of these indicator microbes remain to be clarified.
Conclusion
In this study, the microbes in different parts of citrus trees were investigated by sequencing 16S rRNA genes. More than 2.5 million sequence reads were obtained from 60 samples and were assembled into OTUs. In total, 4733, 3520, 421, and 583 OTUs, respectively, were identified in samples from soil, roots, leaves, and phloem. The dominant microbial phyla of all samples from citrus trees were Proteobacteria, Actinobacteria, and Acidobacteria. The composition and structure of microbial communities in different plant tissues were analyzed using PCoA, CAP, Anosim and MRPP methods, and the species with a significant difference between groups were identified according to the P and Q values. Our results indicated that the microbial community in different groups were heterogeneous and complex. Indicator microbes for each group were identified based on their uniqueness among different sample groups. The microbial communities in different parts of citrus trees revealed in this study laid a foundation for future studies on microbial diversity and impact on citrus trees.
Materials and methods
Sampling sites and collection
The citrus orchard for sampling in this study was in Xinhui, Guangdong Province, China (22°47′N, 113°03′E). C. reticulata cv. Chachiensis was planted in this orchard with strict water and fertilizer management and pest control for three years. A total of 60 samples were obtained from citrus tissues including roots, leaves, and phloem as well as surrounding soil. Fifteen samples were obtained from each tissue, and samples from each tissue were referred to as a group. Samples were collected in the spring (March 28) of 2017. Fresh leaf and phloem samples were obtained from trees selected randomly. After collection, samples were immediately frozen in liquid nitrogen and stored at −80 °C. For root samples, roots were cut from trees and washed with sterilized water to remove the sediment for 5 times. After removing water with paper towels, roots were frozen liquid nitrogen. Soil was collected from 3–5 cm underground and immediately frozen in liquid nitrogen.
DNA extraction and PCR amplification
Citrus tissues were ground with liquid nitrogen to powder for DNA extraction. DNA was isolated and purified using an E.Z.N.A. ®Stool DNA Kit (Omega Bio-tek, Norcross, GA, U.S.) according to the manufacturer’s protocols56. The DNA quality and concentration were checked on a 1.0% agarose gel after electrophoresis and a NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, United States), respectively. For the amplicon library preparation, DNA was used as the template and the amplification of 16 S rRNA genes was performed by an amplified method57 of prokaryotic 16 S rDNA V4-V5 region with the following primers 515 F (5′-GTGCCAGCMGCCGCGG-3′) and 907 R (5′-CCGTCAATTCMTTTRAGTTT-3′), where barcode is an eight-base sequence unique to each sample. PCR reactions were performed in a 20 μL solution containing 4 μL of 5× FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase, and 10 ng of template DNA. The reaction was run under a program with one cycle at 95 °C for 5 min, followed by 27 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s, and a final extension at 72 °C for 5 min.
Amplicon purification and high-throughput sequencing
After electrophoresis, PCR fragments were extracted from 2% agarose gels using an AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, U.S.) according to the manufacturer’s instructions. The purified DNA fragments were quantified using QuantiFluor: trademark: -ST (Promega, U.S.). Purified 16 S rDNA amplicons were pooled in equimolar and paired-end sequenced (2 × 250) on HiSeq 2500 platform according to the standard protocol58. The sequencing was performed by Health Time Gene Institute in Shenzhen city, China.
Bioinformatics analysis
After removing the adaptors, primers and low-quality reads, the pair-end reads were assembled into final sequences based on overlapping alignments. The criterion for overlapping was at least 10 bp overlap with a mismatch ratio of less than 0.2. Chimera tags were filtered out using the Gold database by UCHIME (version 4.2.40)59. Operational taxonomic unit (OTU) analysis was performed using the Uparse package (version 7.0.1001) with a 97% sequence identity60. Each OTU was taxonomically assigned based on the silva database using the RDP classifier61. OTUs matched to chloroplast sequences, chondriosome sequences, and unclassified sequences were removed. Only those OTUs with relative abundance > 0.001% (above three tags in at least one sample) in at least one sample were retained. Similarities and differences among microbial communities from different groups were analyzed by principal co-ordinates analysis based on the distance of Unweighted Unifrac62. Canonical analysis of principal coordinates was chosen for diversity analyses among different groups63. Differences in microbial community structures between groups were examined with the methods of Anosim and Multi Response Permutation Procedure64. Differences in microbial species between groups were identified using metastat software based on the Fisher exact test65.
References
Turner, T. R. et al. The plant microbiome. Genome Biol. 14, 209 (2013).
Mendes, R. et al. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–63 (2013).
van der Heijdena, M. G. A. et al. Root surface as a frontier for plant microbiome research. Proc. Natl Acad. Sci. USA 112, 2299–2300 (2015).
Kembel, S. W. et al. Relationship between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc. Natl Acad. Sci. USA 111, 13715–13720 (2014).
Zhao, Y. et al. Endosphere microbiome comparison between symptomatic and asymptomatic roots of Brassica napus infected with Plasmodiophora brassicae. PLoS One 12, e0185907 (2017).
