Abstract
The clinical manifestation of cystic fibrosis (CF) is heterogeneous also in patients with the same cystic fibrosis transmembrane regulator (CFTR) genotype and in affected sibling pairs. Other genes, inherited independently of CFTR, may modulate the clinical manifestation and complications of patients with CF, including the severity of chronic sinonasal disease and the occurrence of chronic Pseudomonas aeruginosa colonization. The T2R38 gene encodes a taste receptor and recently its functionality was related to the occurrence of sinonasal diseases and upper respiratory infections. We assessed the T2R38 genotype in 210 patients with CF and in 95 controls, relating the genotype to the severity of sinonasal disease and to the occurrence of P. aeruginosa pulmonary colonization. The frequency of the PAV allele i.e., the allele associated with the high functionality of the T2R38 protein, was significantly lower in i) CF patients with nasal polyposis requiring surgery, especially in patients who developed the complication before 14 years of age; and ii) in CF patients with chronic pulmonary colonization by P. aeruginosa, especially in patients who were colonized before 14 years of age, than in control subjects. These data suggest a role for T2R38 as a novel modifier gene of sinonasal disease severity and of pulmonary P. aeruginosa colonization in patients with CF.
Similar content being viewed by others
Introduction
Cystic fibrosis is a severe autosomal recessive disease due to mutations in the cystic fibrosis transmembrane regulator (CFTR) gene. The clinical manifestation of cystic fibrosis (CF) is highly heterogeneous also depending on the functional effect of the CFTR genotype1,2. However, patients with the same CFTR genotype may display a clinical discordance3 and a percentage of sibling pairs affected by CF display a discordant clinical manifestation3,4 reinforcing the concept that complex alleles (i.e., additional mutations on the same allele5), non-coding regions of CFTR6,7 or other genes, inherited independently of CFTR, modulate the clinical manifestation and complications of patients with CF8. Patients with CF frequently show different degrees of chronic rhinosinusitis9,10 with poor perception of sinonasal symptoms11 and pulmonary colonization by Pseudomonas aeruginosa and other opportunistic bacteria12,13. Chronic rhinosinusitis is common in patients affected by CF and it causes smell alterations14 that may impair taste and cause alterations in nutrition, thus, potentially impacting overall therapy outcome15. A percentage of patients develop nasal turbinate hypertrophy (NTH) or nasal polyposis (NP)10. However, according to the US CFF registry16, only 2–3% of patients every year require surgery for NP, which is considered the final, most advanced step of sinonasal disease. It is still unclear why only a small subgroup of patients with CF progress to such phase of the sinonasal disease17,18,19,20, even though the severity of CFTR mutations as well as previous sinus surgery may predict an increased risk11. Interestingly, in a recent study on 101 CF sibling-pairs4 we found NP requiring surgery in both the siblings of 13 pairs and in only one sibling from 12 sibling pairs. The poor correlation of NP to the CFTR genotype and to other clinical manifestations such as the pancreatic status and the severity of lung disease suggest that modifier genes play a role in determining nasal polyposis21.
P. aeruginosa is an opportunistic pathogen that frequently infects the lungs of patients with CF contributing to the decline in pulmonary function. P. aeruginosa expresses a series of specific virulence factors and, through adaptive mutations and antimicrobial resistance, causes chronic colonization via the development of biofilms22. In our study on CF sibling pairs4, we found chronic colonization by P. aeruginosa in 92/208 (44.2%) patients with CF and 21/101 cases in which only one sibling was colonized; 11/21 of these patients lived in the same environment suggesting that the environment has a limited role in P. aeruginosa colonization. Modifier genes, inherited independently by CFTR, may predispose to colonization of P. aeruginosa23.
