Abstract
Pedobacter schmidteae sp. nov. strain EGT (Collection de Souches de l’Unité des Rickettsie CSUR P6417 = Colección Española de Cultivos Tipo CECT 9771) is a new Pedobacter species isolated from the planarian Schmidtea mediterranea. Schmidtea mediterranea are flatworms living in freshwater and exhibiting an unusual ability to regenerate amputated parts. To date, the gut microbiota of Schmidtea mediterranea remains poorly studied. Here, via the culturomics strategy that consists in using diversified culture conditions, we isolated a new bacterium, strain EG, that we characterized using the taxono-genomics approach that combines phenotypic assays and genome sequencing and analysis. Strain EG exhibits a 16S rRNA sequence similarity of 98.29% with Pedobacter nyackensis strain NWG-II14T, its closest neighbour with standing in nomenclature. It is an aerobic bacterium belonging to the family Sphingobacteriaceae. Colonies are small, round, smooth and transparent. Bacterial cells are Gram-negative, rod-shaped, motile and non-spore-forming bacilli with positive catalase and oxidase activities. The genome sequence is 6,198,518 bp–long with a G + C content of 41.13%, and the Ortho-ANI and dDDH values when compared to P. nyackensis are 77.34% and 21.50%, respectively. Strain EGT exhibits unique characteristics that classify it as the type strain of new bacterial species for which we propose the name Pedobacter schmidteae sp. nov.
Similar content being viewed by others
Introduction
Schmidtea mediterranea is an invertebrate living in environmental water. This flatworm is used as a model of development, because of its extraordinary abilty to regenerate after amputation, because of his high contents in stem cells known as neoblasts1. It has been shown that S. mediterranea is an excellent model to investigate host-pathogen relationships2,3, notably in the context of human pathogens. To date, the gut microbiota of S. mediterranea remains poorly studied4,5. Using the microbial culturomics approach6, we investigated the S. mediterranea microbiota. Culturomics is a concept in which diversified culture conditions are used to enable isolation of a maximum of bacterial species from the human microbiota7,8,9,10. During this analysis, we isolated a bacterial strain from S. mediterranea that could not be identified using Matrix Assisted Laser Desorption-Ionisation Time of Flight-Mass Spectrometry (MALDI-TOF-MS). We used the taxono-genomics strategy that combines phenotypic assays and genome sequencing to further characterize this bacterium11,12,13,14. This enabled us to describe a new bacterial strain that exhibited enough genetic and phenotypic differences with closely related bacteria. We propose it as a new species named Pedobacter schmidteae sp. nov.
Materials and Methods
Culture of Schmidtea mediterranea
Schmidtea mediterranea asexual clonal line ClW42 is a laboratory planarian strain that has been preserved in our laboratory for the past 10 years, by cutting animals in tree fragments each month. S. mediterranea were kept in the dark, in filtered tap water, at 19 °C without antibiotics. The animals were fed once per week with homogenized calf liver and were starved for at least two weeks prior to studying them. Filtered water was obtained using a device consisting of two 0.2 µm filters, one containing charcoal and ceramics (Fairey Industrial Ceramics limited, England), and the other being a 0.20 µm membrane (Thermo Scientific Nalgene filtration Products, Mexico). Filtered water was checked for sterility prior to be used for planarian culture.
Isolation and identification of bacteria from Schmidtea mediterranea
Following two weeks of starvation, S. mediterranea were washed in filter-sterilized water and then one ground worm was inoculated in Buffered Charcoal Yeast Extract (BCYE) (Oxoid Deutschland GmbH, Wesel, Germany), Luria Bertani (LB) and 5% sheep blood-enriched Columbia agar (bioMérieux, Marcy l’étoile, France). All inoculated media were incubated at 19, 28 and 37 °C. Each individual bacterial colony was harvested and identified by MALDI-TOF-MS (Microflex spectrometer; Bruker Daltonics, Bremen, Germany)15. The obtained spectra were imported into the MALDI Biotyper 3.0 software (Bruker Daltonics) and analysed against the reference spectra of bacteria included in the database (Bruker database constantly updated with the Mediterranee-Infection database (http://www.mediterranee-infection.com/article.php?larub=280&titre=urms-database). The MALDI Biotyper RTC software was used to interpret the results according to the obtained score values: a colony was likely identified at the species level for a score > 2.0, probably identified for a score between 1.99 and 1.7, but not identified for a score <1.7.