Panke-Buisse, K. et al. Selection on soil microbiomes reveals reproducible impacts on plant function. ISME J. 9, 980–989 (2015).
Berg, G. Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84, 11–18 (2009).
Fijan, S. Microorganisms with claimed probiotic properties: An overview of recent literature. Int. J. Envorn Res. Public. Health 11, 4745–4767 (2014).
Hill, C. et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastro Hepat. 11, 506–14 (2014).
Orozco-Mosqueda, M. C. et al. Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms. Microbiol. Res. 208, 25–31 (2018).
Bulgarelli, D. et al. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17, 392–403 (2015).
Lundberg, D. S. et al. Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90 (2012).
de Souza, R. S. C. et al. Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome. Sci. Rep. 6, 28774 (2016).
Edwards, J. et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl Acad. Sci. USA 112, E911–E920 (2015).
Larousse, M. et al. Tomato root microbiota and Phytophthora parasitica-associated disease. Microbiome 5, 56 (2017).
Peiffer, J. A. et al. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl Acad. Sci. USA 110, 6548–6553 (2013).
Schlemper, T. R. et al. Rhizobacterial community structure differences among sorghum cultivars in different growth stages and soils. FEMS Microbiol. Ecol. 93, fix096 (2017).
Sugiyama, A. et al. Changes in the bacterial community of soybean rhizospheres during growth in the field. PLoS One 9, e100709 (2014).
Rai, S. et al. Dynamics of soil diazotrophic community structure, diversity, and functioning during the cropping period of cotton (Gossypium hirsutum). J. Basic. Microbiol. 55, 62–73 (2015).
Arrigoni, E. et al. Tissue age and plant genotype affect the microbiota of apple and pear bark. Microbiol. Res. 211, 57–68 (2018).
Suhaimi, N. S. M. et al. Diversity of microbiota associated with symptomatic and non-symptomatic bacterial wilt-diseased banana plants determined using 16S rRNA metagenome sequencing. World J. Microbiol. Biotechnol. 33, 168 (2017).
Liu, J. et al. Apple endophytic microbiota of different rootstock/scion combinations suggests a genotype-specific influence. Microbiome 6, 18 (2018).
Wang, N. et al. Tale of the huanglongbing disease pyramid in the context of the citrus microbiome. Phytopathology 107, 380–387 (2017).
Sagaram, U. S. et al. Bacterial diversity analysis of Huanglongbing pathogen-infected citrus, using PhyloChip arrays and 16S rRNA gene clone library sequencing. Appl. Env. Microbiol. 75, 1566–1574 (2009).
Tyler, H. L. et al. Confirmation of the sequence of ‘Candidatus Liberibacter asiaticus’ and assessment of microbial diversity in Huanglongbing-infected citrus phloem using a metagenomic approach. Mol. Plant. Microbe Interact. 22, 1624–1634 (2009).
Trivedi, P. et al. Isolation and characterization of beneficial bacteria associated with citrus roots in Florida. Microb. Ecol. 62, 324–336 (2011).
Zhang, M. Q. et al. Deciphering the bacterial microbiome of citrus plants in response to ‘Candidatus liberibacter asiaticus’-infection and antiobiotic treatments. PLoS One 8, e76331 (2013a).
Zhang, M. Q. et al. Characterization of the microbial community structure in ‘Candidatus Liberibacter asiaticus’-infected citrus plants treated with antibiotics in the field. BMC Microbiol. 13, 112 (2013b).
Blaustein, R. A. et al. Defining the core citrus leaf- and root-associated microbiota: factors associated with community structure and implications for managing Huanglongbing (Citrus Greening) disease. Appl. Env. Microbiol. 83, e00210–17 (2017).
Wang, F. et al. Studying safe storage time of orange peel (Citrus reticulata) using high-throughput sequencing and conventional pure culture. Food Sci. Nutr. 6, 2545–2552 (2018).
Wang, F. et al. Analysis of Flavonoid Metabolites in Citrus Peels (Citrus reticulata “Dahongpao”) Using UPLC-ESI-MS/MS. Molecules 15, E2680 (2019).
Wang, H. et al. Influence of the stage of ripeness on the phytochemical profiles, antioxidant and antiproliferative activities in different parts of Citrus reticulata Blanco cv. Chachiensis. LWT - Food Sci. Technol. 69, 67–75 (2016).
Mitter, B. et al. Next generation microbiome applications for crop production - limitations and the need of knowledge-based solutions. Curr. Opin. Microbiol. 49, 59–65 (2019).
Lebeis, S. L. Greater than the sum of their parts: characterizing plant microbiomes at the community-level. Curr. Opin. Plant. Biol. 24, 82–6 (2015).
Vandenkoornhuyse, P. et al. The importance of the microbiome of the plant holobiont. N. Phytol. 206, 1196–1206 (2015).
Padhi et al. Metabolome and Microbiome Signatures in the Roots of Citrus Affected by Huanglongbing. Phytopathology 109, 2022–2032 (2019).