Bitter receptors are G protein-coupled proteins that detect bitter compounds ingested with the diet24. The T2R38 gene, originally identified in type II taste receptor cells of the tongue, encodes one of these proteins, which exerts a main role as bitter taste receptors with the aim of protecting the individual against the ingestion of toxic substances present in spoiled foods. Among these substances, the receptor recognizes bacterial products such as acyl-homoserine lactones secreted by several gram-negative bacteria including P. aeruginosa, and it is now known that T2R38 and other bitter and sweet taste receptors are widely expressed by the upper respiratory tract ciliated cells and by solitary nose chemosensory cells25,26. These receptors, once activated by bacterial products, modulate ciliary beat frequency, promote the production of NO, and stimulate the release of immune peptides27. Such responses are strongly reduced in subjects who carry the nonfunctional “AVI” allele, while they are more effective in subjects carrying the “PAV” allele25,27 of the T2R38 gene. Furthermore, this polymorphism produces changes in the amino acid residues at positions 49, 262, and 296, generating two common alleles: the functional taster allele encodes proline, alanine, and valine (PAV); and the nonfunctional non-taster allele encodes alanine, valine, and isoleucine (AVI). These two common alleles generate three common genotypes (i.e., PAV/PAV, PAV/AVI, and AVI/AVI). Various studies have described the increased occurrence of sinonasal diseases and upper respiratory infections in patients with altered T2R38 activity, including in patients with CF28,29,30,31,32.
Results
Correlation between TAS2R38 and sinonasal complications
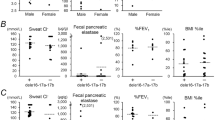
As shown in Table 1, we evaluated the frequency of the PAV allele and, as shown in Fig. 1a, we evaluated the frequency of the PAV homozygous genotype of the TAS2R38 gene in various subgroups of CF patients with different levels of sinonasal complications, in comparison with those of control subjects. The PAV allele was found in 33/98 (33.7%) alleles from CF patients with NP requiring surgery and these figures were significantly lower than those for control subjects (p < 0.01). This frequency was even lower in CF patients who developed NP requiring surgery before 14 years of age (18/68 alleles, 26.4%) and again significantly lower than that in control subjects (p < 0.01). However, the allele frequency of PAV was not significantly different between CF patients with NTH and control subjects. The same was true for CF patients with no NP or NTH.
These results were partially mirrored by the comparison of the frequency of the PAV homozygous genotype (Fig. 1a). It was significantly less frequent in patients with NP requiring surgery (5/49, 10.2%) than in control subjects (29/95, 30.5%; p < 0.01) and even lower in CF patients who developed NP requiring surgery before 14 years of age (3/34, 8.8%); the difference with control subjects was significant (p < 0.01). Again, the frequency of the PAV homozygous genotype did not differ between either CF patients with NTH and patients with no NP or NTH compared to that for control subjects.
Correlation between TAS2R38 and P. aeruginosa chronic colonization
Table 1 shows that the frequency of the PAV allele of the TAS2R38 gene was significantly lower in CF patients with P. aeruginosa chronic colonization (CC) (53/138, 38.4%) than in control subjects (p < 0.01), and it was even lower in patients colonized before 14 years of age (28/90, 31.1%), which was significantly lower (p < 0.01) than in control subjects. However, the frequency of the PAV allele did not differ between CF patients with no P. aeruginosa CC and control subjects. Again, the comparison of the frequency of the homozygous PAV allele (Fig. 1b) mirrored the results, being lower (though not significantly) in patients with P. aeruginosa CC (12/69 patients, 17.4%) than in control subjects, and even lower in CF patients colonized before 14 years of age (5/45, 11.1%), which was significantly lower than in control subjects (p < 0.01). No significant difference was observed between CF patients with no CC and control subjects.