Phylogenetic analysis
Bacterial colonies that were not identified at the species level using MALDI-TOF MS were further tested using 16S rRNA sequencing. Genomic DNA was extracted using an EZ1 automate and the DNA tissue kit (Qiagen, Hilden, Germany). The complete 16S rRNA gene amplification and sequencing was performed using eight primers on an ABI Prism 3130xl Genetic Analyzer capillary sequencer (Applied Bio systems, Bedford, MA, USA). The primers used were Fd1 (5′-AGAGTTTGATCCTGGCTCAG-3′), Rp2 (5′-ACGGCTACCTTGTTACGACTT-3′), F536 (5′-CAGCAGCCGCGGTAATAC-3′), R536 (5′-GTATTACCGCGGCTGCTG-3′), F800 (5′-ATTAGATACCCTGGTAG-3′), R800 (5′-CTACCAGGGTATCTAAT-3′), F1050 (5′-TGTCGTCAGCTCGTG-3′) and R1050 (5′-CACGAGCTGACGACA-3′) (Eurogentec, Angers, France). The CodonCode Aligner software was used for sequence alignment, assembly and correction (https://www.codoncode.com/). For taxonomic assignation, a BLASTn search was performed against the nr database16. A sequence similarity threshold of 98.65% by comparison with the phylogenetically closest species with standing in nomenclature was used to delineate a putative new species17. Phylogenetic relationships were inferred from the comparison of 16S rRNA sequences using the MEGA7 software18.
Phenotypic, biochemical and chemical characteristics of strain EG
Culture of strain strain EG and P. nyackensis strain NWG-II14T (DSM19625) was attempted at various growth temperatures (6, 19, 30, 37 and 45 °C) in 5% sheep blood-enriched Columbia agar (bioMérieux) under anaerobic, aerobic and microaerophilic atmospheres using GasPak™ EZ generators (Becton- Dickinson, Maryland, USA). Sporulation was tested by thermal shock, which consists in exposing bacteria to a temperature of 80 °C for 30 minutes and then monitoring their growth for 4 days. Various salinity (0, 8.5, 25, 50, 100 and 200 g/l) and pH (4, 5.5, 6, 7.5, 8, 9 and 10) conditions were tested. Gram staining and motility from fresh colonies were observed using a DM1000 photonic microscope (Leica Microsystems, Nanterre, France) with a 40 × objective lens. Catalase and oxidase activities were tested by using a BBL DrySlide according to the manufacturer’s instructions (Becton Dickinson, Le Pont de Claix, France). The size of bacterial cells was measured using transmission electron microscopy. API strips (API ZYM19,20,21, API 20NE22,23, API 20E24,25 and API 50CH26,27,28,29, bioMérieux) were used to study the biochemical characteristics of the strains. Bacterial susceptibility to benzylpenicillin, amoxicillin, ampicillin, ceftriaxone, imipenem, ciprofloxacin, amikacin, gentamicin, streptomycin, daptomycin, doxycycline, metronidazole, rifampicin, and vancomycin was assessed using E-tests and a 0.5 McFarland concentration of strains EG and NWG-II14T. Cellular fatty acid methyl ester (FAME) analysis was performed by GC/MS. Two samples were prepared with approximately 120 mg of bacterial biomass per tube harvested from several culture plates. Fatty acid methyl esters were prepared as described by Sasser30 and GC/MS analysis was carried out as previously described31. Briefly, FAMES were separated using an Elite 5-MS column and monitored by mass spectrometry (Clarus 500 - SQ 8S, Perkin Elmer, Courtaboeuf, France). Spectral database search was performed using MS Search 2.0 operated with the following Standard Reference Database 1 A (NIST, Gaithersburg, USA) and FAMEs mass spectral database (Wiley, Chichester, UK).