Ascunce et al. Penicillin trunk injection affects bacterial community structure in citrus trees. Microb. Ecol. 78, 457–469 (2019).
Sadeghi, F. et al. Diversity and spatiotemporal distribution of fungal endophytes associated with Citrus reticulata cv. Siyahoo. Curr. Microbiol. 76, 279–289 (2019).
He, J. et al. The central bacterial community in Pericarpium Citri Reticulatae ‘Chachiensis’. Food Res. Int. 125, 108624 (2019).
Le Nguyen, D. D. et al. Determination of citrus fruit origin by using 16S rDNA fingerprinting of bacterial communities by PCR- DGGE: an application to clementine from Morocco and Spain. Fruits 63, 75–84 (2008).
Li, Y. et al. Microbial taxonomical composition in spruce phyllosphere, but not community functional structure, varies by geographical location. PeerJ 7, e7376 (2019).
Kaur, M. et al. The fungal community structure of barley malts from diverse geographical regions correlates with malt quality parameters. Int. J. Food Microbiol. 215, 71–78 (2015).
Shade, A. et al. Unexpected diversity during community succession in the apple flower microbiome. MBio 4, e00602–e00612 (2013).
Ren, F. et al. Tissue Microbiome of norway spruce affected by heterobasidion-induced wood decay. Microb. Ecol. 77, 640–650 (2018).
Hassani, M. A. et al. Microbial interactions within the plant holobiont. Microbiome 6, 58 (2018).
Kinkel, L. L. et al. A coevolutionary framework for managing disease-suppressive soils. Annu. Rev. Phytopathol. 49, 47–67 (2011).
Berg, G. et al. Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 93, fix050 (2017).
Little, A. E. F. et al. Rules of engagement: interspecies interactions that regulate microbial communities. Annu. Rev. Microbiol. 62, 375–401 (2008).
Zhang, Y. et al. Huanglongbing impairs the rhizosphere-torhizoplane enrichment process of the citrus root-associated microbiome. Microbiome 5, 97 (2017).
Xu, J. et al. The structure and function of the global citrus rhizosphere microbiome. Nat. Commun. 9, 4894 (2018).
Vandenkoornhuyse, P. et al. Active rootinhabiting microbes identified by rapid incorporation of plantderived carbon into RNA. Proc. Natl Acad. Sci. USA 104, 16970–16975 (2007).
Bai, Y. et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369 (2015).
Finkel, O. M. et al. Geographical location determines the population structure in phyllosphere microbial communities of a salt-excreting desert tree. Appl. Env. Microbiol. 77, 7647–7655 (2011).
Grafton-Cardwell, E. E. et al. Biology and management of Asian citrus psyllid, vector of the huanglongbing pathogens. Annu. Rev. Entomol. 58, 413–32 (2013).
Li, J. et al. Endophytic bacterial community analysis of Catharanthus roseus and its association with huanglongbing pathogen. Acta microbiol. Sin. 52, 489–497 (2012).
Li, T. et al. Multi-omics analysis reveals a correlation between the host phylogeny, gut microbiota and metabolite profiles in cyprinid fishes. Front. Microbiol. 8, 454 (2017).
Liu, H. et al. The gut microbiome and degradation enzyme activity of wild freshwater fishes influenced by their trophic level. Sci. Rep. 6, 24340 (2016).
Pan, H. et al. Berberine influences blood glucose via modulating the gut microbiome in grass carp. Front. Microbiol. 10, 1066 (2019).
Edgar, R. C. et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–200 (2011).
Edgar, R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998 (2013).
Pruesse, E. et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–96 (2007).
Hamady, M. et al. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 4, 17–27 (2010).
Marti, J. et al. Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology 84, 511–525 (2003).
Mota, J. F. et al. Use of the multi-response permutation procedure and indicator species value for the statistical classification of the gypsicolous iberian scrub communities. Candollea 65, 117–134 (2010).
White, J. R. et al. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 5, e1000352 (2009).
Acknowledgements
This work was financially supported by a grant from the National Natural Science Foundation of China (Grant 31672063), a grant from the Guangdong Province Science and Technology Plan Projects (Grant 2016B02020009), and funds from the Innovation Team Project in Guangdong Provincial Department of Education (2017KCXTD018) and Guangzhou Science and Technology Plan Projects (Grants 201704020190, 201805010008 and 201803020009). The authors sincerely thank Prof. Ming-Shun Chen of Kansas State University for editing this manuscript for grammar mistakes.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: B.S. and Y.W.; Performance of the experiments: Y.W., M.Q. and X.P.; Analysis of the data: Y.W. and B.S.; Manuscript writing: B.S.; Manuscript editing: J.L. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wu, Y., Qu, M., Pu, X. et al. Distinct microbial communities among different tissues of citrus tree Citrus reticulata cv. Chachiensis. Sci Rep 10, 6068 (2020). https://doi.org/10.1038/s41598-020-62991-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-62991-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.