TAS2R38 genotype in CF sibling pairs discordant for P. aeruginosa CC or NP requiring surgery
We included 34 pairs and two triplets of siblings affected by CF in the study. In 23 pairs of siblings we observed concordance between siblings of each pair for P. aeruginosa CC and NP requiring surgery. In particular, in 12 sibling pairs both members were colonized and in 11 pairs of siblings both members were not colonized; in 4 sibling pairs, both members experienced NP requiring surgery, and in 19 sibling pairs, both members did not experience such complications. In these 23 sibling pairs the PAV genotype was concordant (data not shown). As shown in Table 2, in 9 pairs and in two triplets of CF siblings we observed discordance between siblings for P. aeruginosa CC (5 sibling pairs) or NP requiring surgery (6 cases). In 7/11 discordant sibling pairs, both members had the same TAS2R38 genotype. In 4/11 pairs (pairs # 2, 4, 5 and 6) we observed a different genotype and in all cases the PAV allele was present (homozygous or heterozygous) in the member not affected by the complication.
Discussion
Our study, performed on the largest population of patients studied so far in this field, demonstrates that the frequency of the PAV allele of the TAS2R38 gene, i.e., the allele associated with the high functionality of the protein, was significantly reduced in CF patients with NP requiring surgery, the most advanced phase of CF sinonasal complications18. This finding confirms the relationship between the altered function of the TAS2R38 protein and the risk of upper airway infections31 and chronic rhinosinusitis28. The frequency of the PAV allele of the TAS2R38 gene was also significantly reduced in CF patients with chronic pulmonary colonization by P. aeruginosa, suggesting - for the first time - a role of the altered protein as a risk factor for lower respiratory infections.
Concerning the correlation between TAS2R38 and sinonasal diseases, in 2013, a pilot study on 28 patients with chronic rhinosinusitis demonstrated that supertaster patients (i.e., those with the highest activity of the TAS2R38 receptor being homozygous for the PAV allele) had a reduced risk of progressing to a disease requiring surgery. The result was later confirmed with a higher number of cases30. The same group studied 49 patients with CF, demonstrating that the patients homozygous for the PAV allele had a less severe sinonasal disease based on the SNOT score29 and the same results were reported in 123 non-CF patients with rhinosinusitis28. A single study that included only 53 patients with chronic rhinosinusitis and 39 healthy controls did not find associations between TAS2R38 genotypes and sinonasal disease33. Finally, a study by Cantone et al.34 on 100 subjects confirmed the inverse correlation between TAS2R38 functionality and the risk of developing sinonasal infections caused by gram-negative bacteria. The present study, which included 210 patients with CF and 95 control subjects, confirms that CF patients with both functional alleles of TAS2R38 have a significantly reduced risk of developing to the most severe phase of sinonasal disease, i.e., NP requiring surgery. Such complications occur only in 2–3% of patients with CF per year and it is still unclear why only a small subset of patients will progress to this phase of sinonasal disease16,18. Regarding surgical treatment, endoscopic sinus surgery for management of paranasal sinus diseases in CF patients is undoubtedly a challenge for the surgeon35. In the literature, surgery is indicated when CRS does not respond to conservative maximal medical therapy and is performed in 20–60% of patients with CF36. This variable frequency of surgically treated patients mirrors the absence of a consensus on indications and timing for surgery. In fact, no universally accepted guidelines for the surgical management of CRS in CF patients are available36.
Interestingly, our data relate the TAS2R38 genotype to the age of onset of NP requiring surgery, indicating that CF patients with nonfunctional alleles would develop NP requiring surgery at a younger age than CF patients with functional alleles. Thus, TAS2R38 can be considered a novel CF modifier gene, that modulates the severity of sinonasal disease contributing to explaining the known discordance for sinonasal severity observed in CF patients with the same CFTR genotype and in some pairs of CF patients4.