Sequencing, assembly, annotation and genomic comparison
The bacterial genomic DNA (gDNA) of strain EG was extracted using an EZ1 automate and the DNA tissue kit (Qiagen, Hilden, Germany) and then was quantified using a Qubit assay (Life Technologies, Carlsbad, CA, USA) at 82.6 ng/μL. The bacterial gDNA was prepared and sequenced as previously described32. Briefly, sequencing was performed using the Mate-Pair strategy and a Miseq sequencer (Illumina, San Diego, CA, USA). A concentration of 1.5 μg of gDNA prepared following the Nextera Mate-Pair Illumina guide was used to prepare the Mate-Pair library. The gDNA was simultaneously fragmented and tagged using a Mate-Pair junction adapter. Then the fragmentation pattern was validated using a DNA 7500 labchip on a BioAnalyzer (Agilent 2100, Agilent Technologies, Santa Clara, CA, USA). The size of the DNA fragments ranged from 1.5 kb to 11 kb. No size selection was performed and 662 ng of labelled fragments were circularized. Next, circularized DNA was mechanically sheared using a Covaris device S2 (Covaris, Woburn, MA, USA) into small fragments with an optimal size at 1200 bp. The library profile was analysed on a High Sensitivity Bioanalyzer LabChip (Agilent Technologies) and the concentration library was measured at 61.4 nmol/l. Then, the library was loaded on the sequencer after a denaturation step and a dilution at 15 pM. Automated cluster generation and sequencing were performed in a single 39-h run in a 2 × 251-bp and sequencing reads were assembled using the A5 pipeline. Genomic annotation was obtained using the Prokka software. A search for virulence factors was performed by comparaison with the VFDB database (http://www.mgc.ac.cn/VFs/) using BLASTn33,34.
The genome from strain EG was compared to those of Pedobacter africanus strain DSM 12126 T (NZ_FWXT00000000.1), P. antarcticus strain 4BYT (NZ_JNFF00000000.1), P. ginsengisoli strain T01R-27 (NZ_CP024091.1), P. heparinus strain DSM 2366 T (NC_013061.1), P. nutrimenti strain DSM 27372 (NZ_QKLU00000000.1), P. nyackensis strain DSM 19625 (NZ_FWYB00000000.1), P. panaciterrae strain 048 (NZ_LGEL00000000.1) and P. steynii strain DX4 (NZ_CP017141.1). Degrees of genomic similarity between strain EG and compared genome were evaluated using the GGDC (http://ggdc.dsmz.de/ggdc.php#)16 and Orthologous Average Nucleotide Identity (Ortho-ANI) (https://www.ezbiocloud.net/tools/orthoani)35 softwares.
Results and Discussion
Strain isolation, 16S rRNA gene sequencing and phylogenetic analysis
Strain EG was isolated on 5% sheep blood-enriched Columbia agar (bioMérieux) after 2 days at 28 °C in aerobic atmosphere at pH 7. The 16S rRNA-based phylogenetic tree demonstrated that strain EG was most closely related to Pedobacter nyackensis strain NWG-II14T with which it exhibited a sequence similarity of 98.29%. When compared to other Pedobacter species, strain EG exhibited 16S rRNA similarity values of 97.84%, 97.69%, 97.63%, 97.93%, 97.43%, 97.28%, 97.33%, 97.62%, 97.93%, 97.32%, 97.32% and 97.0% with P. heparinus strain DSM 2366 T, P. steynii strain WB2.3–45 T, P. metabolipauper strain WB2.3–71 T, P. nutrimenti strain J22T, P. duraquae strain WB2.1–25 T, P. africanus strain NBRC 100065 T, P. panaciterrae strain Gsoil 042 T, P. ginsengisoli strain Gsoil 104 T, P. seoulensis strain THG-G12T, P. trunci strain THG-DN3.19 T, P. humi strain THG S15–2T and P. antarcticus strain 4BYT 36,37,38,39,40, respectively (Fig. 1).
: 16S rRNA-based maximum likelihood phylogenetic tree highlighting the position of Pedobacter schmidteae strain EGT relative to other closely related species. The respective GenBank accession numbers for 16S rRNA genes are indicated in parentheses. Sequences were aligned using CLUSTAL W with default parameters and phylogenies were inferred by the software MEGA 718. Numbers at the nodes are percentages of bootstrap values obtained by repeating the analysis 500 times to generate a majority consensus tree. Only bootstrap values ≥50% were retained. The scale bare indicates a 0.5% sequence divergence.
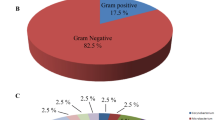
The analysis of 13 S. mediterranea revealed the presence of strain EG in 11 S. mediterranea sp. nov. Strain EG is a member of the family Sphingobacteriaceae within the phylum Bacteroidetes (Table 1).
Phenotypic, enzymatic and biochemical characteristics
After 4 days of culture on blood-enriched Columbia agar, colonies from strain EG were small (0.4 mm of diameter), transparent, round with a convex shape and smooth. Bacterial cells were Gram-negative (Fig. 2), rod-shaped, motile, and non-spore-forming bacilli. Using the Image J software, their mean length and width were 1.98 µm and 0.69 µm, respectively (Fig. 3), without any flagellum. For the two strains EG and P. nyackensis NWG-II14T, no growth was obtained in anaerobic or microaerophilic conditions. Both strains grew at temperatures ranging from 6 to 30 °C in aerobic atmosphere at pH values ranging from 5.5 to 9; the strains also grew at salinity concentrations lower than 25 g/L. Catalase and oxidase activities were positive for both strains.
Strain EG and P. nyackensis strain NWG-II14T were susceptible to benzylpenicillin, ampicillin, ceftriaxone, imipenem, gentamicin, doxycycline and rifampicin (Table 2). Both strains were resistant to amoxicillin, amikacin, daptomycin, metronidazole and vancomycin. Pedobacter nyackensis, but not P. schmidteae, was susceptible to ciprofloxacin and streptomycin.
Positive and negative reactions obtained using API 50CH, API 20NE, API Zym, and API 20E strips are presented in Table 3. By comparison with P. nyackensis NWG-II14T, strain EG differed by exhibing of Esterase (C4), Esterase lipase (C8), Valine arylamidase, β-galactosidase and β-glucosidase activities (Table 3). By comparaison with all other tested species, strain EG differed in production of esterase (C4) and β-glucosidase.
The analysis of the composition in fatty acid of the strain EG revealed that the major fatty acids were 13-methyl-tetradecanoic acid (49.8%), 9-Hexadecenoic acid (27.7%) and 3-hydroxy-15-methyl-Hexadecanoic acid (7.5%). Other fatty acids included 15-methyl-Hexadecenoic acid (3.6%), 14-Methylpentadec-9-enoic acid (3.4%), Hexadecanoic acid (3.3%), 3-hydroxy-13-methyl-Tetradecanoic acid (1.9%), Tetradecanoic acid (1.4%) and 3-hydroxy-Hexadecanoic acid (1.0%). Minor amounts (<1%) of the fatty acids were also detected as Pentadecanoic acid and 9,12-Octadecadienoic acid (Table 4). 15-methyl-Hexadecenoic acid, 14-Methylpentadec-9-enoic acid is detected only in strain EGT. In contrast to P. nyackensis NWG-II14T, the strain EG have in its membrane composition the 3-hydroxy-Hexadecanoic acid, 13-methyl-Tetradecanoic acid, 3-hydroxy-13-methyl-Tetradecanoic acid, 3-hydroxy-15-methyl-Hexadecanoic acid, 15-methyl-Hexadecenoic acid and 14-Methylpentadec-9-enoic acid; and for the absence of Dodecanoic acid, 3-hydroxy-8-methyl-nonanoic acid, 9-Heptadecenoic acid, 9-Octadecenoic acid.