In addition, we also found a clear relationship between the TAS2R38 genotypes and chronic pulmonary colonization by P. aeruginosa. Our data indicate that CF patients with functional alleles of the gene (including some discordant sibling pairs) have a reduced risk of developing chronic colonization by P.aeruginosa, and have a decreased risk of developing colonization in childhood (i.e., before 14 years of age). There are no data so far with which compare our results. All the studies performed so far relate the TAS2R38 genotype only to upper respiratory infections, while this report is the first study that investigated patients with pulmonary colonization. However, considering that P. aeruginosa is among the bacteria that produce large amounts of acyl-homoserine lactones (known activators of the TAS2R38 receptor) we may speculate on the role of the TAS2R38 protein in the lower respiratory tract. Thus, if future studies confirm the present results, TAS2R38 would become a modifier gene that can impact P. aeruginosa infection in patients with CF. This result would help to improve management strategies to prevent P. aeruginosa colonization in CF patients with an elevated risk.
Materials and Methods
Study population
We recruited 210 patients with CF (105 males; age: 5–65 years; median: 20 years) followed at the Regional Cystic Fibrosis Reference Center. All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of Federico II University Hospital (244–2015). All the subjects (guardians in the case of minors) provided written informed consent to anonymously use their DNA samples and clinical data for research purposes Among these subjects, we studied 34 pairs and 2 triplets of siblings affected by CF. The data were anonymized. All patients met the criteria for the diagnosis of CF37. Airway colonization by P. aeruginosa was identified by sputum or oropharyngeal swab culture. Chronic colonization (CC) was defined according to the modified Leeds criteria38. The history of sinonasal disease including NTH and NP requiring surgery was evaluated by an otolaryngologist.
All subjects underwent the ENT (ear nose throat) examination by nasal endoscopy to assess the endoscopic appearance of the nose standardized by the Lund and Kennedy scale, which evaluates the clinical status of the nasal cavities (i.e., the presence of nasal polyps, oedema, discharge, scarring, and crusting)14. The ENT examination was performed with a 2.7-mm 30° rigid endoscope (Storz, Tuttlingen, Germany). All individuals enrolled in this study presented an endoscopic nasal score between 2 and 13, mean 5.65 ± 2.87, characterized by the presence of purulent discharge and diffuse oedema of the nasal mucosa in most of the cases. No sinonasal samplings (e.g. nasal lavage, nasal swabs) were performed because recent studies reported a relationship between upper and lower airway infection revealing concordant S. aureus and P. aeruginosa strains in upper and lower airways of the same patients39. No health related QoL (quality of life) assessment (e.g. SNOT-20 or SNOT-22) was performed because it is well known that SNOT-22 is frequently normal in patients with CF9,14.
In addition, as control we studied, 95 healthy subjects (38 males; age: 7–66 years; median: 18 years) who presented an endoscopic nasal score of 0 and who were characterized by the absence of any endoscopic sign of CRS.
Sequencing analysis
Sweat chloride levels were tested according to the guidelines40 under quality control procedures41. In all patients, the CFTR genotype was defined by screening a panel of the most frequent point mutations42 and gene rearrangements43. Moreover, CFTR Sanger gene sequencing44 or NGS45 was performed when mutations were not detected in one or both alleles by first-level analysis. Furthermore, we analysed 7 intragenic CFTR short tandem repeats46 to verify that both members of three sibling-pairs carrying only one known mutation had the same CFTR genotype. DNA was extracted from blood samples with standard proteinase K digestion followed by phenol/chloroform extraction and ethanol precipitation. Following extraction, samples were quantified using a spectrophotometer (ND-1000, NanoDrop) and analysed by PCR followed by Sanger sequencing for TAS2R38 polymorphism assessment. We compared the frequency of the alleles and of the genotypes in the different subgroups using the chi-square test (with Fisher correction); we considered a p value <0.01 to be significant.
References
Koch, C. et al. European Epidemiologic Registry of Cystic Fibrosis (ERCF): comparison of major disease manifestations between patients with different classes of mutations. Pediatr Pulmonol 31, 1–12, https://doi.org/10.1002/1099-0496(200101)31:1<1::AID-PPUL1000>3.0.CO;2-T (2001).