Genomic characteristics
The genome sequence from strain EG was assembled into one contig of 6,198,518 bp with a G + C content of 41.13%. We identified a total of 5,012 predicted protein-coding genes, in addition to 3 complete rRNA operons, 52 tRNAs and 1 tmRNA. Comparison of these genomic data with those from of closely related species is presented in Table 5. The distribution of genes in functional categories (COGs) is shown within Table 6. In addition, using the Prokka software, we identified a two-component sensor histidine kinase system. This system exhibited an identity of 92% (E-value of 10−3) and a score of 52 with the GacS [GacS/GacA two-component system]41,42 sensor histidine kinase/response regulator in Pseudomonas putida strain KT2440 according to the VFDB database. It has been reported that the two-component system signaling pathways are the major signaling mechanisms in bacteria and as well in Archaea43 to monitor external and internal stimuli (including concentration of ions and gas, redox states, levels of nutrients and cell density)44. This pathway is also found in simple eukaryota and higher plants45. In opportunistic bacterial pathogens, the use of two-components systems is required to regulate the expression of genes necessary for the transition from the environmental reservoir to the host46. Digital DNA-DNA hybridization values (dDDH) obtained using the GGDC software are reported in Table 7. For strain EG, these values ranged from 18.90 with P. nutrimenti to 21.50% with P. nyackensis. Such values were lower than the 70% threshold recognized to delineate distinct species. Similarly, Ortho-ANI values (Fig. 4) ranged from 70.98% with P. nutrimenti to 74.65% with P. nyackensis, which is lower than the 95% threshold used to discriminate bacterial species. Thus, we could confirm that strain EG belonged to a separate Pedobacter species for which we propose the name Pedobacter schmidteae sp. nov.
Heatmap generated with OrthoANI values calculated using the OAT software35 between Pedobacter schmidteae and other closely related species with standing in nomenclature. The colour code indicates the closest species in green to the farthest in red.
Conclusion
Using the taxono-genomic approach, we concluded that strain EG is the representative strain of the new species P. schmidteae sp. nov. Interestingly, P. schmidteae sp. nov. is present in 84.6% of S. mediterranea worms tested. To best of our knowledge, P. schmidteae sp. nov has never been identified anywhere else than in S. mediterranea. Indeed, no nucleotide sequence linked to this new strain has been found in the nr database (BLASTn https://blast.ncbi.nlm.nih.gov). To date, we assume that P. schmidteae sp. nov is unique to S. mediterranea, but we cannot exclude formally that it might be present in other living organisms of the environment.
Protologue
The protologue is to standardize the format of descriptions of new taxa, supported by the Judicial Commission of the International Committee on Systematic Bacteriology47. Pedobacter schmidteae (schmid.te’ae. N.L. gen. n. schmidteae of the planarian genus Schmidtea, from which strain EG was isolated). The bacterium belongs to the family Sphingobacteriaceae within the phylum Bacteroidetes. The type strain EGT (CSUR P6417 = CECT9771) was isolated on 5% sheep blood-enriched Columbia agar after 2 days at 28 °C in aerobic atmosphere at pH 7 from the microbiota of the planarian S. mediterranea. Colonies are small, round, smooth, transparent and convex. Bacterial cells are Gram-negative, rod-shaped, motile and non-spore-forming bacilli with positive catalase and oxidase activities. The main cellular fatty acids detected are 13-methyl-tetradecanoic acid (49.8%), 9-Hexadecenoic acid (27.7%), 3-hydroxy-15-methyl-Hexadecanoic acid (7.5%), 15-methyl-Hexadecenoic acid (3.6%), 14-Methylpentadec-9-enoic acid (3.4%), Hexadecanoic acid (3.3%), 3-hydroxy-13-methyl-Tetradecanoic acid (1.9%), Tetradecanoic acid (1.4%) and 3-hydroxy-Hexadecanoic acid (1.0%). We found traces (<1%) of Pentadecanoic acid and 9,12-Octadecadienoic acid.
Using an APIZYM strip, strain EGT exhibits positive reactions for catalase, oxidase, alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, acid phosphatase, Naphtol-AS-BI-phosphohydrolase, β-galactosidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, esculin ferric citrate, glucose, D-mannose, N-Acetyl-glucosamine and D-maltose, but negative reaction for lipase (C14), cystine arylamidase, trypsin, α-chymotrypsin, α-galactosidase, β-glucuronidase, α-mannosidase, α-fucosidase.
Using a API 50CH strip, strain EGT is unable to metabolize glycerol, starch, amygdalin, arbutin, D-adonitol, D-arabinose, D-arabitol, D-cellobiose, D-fructose, D-fucose, D-galactose, D-glucose, D-lactose, D-melezitose, D-melibiose, D-raffinose, D-ribose, D-saccharose, D-sorbitol, D-tagalose, D-trehalose, D-turanose, dulcitol, D-xylose, erythritol, gentiobiose, glycogen, inositol, inulin, L-arabinose, L-arabitol, L-fucose, L-rhamnose, L-sorbose, L-xylose, methyl-αD-Glucopyranoside, methyl-αD-Mannopyranoside, methyl-βD-xylopyranoside, potassium 2-CetoGluconate, potassium 5-Cetogluconate, potassium gluconate, salicin, xylitol.
In addition, strain EGT use potassium nitrate, L-tryptophane, L-arginine, urea, gelatin, D-mannitol, malic acid, phenylacetic acid, L-lysin, trinatriumcitrate, L-ormithin, Esculin ferric citrate, Glucose, D-mannose, N-Acetyl-glucosamine and D-maltose.
The genome of strain EGT is 6,198,518 bp–long with a G + C content of 41.13%.
The 16S rRNA gene and genome sequence are deposited in GenBank under accession numbers LS453293 and LS999839, respectively.
MALDI-TOF MS spectrum
The MALDI-TOF MS spectrum of Pedobacter schmidteae strain EGT is available at http://www.mediterranee-infection.com/article.php?laref=936
Nucleotide sequence accession number
The 16S rRNA gene sequence and genome sequence were deposited in GenBank under accession numbers LS453293 and LS999839, respectively. The Digital Protologue database Taxonumber for strain EGT is TA00631 in the http://imedea.uib-csic.es/dprotologue/ website.
Deposit in culture collections
Strain EGT (Pedobacter schmidteae sp; nov.) was deposited in the CSUR (Collection de Souches de l’Unité des Rickettsies WDCM 875) and CSCT (Colección Española de Cultivos Tipo) strain collections under numbers CSUR P6417 and CECT9771, respectively.
References
Elliott, S. A. & Sánchez Alvarado, A. The history and enduring contributions of planarians to the study of animal regeneration. Wiley Interdiscip Rev Dev Biol 2, 301–326 (2013).
Abnave, P. et al. Screening in planarians identifies MORN2 as a key component in LC3-associated phagocytosis and resistance to bacterial infection. Cell Host Microbe 16, 338–350 (2014).
Torre, C. & Ghigo, É. Planaria: an immortal worm to clarify human immune response. Med Sci (Paris) 31, 20–22 (2015).
Arnold, C. P. et al. Pathogenic shifts in endogenous microbiota impede tissue regeneration via distinct activation of TAK1/MKK/p38. Elife 5, (2016).
Lee, F. J., Williams, K. B., Levin, M. & Wolfe, B. E. The Bacterial Metabolite Indole Inhibits Regeneration of the Planarian Flatworm Dugesia japonica. Science 10, 135–148 (2018).
Seng, P. et al. Identification of Rare Pathogenic Bacteria in a Clinical Microbiology Laboratory: Impact of Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry. J Clin Microbiol 51, 2182–2194 (2013).
Lagier, J.-C. et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 18, 1185–1193 (2012).
Lagier, J.-C. et al. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin. Microbiol. Rev. 28, 237–264 (2015).
Lagier, J.-C. et al. Current and past strategies for bacterial culture in clinical microbiology. Clin. Microbiol. Rev. 28, 208–236 (2015).
Lagier, J.-C. et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol 1, 16203 (2016).
Ramasamy, D. et al. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. IJSEM 64, 384–391 (2014).
Fournier, P.-E., Lagier, J.-C., Dubourg, G. & Raoult, D. From culturomics to taxonomogenomics: A need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe 36, 73–78 (2015).
Morel, A.-S. et al. Complementarity between targeted real-time specific PCR and conventional broad-range 16S rDNA PCR in the syndrome-driven diagnosis of infectious diseases. Eur. J. Clin. Microbiol. Infect. Dis. 34, 561–570 (2015).
Diop, A. et al. Microbial culturomics unravels the halophilic microbiota repertoire of table salt: description of Gracilibacillus massiliensis sp. nov. Microb. Ecol. Health Dis. 27, 32049 (2016).
Seng, P. et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49, 543–551 (2009).
Auch, A. F., von Jan, M., Klenk, H.-P. & Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci 2, 117–134 (2010).