McKone, E. F., Goss, C. H. & Aitken, M. L. CFTR genotype as a predictor of prognosis in cystic fibrosis. Chest 130, 1441–1447, https://doi.org/10.1378/chest.130.5.1441 (2006).
Mekus, F. et al. Categories of deltaF508 homozygous cystic fibrosis twin and sibling pairs with distinct phenotypic characteristics. Twin Res 3, 277–293, https://doi.org/10.1375/twin.3.4.277 (2000).
Terlizzi, V. et al. Clinical expression of cystic fibrosis in a large cohort of Italian siblings. BMC Pulm Med 18, 196, https://doi.org/10.1186/s12890-018-0766-6 (2018).
Terlizzi, V. et al. Genotype-phenotype correlation and functional studies in patients with cystic fibrosis bearing CFTR complex alleles. J Med Genet 54, 224–235, https://doi.org/10.1136/jmedgenet-2016-103985 (2017).
Amato, F. et al. Gene mutation in microRNA target sites of CFTR gene: a novel pathogenetic mechanism in cystic fibrosis? PLoS One 8, e60448, https://doi.org/10.1371/journal.pone.0060448 (2013).
Giordano, S. et al. Molecular and functional analysis of the large 5’ promoter region of CFTR gene revealed pathogenic mutations in CF and CFTR-related disorders. J Mol Diagn 15, 331–340, https://doi.org/10.1016/j.jmoldx.2013.01.001 (2013).
Salvatore, F., Scudiero, O. & Castaldo, G. Genotype-phenotype correlation in cystic fibrosis: the role of modifier genes. Am J Med Genet 111, 88–95, https://doi.org/10.1002/ajmg.10461 (2002).
Kang, S. H., Meotti, C. D., Bombardelli, K., Piltcher, O. B. & de Tarso Roth Dalcin, P. Sinonasal characteristics and quality of life by SNOT-22 in adult patients with cystic fibrosis. Eur Arch Otorhinolaryngol 274, 1873–1882, https://doi.org/10.1007/s00405-016-4426-2 (2017).
Virgin, F. W. Clinical chronic rhinosinusitis outcomes in pediatric patients with cystic fibrosis. Laryngoscope Investig Otolaryngol 2, 276–280, https://doi.org/10.1002/lio2.78 (2017).
Bock, J. M. et al. Importance to question sinonasal symptoms and to perform rhinoscopy and rhinomanometry in cystic fibrosis patients. Pediatr Pulmonol 52, 167–174, https://doi.org/10.1002/ppul.23613 (2017).
Aanaes, K. Bacterial sinusitis can be a focus for initial lung colonisation and chronic lung infection in patients with cystic fibrosis. J Cyst Fibros 12(Suppl 2), S1–20, https://doi.org/10.1016/S1569-1993(13)00150-1 (2013).
Malhotra, S., Hayes, D., Jr. & Wozniak, D. J. Cystic Fibrosis and Pseudomonas aeruginosa: the Host-Microbe Interface. Clin Microbiol Rev 32, https://doi.org/10.1128/CMR.00138-18 (2019).
Di Lullo, A. M. et al. Cystic Fibrosis: The Sense of Smell. Am J Rhinol Allergy 34, 35–42, https://doi.org/10.1177/1945892419870450 (2019).
Lindig, J. et al. Smell in cystic fibrosis. Eur Arch Otorhinolaryngol 270, 915–921, https://doi.org/10.1007/s00405-012-2124-2 (2013).
Foundation, C. F. Cystic Fibrosis Foundation Patient Registry 2013 Annual Data Report. Bethesda, MD. (2013).
Brook, C. D. et al. Factors influencing the need for endoscopic sinus surgery in adult patients with cystic fibrosis. Am J Rhinol Allergy 31, 44–47, https://doi.org/10.2500/ajra.2017.31.4385 (2017).
Fokkens, W. J. et al. European Position Paper on Rhinosinusitis and Nasal Polyps 2012. Rhinol Suppl 23, 3 p preceding table of contents, 1–298 https://doi.org/10.4193/Rhino50E2 (2012).