Meier-Kolthoff, J. P., Göker, M., Spröer, C. & Klenk, H.-P. When should a DDH experiment be mandatory in microbial taxonomy? Arch. Microbiol. 195, 413–418 (2013).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Unaogu, I. C., Gugnani, H. C. & Boiron, P. The enzymatic profile of some pathogenic aerobic actinomycetes as determined by api-zym method. J. Myco. Med. 9(4), 235 (1999).
Gruner, E., von Graevenitz, A. & Altwegg, M. The API ZYM system: a tabulated review from 1977 to date. J. Microbiolo. Meth. 16, 101–118 (1992).
Humble, M. W., King, A. & Phillips, I. API ZYM: a simple rapid system for the detection of bacterial enzymes. J. Clin. Pathol. 30, 275–277 (1977).
Søgaard, P., Gahrn-Hansen, B., Zhou, H. P. & Frederiksen, W. An investigation of three commercial methods for rapid identification of non-enteric gram-negative rods. Application on Pseudomonas paucimobilis and some other Pseudomonas species. Acta. Pathol. Microbiol. Immunol. Scand. B 94, 357–363 (1986).
MK, B., DA, B., GL, C. & JG, G. Comparison of five commercial methods for the identification of non-fermentative and oxidase positive fermentative gram negative bacilli. NZ J. Med. Lab. Technol. 42, 8–12 (1988).
Swanson, E. C. & Collins, M. T. Use of the API 20E system to identify veterinary Enterobacteriaceae. J. Clin. Microbiol. 12, 10–14 (1980).
Smith, P. B., Tomfohrde, K. M., Rhoden, D. L. & Balows, A. API System: a Multitube Micromethod for Identification of Enterobacteriaceae. Appl. Microbiol. 24, 449–452 (1972).
Véron, M. & Le Minor, L. Nutrition and taxonomy of ‘enterobacteriaceae’ and related bacteria. III. Nutritional characters and differentiation of the taxonomic groups (author’s transl). Ann. Microbiol. (Paris) 126, 125–147 (1975).
Bergey, D. H., Krieg, N. R. & Holt, J. G. Bergey’s manual of systematic bacteriology. (Williams & Wilkins, 1984).
Rogosa, M. & Sharpe, M. E. An approach to the classification of the lactobacilli. Journal of Appl. Bact. 22, 329–40 (1960).
Sharpe, M. E., Hill, L. R. & Lapage, S. P. Pathogenic Lactobacilli. J. Med. Microbiol. 6, 281–286 (1973).
Sasser, M. Identification of bacteria by gas chromatography of cellular fatty acids. USFCC Newsl. 20, 1–6 (1990).
Dione, N. et al. Genome sequence and description of Anaerosalibacter massiliensis sp. nov. NMNI. 10, 66–76 (2016).
Beye, M. et al. Draft genome sequence of Fermentimonas caenicola strain SIT8, isolated from the human gut. Stand. Gen. Sc. 13, 8 (2018).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic. Acids. Res. 25, 3389–3402 (1997).
Liu, B., Zheng, D., Jin, Q., Chen, L. & Yang, J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic. Acids. Res. 47, D687–D692 (2019).
Lee, I., Ouk Kim, Y., Park, S.-C. & Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103 (2016).
Ten, L. N. et al. Pedobacter ginsengisoli sp. nov., a DNase-producing bacterium isolated from soil of a ginseng field in South Korea. Int. J. Syst. Evol. Microbiol. 56, 2565–2570 (2006).
Yoon, M.-H., Ten, L. N., Im, W.-T. & Lee, S.-T. Pedobacter panaciterrae sp. nov., isolated from soil in South Korea. Int. J. Syst. Evol. Microbiol. 57, 381–386 (2007).
Steyn, P. L. et al. Classification of heparinolytic bacteria into a new genus, Pedobacter, comprising four species: Pedobacter heparinus comb. nov., Pedobacter piscium comb. nov., Pedobacter africanus sp. nov. and Pedobacter saltans sp. nov. proposal of the family Sphingobacteriaceae fam. nov. Int. J. Syst. Bacteriol. 48 Pt 1, 165–177 (1998).
Gordon, N. S. et al. Pedobacter nyackensis sp. nov., Pedobacter alluvionis sp. nov. and Pedobacter borealis sp. nov., isolated from Montana flood-plain sediment and forest soil. Int. J. Syst. Evol. Microbiol. 59, 1720–1726 (2009).
Muurholm, S., Cousin, S., Päuker, O., Brambilla, E. & Stackebrandt, E. Pedobacter duraquae sp. nov., Pedobacter westerhofensis sp. nov., Pedobacter metabolipauper sp. nov., Pedobacter hartonius sp. nov. and Pedobacter steynii sp. nov., isolated from a hard-water rivulet. Int. J. Syst. Evol. Microbiol. 57, 2221–2227 (2007).
Brencic, A. et al. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73, 434–445 (2009).
Heeb, S. & Haas, D. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant. Microbe. Interact. 14, 1351–1363 (2001).
Grebe, T. W. & Stock, J. B. The histidine protein kinase superfamily. Adv. Microb. Physiol. 41, 139–227 (1999).
Two-component signal transduction. ASM Press, eds J.A. Hoch and T.J. Silhavy (1995).
Koretke, K. K., Lupas, A. N., Warren, P. V., Rosenberg, M. & Brown, J. R. Evolution of two-component signal transduction. Mol. Biol. Evol. 17, 1956–1970 (2000).
Goodman, A. L. et al. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes. Dev. 23, 249–259 (2009).
De Vos, P. & Truper, H., Judicial Commission of the International Committee on Systematic Bacteriology. IXth International (IUMS) Congress of Bacteriology and Applied Microbiology. IJSEM. 50, 2239–2244 (2000).
Woese, C. R., Kandler, O. & Wheelis, M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 87, 4576–4579 (1990).
Hahnke, R. L. et al. Genome-Based Taxonomic Classification of Bacteroidetes. Front. Microbiol. 7, (2016).
List of new names and new combinations previously effectively, but not validly, published. IJSEM. 62, 1–4 (2012).
Krieg, N. R., Ludwig, W., Euzéby, J. P. & Whitman, W. B. Bacteroidetes phyl. nov. in Bergey’s Manual of Systematics of Archaea and Bacteria 1–2, https://doi.org/10.1002/9781118960608 (American Cancer Society, 2015)..
Kämpfer, P. Sphingobacteriia class. nov. in Bergey’s Manual of Systematics of Archaea and Bacteria 1–1. 10.1002/9781118960608 (American Cancer Society, 2015).
Kämpfer, P. Sphingobacteriales ord. nov. in Bergey’s Manual of Systematics of Archaea and Bacteria 1–1. 10.1002/9781118960608 (American Cancer Society, 2015).
Acknowledgements
The study was funded by the Méditerranée-Infection foundation, the National Research Agency under the program “Investissements d’avenir”, reference ANR-10-IAHU-03 and by Région Provence Alpes Côte d’Azur and European funding FEDER IHUBIOTK. We also thank Aurelia Caputo for submitting the genomic sequences to GenBank. LJK is fellow of the Méditerranée-Infection foundation.
Author information
Authors and Affiliations
Contributions
L.J.K.: conceived the experiments, realised the experiments, analysed the data, prepared figures, wrote the manuscript. D.R., E.G. and P.E.F.: designed the experiments, conceived the experiments, analysed the data, wrote the manuscript
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kangale, L.J., Raoult, D., Ghigo, E. et al. Pedobacter schmidteae sp. nov., a new bacterium isolated from the microbiota of the planarian Schmidtea mediterranea. Sci Rep 10, 6113 (2020). https://doi.org/10.1038/s41598-020-62985-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-62985-x
This article is cited by
-
Chryseobacterium schmidteae sp. nov. a novel bacterial species isolated from planarian Schmidtea mediterranea
Scientific Reports (2021)
-
Culturomics revealed the bacterial constituents of the microbiota of a 10-year-old laboratory culture of planarian species S. mediterranea
Scientific Reports (2021)
-
Complete Genome Sequence of Pedobacter sp. PAMC26386 and Their Low Temperature Application in Arabinose-containing Polysaccharides Degradation
Current Microbiology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.