Liang, J. et al. Medical management of chronic rhinosinusitis in cystic fibrosis: a systematic review. Laryngoscope 124, 1308–1313, https://doi.org/10.1002/lary.24503 (2014).
Tipirneni, K. E. & Woodworth, B. A. Medical and Surgical Advancements in the Management of Cystic Fibrosis Chronic Rhinosinusitis. Curr Otorhinolaryngol Rep 5, 24–34, https://doi.org/10.1007/s40136-017-0139-3 (2017).
Baldan, A. et al. IFRD1 gene polymorphisms are associated with nasal polyposis in cystic fibrosis patients. Rhinology 53, 359–364, https://doi.org/10.4193/Rhin14.229 (2015).
Ciofu, O. & Tolker-Nielsen, T. Tolerance and Resistance of Pseudomonas aeruginosa Biofilms to Antimicrobial Agents-How P. aeruginosa Can Escape Antibiotics. Front Microbiol 10, 913, https://doi.org/10.3389/fmicb.2019.00913 (2019).
Cutting, G. R. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet 16, 45–56, https://doi.org/10.1038/nrg3849 (2015).
Kim, U. K. et al. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science 299, 1221–1225, https://doi.org/10.1126/science.1080190 (2003).
Lee, R. J. et al. Bitter and sweet taste receptors regulate human upper respiratory innate immunity. J Clin Invest 124, 1393–1405, https://doi.org/10.1172/JCI72094 (2014).
Shah, A. S., Ben-Shahar, Y., Moninger, T. O., Kline, J. N. & Welsh, M. J. Motile cilia of human airway epithelia are chemosensory. Science 325, 1131–1134, https://doi.org/10.1126/science.1173869 (2009).
Carey, R. M., Adappa, N. D., Palmer, J. N., Lee, R. J. & Cohen, N. A. Taste Receptors: Regulators of Sinonasal Innate Immunity. Laryngoscope Investig Otolaryngol 1, 88–95, https://doi.org/10.1002/lio2.26 (2016).
Adappa, N. D. et al. TAS2R38 genotype predicts surgical outcome in nonpolypoid chronic rhinosinusitis. Int Forum Allergy Rhinol 6, 25–33, https://doi.org/10.1002/alr.21666 (2016).
Adappa, N. D. et al. T2R38 genotype is correlated with sinonasal quality of life in homozygous DeltaF508 cystic fibrosis patients. Int Forum Allergy Rhinol 6, 356–361, https://doi.org/10.1002/alr.21675 (2016).
Adappa, N. D. et al. The bitter taste receptor T2R38 is an independent risk factor for chronic rhinosinusitis requiring sinus surgery. Int Forum Allergy Rhinol 4, 3–7, https://doi.org/10.1002/alr.21253 (2014).
Cohen, N. A. The genetics of the bitter taste receptor T2R38 in upper airway innate immunity and implications for chronic rhinosinusitis. Laryngoscope 127, 44–51, https://doi.org/10.1002/lary.26198 (2017).
Lee, R. J. et al. T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J Clin Invest 122, 4145–4159, https://doi.org/10.1172/JCI64240 (2012).
Gallo, S. et al. TAS2R38 taste receptor gene and chronic rhinosinusitis: new data from an Italian population. BMC Med Genet 17, 54, https://doi.org/10.1186/s12881-016-0321-3 (2016).
Cantone, E. et al. In Vivo Biofilm Formation, Gram-Negative Infections and TAS2R38 Polymorphisms in CRSw NP Patients. Laryngoscope 128, E339–E345, https://doi.org/10.1002/lary.27175 (2018).
Virgin, F. W. et al. Extensive surgical and comprehensive postoperative medical management for cystic fibrosis chronic rhinosinusitis. Am J Rhinol Allergy 26, 70–75, https://doi.org/10.2500/ajra.2012.26.3705 (2012).
Aanaes, K. et al. Extensive endoscopic image-guided sinus surgery decreases BPI-ANCA in patients with cystic fibrosis. Scand J Immunol 76, 573–579, https://doi.org/10.1111/j.1365-3083.2012.02775.x (2012).
Farrell, P. M. et al. Diagnosis of Cystic Fibrosis: Consensus Guidelines from the Cystic Fibrosis Foundation. J Pediatr 181S, S4–S15 e11, https://doi.org/10.1016/j.jpeds.2016.09.064 (2017).
Lee, T. W., Brownlee, K. G., Conway, S. P., Denton, M. & Littlewood, J. M. Evaluation of a new definition for chronic Pseudomonas aeruginosa infection in cystic fibrosis patients. J Cyst Fibros 2, 29–34, https://doi.org/10.1016/S1569-1993(02)00141-8 (2003).
Fischer, N. et al. Non-invasive assessment of upper and lower airway infection and inflammation in CF patients. Pediatr Pulmonol 49, 1065–1075, https://doi.org/10.1002/ppul.22982 (2014).
LeGrys, V. A., Yankaskas, J. R., Quittell, L. M., Marshall, B. C. & Mogayzel, P. J. Jr. Diagnostic sweat testing: the Cystic Fibrosis Foundation guidelines. J Pediatr 151, 85–89, https://doi.org/10.1016/j.jpeds.2007.03.002 (2007).
Salvatore, M. et al. The Italian pilot external quality assessment program for cystic fibrosis sweat test. Clin Biochem 49, 601–605, https://doi.org/10.1016/j.clinbiochem.2015.12.014 (2016).
Tomaiuolo, R., Spina, M. & Castaldo, G. Molecular diagnosis of cystic fibrosis: comparison of four analytical procedures. Clin Chem Lab Med 41, 26–32, https://doi.org/10.1515/CCLM.2003.006 (2003).
Tomaiuolo, R. et al. Epidemiology and a novel procedure for large scale analysis of CFTR rearrangements in classic and atypical CF patients: a multicentric Italian study. J Cyst Fibros 7, 347–351, https://doi.org/10.1016/j.jcf.2007.12.004 (2008).
Amato, F. et al. Extensive molecular analysis of patients bearing CFTR-related disorders. J Mol Diagn 14, 81–89, https://doi.org/10.1016/j.jmoldx.2011.09.001 (2012).
Bergougnoux, A. et al. Multicenter validation study for the certification of a CFTR gene scanning method using next generation sequencing technology. Clin Chem Lab Med 56, 1046–1053, https://doi.org/10.1515/cclm-2017-0553, (2018).
Elce, A. et al. Three novel CFTR polymorphic repeats improve segregation analysis for cystic fibrosis. Clin Chem 55, 1372–1379, https://doi.org/10.1373/clinchem.2008.119545 (2009).
Acknowledgements
This work was supported by Grants from Ministero della Salute, L.548/93, annualità 2012–15.
Author information
Authors and Affiliations
Contributions
Conceptualization, Writing – review & editing and Project administration: F.A., Investigation and methodology: G.C., M.C., Data curation (Clinical): A.C., P.I., A.T., C.C., Data curation (Otolaryngology): A.M.D.L., Validation: M.G., Supervision: V.C., V.R.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Castaldo, A., Cernera, G., Iacotucci, P. et al. TAS2R38 is a novel modifier gene in patients with cystic fibrosis. Sci Rep 10, 5806 (2020). https://doi.org/10.1038/s41598-020-62747-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-62747-9
This article is cited by
-
Lung Microbiome as a Treatable Trait in Chronic Respiratory Disorders
Lung (2023)
-
Oxylipin profile in saliva from patients with cystic fibrosis reveals a balance between pro-resolving and pro-inflammatory molecules
Scientific Reports (2022)
-
An update on extra-oral bitter taste receptors
Journal of Translational Medicine (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.